Abstract
Aging can be conceptualized as the stochastic accumulation of damage and loss of resilience leading to organism demise. Resilience mechanisms that repair, recycle or replace damaged molecules and organelles are energy-demanding, therefore energy availability is essential to healthy aging. We propose that changes in mitochondrial and energy status regulate RNA splicing and that splicing is a resilience strategy that preserves energetic homeostasis with aging.
Aging is broadly defined as a progressive degenerative process that involves the gradual decline of physiological function and a rising mortality risk with time1. Most aging theories integrate the gradual build-up of stochastic macromolecular damage that leads to a global failure in normal cellular operations, conceptualized as the ratio of damage accumulation versus repair, compensation and regeneration. The so-called hallmarks of aging are connected to resilience: that is, the capacity of complex systems to react to stressful perturbations and regain a stable homeostasis through recycling of damaged molecules or organelles and ‘de novo’ biogenesis2. Researchers have proposed that the pace of aging is dictated in part by the efficiency of overall resilience. Because all resilience mechanisms require energy to be effective, a progressive decline in energy availability may also contribute to the loss of resilience with aging.
The production of ATP from nutrients, aerobically or anaerobically, provides energy for nearly all essential biological activities3. Oxidative phosphorylation in mitochondria is by far the most efficient mechanism to generate ATP from multiple substrates. In conditions of relative hypoxia or mitochondrial dysfunction, energy availability drops, which triggers the activation of a complex signaling network that aims to increase energy production and minimize energy consumption by nonessential functions4. Some of the features of this energy regulatory network are modulated by AMP-activated protein kinase (AMPK), a protein that is widely considered to be the hub of energy distribution as it senses the energy equilibrium in terms of the AMP:ATP ratio. The complexity of the biological system that responds to energy perturbations, including AMPK-activated mechanisms, is only partially understood. Recent evidence indicates that challenges to energetic homeostasis with aging increase the diversity of mRNA isoform variation. We explore the hypothesis that an ‘energy–splicing resilience axis’ is triggered in physiological conditions that challenge mitochondrial oxidative capacity and energy status and, by affecting the spliceosome, leads to the production of alternative protein variants that support resilience and longevity.
RNA splicing in aging
Pre-mRNA splicing involves the removal of noncoding introns and joining of coding exons to generate mature mRNAs that are exported to the cytoplasm for translation into proteins. The spliceosome, a mega-dalton complex composed of about 300 proteins and several small nuclear RNAs, is responsible for carrying out pre-mRNA splicing5. Constitutive splicing is the removal of introns and ligation of exons as they appear in the pre-mRNA, whereas alternative splicing entails the ligation of introns-exons in various combinations such that multiple mature mRNA isoforms are produced from a single pre-mRNA product6. Many factors, including the expression of proteins that make up or interact with the spliceosome, influence the sequences that are excised, retained and combined7. Alternative splicing endows organisms with the capacity to adapt their expression profile and protein arsenal in response to different situations, such as developmental stage, cell type, intracellular conditions, external environment and the health and/or age of the cell or organism8.
Studies have generally found that inflammation and stress responses are upregulated with aging, whereas pathways connected to energetic metabolism and membrane integrity are downregulated9. Gene-expression analysis of circulating leukocytes from 30–104-year-old individuals also revealed a dysregulation of mRNA processing with aging10. Follow-up studies uncovered that approximately one-third of splicing factors exhibit age-related changes; these findings were replicated in two human cohorts, and have also been seen in senescent cells11. Consistent with splicing regulation being intimately involved in aging and lifespan, specific differences in expression of splicing factors and distinct alternative splicing of key genes were found to be associated with mouse-strain longevity12. In mouse skin, skeletal muscle and bone, the variety of alternatively spliced mRNA isoforms increases with age, some of which affect RNA processing13. Age-related splicing changes arising independently of transcript-level changes have also been observed in the brains of cognitively normal individuals, mostly affecting metabolism and DNA repair14. About 40% of genes expressed in the human prefrontal cortex and cerebellum undergo splicing changes over the postnatal lifespan, with about one-third of these accounted for by aging15. An analysis of about 8,500 RNA-sequencing (RNA-seq) samples across 48 tissues in 544 individuals found that the genome-wide splicing profile is a better predictor of biological age than transcript expression, and that it strongly correlates with age-associated chronic diseases16.
