Abstract
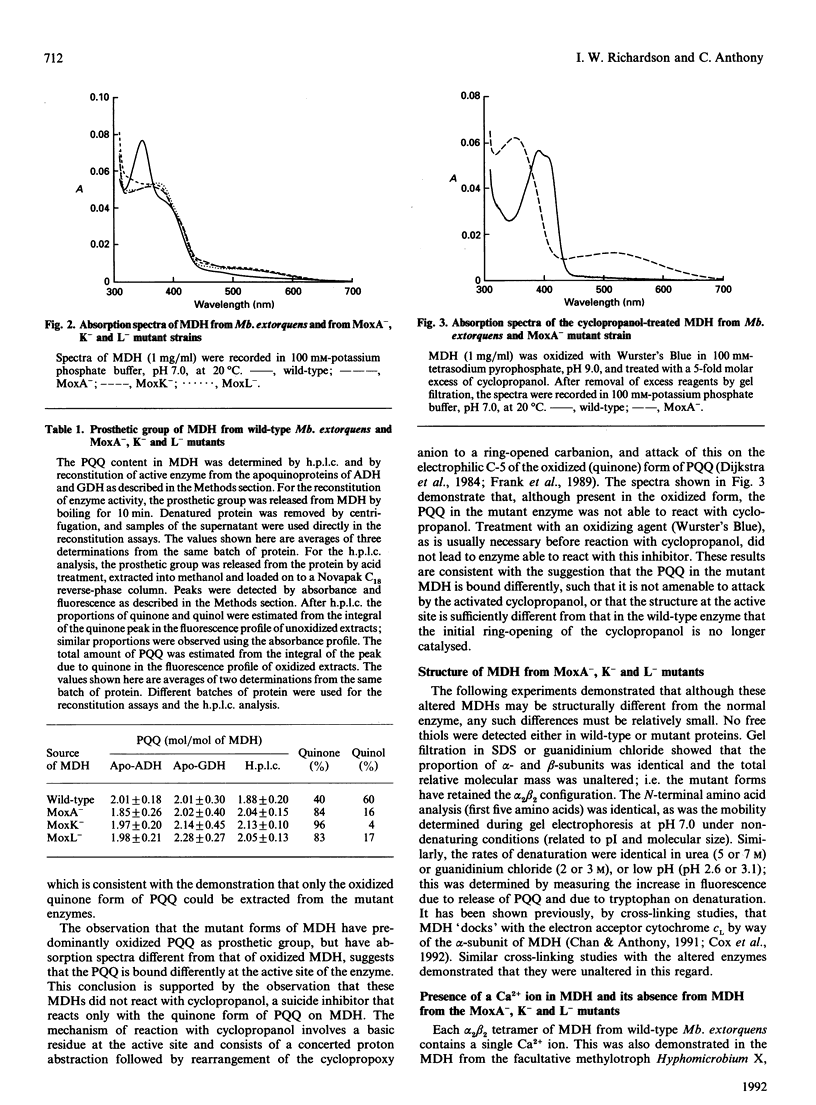