Extensive literature indicates that several human diseases are directly linked to defects in RNA splicing, including Hutchinson–Gilford progeria syndrome (HGPS), a disorder that manifests features of clinical aging17. The expression of three splicing-factor genes — HNRNPM, HNRNPA0 and AKAP17A — has been associated with accelerated cognitive decline, and AKAP17A expression has also been associated with a decline of hand-grip strengthl18. This certainly incomplete snapshot of the literature, which documents that age-related changes of spliceosomal components are tied to aging and longevity both in model organisms and humans, suggests that RNA splicing has a role in the aging process.
Alternative splicing and mitochondria
Mitochondria are the main source of biological energy, and as their function declines with aging, less energy (ATP) is available for maintenance, repair, regeneration and function3. A recent proteomic study showed that mitochondrial proteins and proteins related to energy metabolism — including those connected to the tricarboxylic acid cycle, electron transport chain complexes and glycolysis — are significantly underrepresented in skeletal muscle from older (versus younger) healthy individuals19. An unexpected observation from this study was that most proteins that regulate RNA processing and alternative splicing were overrepresented in older participants (Fig. 1a middle). As previous authors had suggested that alternative splicing evolved as a platform for phenotypic novelty (in particular to refine the adaptive response to stress20), we hypothesize the existence of a programmed energy–splicing resilience axis that is activated in situations of energetic stress.
Fig. 1 |. Skeletal muscle proteomics.
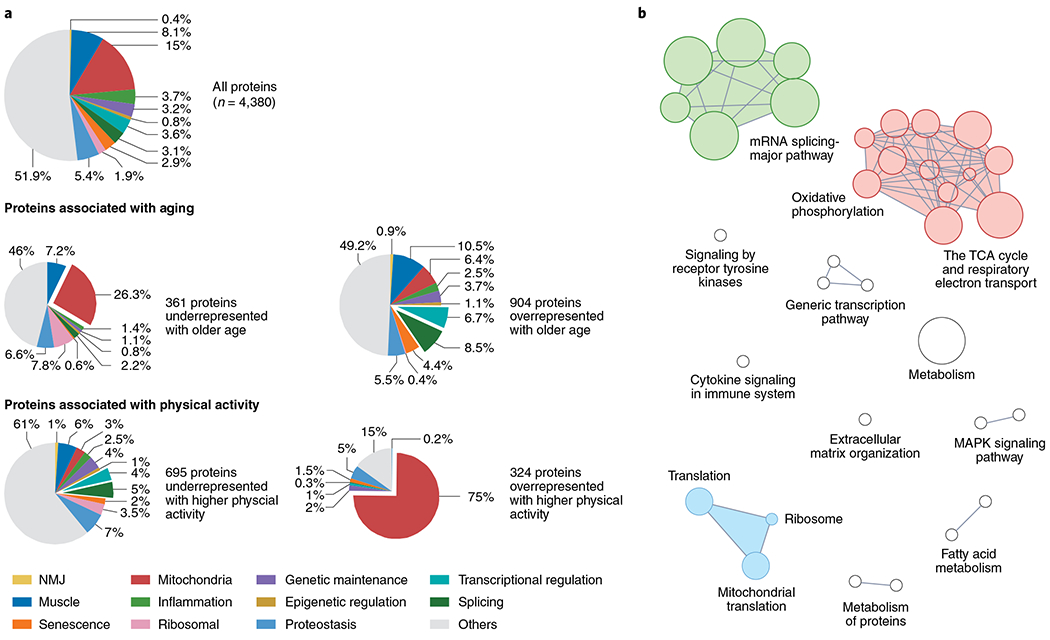
a, Analysis of aging and physical activity specimens. Vastus lateralis muscle biopsies were analyzed for protein expression in 58 healthy participants in the GESTALT study (aged between 20 and 80+ years); 4,380 proteins were detectable and quantifiable. Top, pie chart showing the distribution of age-relevant proteins in the entire dataset for comparison. Middle, pie charts reporting classes of proteins that are underrepresented and overrepresented with aging. Bottom, pie charts of proteins differentially regulated by higher levels of physical activity. after statistically accounting for age and sex. The results of physical activity are opposite to the effects of aging, with mitochondrial proteins representing 75% of the overrepresented proteins with high levels of physical activity, whereas spliceosome and transcriptional regulation proteins are underrepresented21. b, Proteins associated with higher muscle oxidative capacity. Overall, 253 proteins were significantly overrepresented in samples exhibiting better muscle oxidative capacity, as assessed by phosphorus-31 MRS. Enrichment was seen for proteins in the pathway of electron transport chain and tricarboxylic acid (TCA) cycle; proteins involved in mitochondria translation; and spliceosome proteins22. NMJ, neuromuscular junction.
To challenge this hypothesis, we reanalyzed muscle proteomic data in relationship to physical activity level after statistically accounting for age, sex and technical covariates21. It is well known that physical activity improves mitochondrial function even in older individuals. Not surprisingly, mitochondrial proteins made up about 75% of the proteins that were significantly overrepresented in samples from individuals with higher physical activity relative to more sedentary individuals. Conversely, higher activity was associated with reduced spliceosome protein expression, consistent with an inverse relationship between mitochondria function and RNA splicing (Fig. 1a bottom). Thus, aging and physical activity, independent of each other, have opposing effects on mitochondrial and splicing proteins, supportive of the energy–splicing resilience axis.
As physical activity is only a proxy measure of mitochondrial function, we searched for skeletal muscle proteins that were overrepresented in participants with higher maximal oxidative capacity as assessed ‘in vivo’ by phosphorus-31 magnetic resonance spectroscopy (MRS)22. After adjusting for age, sex, body mass index and physical activity, we found that as many as 253 proteins were significantly overrepresented, falling into three major cluster types: (1) key proteins of the electron transport chain and tricarboxylic acid cycle; (2) proteins that regulate mitochondrial translation; and (3) spliceosome proteins (Fig. 1b). Thus, when the effect of confounders is accounted for upregulation of the splicing machinery is associated with better skeletal-muscle oxidative capacity (that is, mitochondrial function), further supporting the energy–splicing resilience axis. Taken together, our observations suggest that, when mitochondrial aerobic metabolism is compromised or a higher energetic need is detected, cells overexpress spliceosome proteins, seemingly as a compensatory mechanism to produce novel splicing variants that promote improved mitochondrial function and resolution of the energetic crisis. Conversely, when mitochondria are healthy and ATP production is robust (such as in individuals who are physically active), constitutively expressed proteins prevail, less alternative splicing is required and spliceosome proteins are downregulated.
The energy–splicing resilience axis
A common thread among many of the lifespan-extending interventions (dietary, genetic or otherwise) is their connection to low-energy status, nutrient deprivation, and optimization and preservation of mitochondrial function23. In brief, increased longevity seems to arise in part by reducing molecular energy (that is, ATP), resulting in the activation of various pathways that promote survival and resilience. Yet, to date, the precise molecular strategies used to improve lifespan and healthspan — particularly in scenarios involving low energy — remain incompletely defined. For the energy–splicing resilience axis (Fig. 2), we envision at least five major components: mitochondria function, ATP availability, signaling response pathways, alternative splicing mechanisms and the resilience outcome.
Fig. 2 |. Energy–splicing resilience axis.
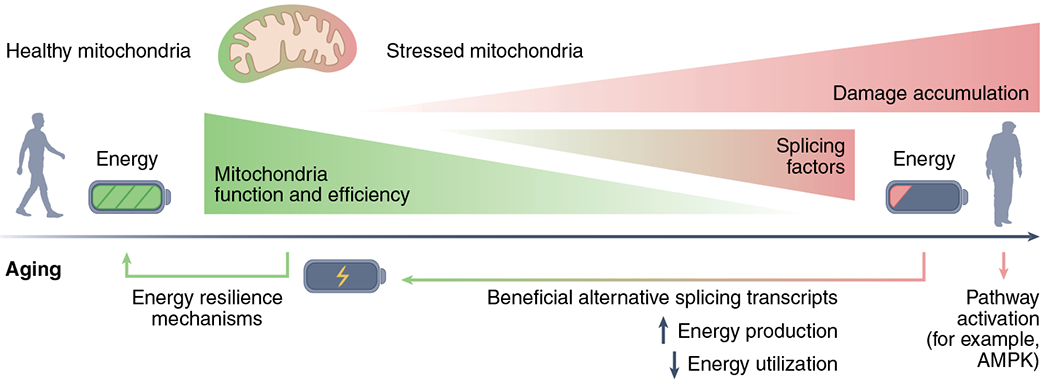
When mitochondria are healthy and generation of ATP matches demand, there is almost no energetic stress and little need to activate the splicing machinery. When mitochondrial function is impaired and energy production falls below a critical level, changes occur that are aimed at increasing energy production and reducing energy consumption for nonessential activities, promoting resilience and energetic homeostasis. Some of the molecular changes that occur involve the production of alternative RNA splicing variants (directly or indirectly) through various signaling mechanisms, including those that engage AMPK (a master regulator of the energy crisis response).
Consistent with the proposed axis, caloric restriction restrains mitochondria function, but even more so curbs energetic demands — tilting the energetic equation toward a positive balance in part by invoking the splicing machinery. For example, hepatic transcriptome, proteome and metabolome studies in rhesus monkeys have found that alternative pre-RNA processing is a central feature of the response to caloric restriction24. Similarly, a role for splicing factors was identified in the lifespan modulatory effects of dietary restriction across different mouse strains and in different tissues25. Previous studies in Caenorhabditis elegans have found that pre-mRNA splicing homeostasis is a predictor of life expectancy and that dietary restriction reverses splicing defects associated with aging specifically via splicing factor 1 through modulation of the TORC1–AMPK pathway26.
The energy–splicing resilience axis in Fig. 2 is supported by a growing body of literature, and appears to engage at minimum AMPK signaling. First, a recent study has identified splicing factor serine/arginine-rich splicing factor 1 (SRSF1) as a novel substrate of AMPK: AMPK-dependent phosphorylation of SRSF1 at Ser133 inhibits the ability of the splicing factor to bind RNA and affects alternative pre-mRNA processing, providing evidence of a direct link between AMPK and splicing mechanisms27. Second, the addition of metformin (a drug that activates AMPK) has been found to suppress the effects of a 14-week program of resistance exercise training (that is, increased muscle mass and reduced expression of transcripts related to RNA processing pathways), implicating splicing as part of the AMPK response28. Third, AMPK activity, which is directly regulated by mitochondrial oxidative capacity, decreases with aging, suggesting a potential mechanism by which splicing undergoes its broad age-dependent alterations29. Fourth, the premature aging disorder HGPS is caused by a de novo mutation in the lamin A (LMNA) gene that results in the production of a truncated splice variant of prelamin A known as progerin, a protein that increase with aging even in healthy persons17. Recent evidence indicates that metformin alleviates the pathological defects in HGPS cells and normalizes gene expression, perhaps through the upregulation of SRSF1 via AMPK30. Finally, analysis of the RNA-seq data from the skeletal muscle of participants in the ‘Genetic and Epigenetic Signatures of Translational Aging Laboratory Testing’ (GESTALT) study reveals that overrepresentation of the spliceosome proteins was associated with a higher number of splicing events detected by RNA-seq, supporting a functional consequence of the molecular adaptations31. Notably, this differential exon utilization affects many biological pathways that are transcriptionally modulated by AMPK, such as oxidative phosphorylation, adipogenesis, MTORC1 signaling and fatty acid metabolism4. The collective results begin to draw a picture that links mitochondrial function, ATP availability, AMPK, aging mechanisms and specific spliceosome factors, although additional studies are clearly required to fully elucidate the molecular composition of the energy–splicing resilience axis.
The specific mechanisms by which mitochondrial dysfunction modulates splicing remain unclear, yet — in addition to a central role for AMPK — the literature provides a few possibilities. Previous studies in neuroblastoma cell lines have found that paraquat treatment, which interferes with mitochondrial function and causes oxidative stress and energy deficits, uniquely affects alternative splicing in select genes, a phenomenon that was revealed (by using a set of defined treatment strategies) to stem from ATP depletion and not elevated levels of reactive oxygen species32. In addition, p32, a protein that normally resides in the mitochondrial matrix, is released into the cytoplasm under conditions of mitochondrial stress. p32 promotes autophagy by stabilizing UNC-51-like autophagy activating kinase (ULK1), and through a physical interaction facilitates the nuclear translocation of the splicing factor U2AF26, which is critical for the early steps of spliceosome assembly33. Interestingly, haploinsufficiency in p32 impairs glucose oxidation, resulting in a compensatory increase in fatty acid oxidation and glycolysis, and, by increasing energy expenditure, protects mice from age- and diet-induced obesity34.
A role for alternative splicing in modulating resilience as a function of energy metabolism is more firmly established in cancer. Most notably, in cancer cells, a specific RNA splicing event modulates the ratio between M1 and M2 pyruvate kinase isoforms, shifting the metabolic phenotype from oxidative phosphorylation to aerobic glycolysis35. This molecular change accounts for the Warburg effect, a prominent adaptive strategy that provides a selective growth advantage (particularly during hypoxia). Furthermore, intermediate metabolites of the Krebs cycle affect different splicing factors by modulating the activity of 2-oxoglutarate-dependent oxidases that function as master sensors of energetic stress, stabilizing and activating hypoxia-inducible factor 1 (HIF1) during hypoxia and controlling histone demethylases that are relevant to splicing processes36. The above, as well as additional literature37, strongly suggest that splicing factors are primary movers of metabolic stress resilience in cancer and the HIF-1-mediated response to hypoxia. The relevance of these pathways and the identification of the resilience mechanisms related to aging are central to future studies aimed at unraveling the composition of the energy–splicing resilience axis.
Closing thoughts and research vision
It is well established that in situations of severe ATP starvation, life is unsustainable and cell death is activated through different mechanisms (primarily apoptosis). However, in conditions of intermediate scarcity of ATP, cells unleash resilience strategies that attempt to increase ATP production and curtail any use of ATP that is not essential. Transcriptome plasticity provided by pre-mRNA splicing is emerging as a module of the resilience strategy that counteracts damage accumulation with aging and circumvents associated phenotypic and functional manifestations — a biological mechanism that undoubtedly involves other regulatory features, such as noncoding RNA control of gene expression, modulation of mRNA decay or translation efficiency, and the activity of small RNAs. Although it might seem counterintuitive to activate a mechanism that consumes substantial ATP resources at a time of energy scarcity (that is, RNA splicing), such an investment may be worthwhile if it is followed by improved energetic homeostasis and cell survival.A notable example of such a seemingly inharmonious situation is the activation of autophagy or mitophagy during hypoxia via stimulation of AMPK and ULK1 activities, a stress response that similarly entails complex alternative splicing events29.
To better define the energy–splicing resilience axis, experiments are now needed in which (1) mitochondrial function is specifically modulated and (2) the identity of unique differentially spliced proteins is determined. For example, mitochondrial function can be impaired or enhanced by specific molecules in cell cultures of myocytes, providing a simple platform for characterizing downstream events. In animal models and in humans, the study of transcriptome changes — with a focus on splice variants (possibly using long-range RNA-seq) — in skeletal muscle following alteration of one’s exercise regimen or restriction of mobility may provide additional clues to key factors involved in the energy–splicing resilience axis. Identification of splicing protein isoforms or components of the spliceosome that are central to the resilience response will permit quantification in fluids and tissues using appropriately developed targeting techniques (for example, selective monoclonal antibodies or PCR primers) to better understand their physiological and pathological roles. That, coupled with CRISPR or RNA interference tools that permit selective gene-expression silencing, will facilitate the evaluation of the functions of central factors to the energy–splicing resilience axis in model organisms. Studies that target the transducers and molecular mechanisms that drive the energy availability response and subsequently modulate splicing, as well as the downstream factors that execute the resilience response, are urgently needed. Such studies will define the role of alternative splicing in regulating energy homeostasis and its effects on both survival and longevity, perhaps revealing RNA splicing as a novel hallmark of aging while also uncovering new therapeutic targets for the promotion of a healthy lifespan.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institute on Aging, NIH.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Ferrucci L et al. Aging Cell 19, e13080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy BK et al. Cell 159, 709–713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang JY, Blum A, Liu J & Finkel T J. Clin. Invest 128, 3662–3670 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herzig S & Shaw RJ Nat. Rev. Mol. Cell Biol 19, 121–135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahl MC, Will CL & Lührmann R Cell 136, 701–718 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Lee Y & Rio DC Annu. Rev. Biochem 84, 291–323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z & Burge CB RNA 14, 802–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhadra M, Howell P, Dutta S, Heintz C & Mair WB Hum. Genet 139, 357–369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng L et al. Aging Cell 19, e13121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harries LW et al. Aging Cell 10, 868–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holly AC et al. Mech. Ageing Dev 134, 356–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee BP et al. Aging Cell 15, 903–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez SA et al. Aging Cell 15, 267–278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tollervey JR et al. Genome Res. 21, 1572–1582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazin P et al. Mol. Syst. Biol 9, 633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K et al. Sci. Rep 8, 10929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalo S, Kreienkamp R & Askjaer P Ageing Res. Rev 33, 18–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee BP et al. Biogerontology 20, 649–663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ubaida-Mohien C et al. eLife 8, e49874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kourtis N & Tavernarakis N EMBO J. 30, 2520–2531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ubaida-Mohien C et al. Front. Physiol 10, 312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adelnia F et al. Aging Cell 19, e13124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruetenik A & Barrientos A Biochim. Biophys. Acta 1847, 1434–1447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhoads TW et al. Cell Metab. 27, 677–688 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee BP et al. Exp. Gerontol 128, 110736 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Heintz C et al. Nature 541, 102–106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto E et al. Biochem. J 477, 2237–2248 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Kulkarni AS et al. Aging 12, 19852–19866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salminen A & Kaarniranta K Ageing Res. Rev 11, 230–241 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Egesipe AL et al. NPJ Aging Mech. Dis 2, 16026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tumasian RA III et al. Nat. Commun 12, 2014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maracchioni A et al. J. Neurochem 100, 142–153 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Heyd F, Carmo-Fonseca M & Möröy T J. Biol. Chem 283, 19636–19645 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Liu Y et al. Sci. Rep 7, 5754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christofk HR et al. Nature 452, 230–233 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Salminen A, Kauppinen A & Kaarniranta K Cell. Mol. Life Sci 72, 3897–3914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farina AR et al. J. Exp. Clin. Cancer Res 39, 110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


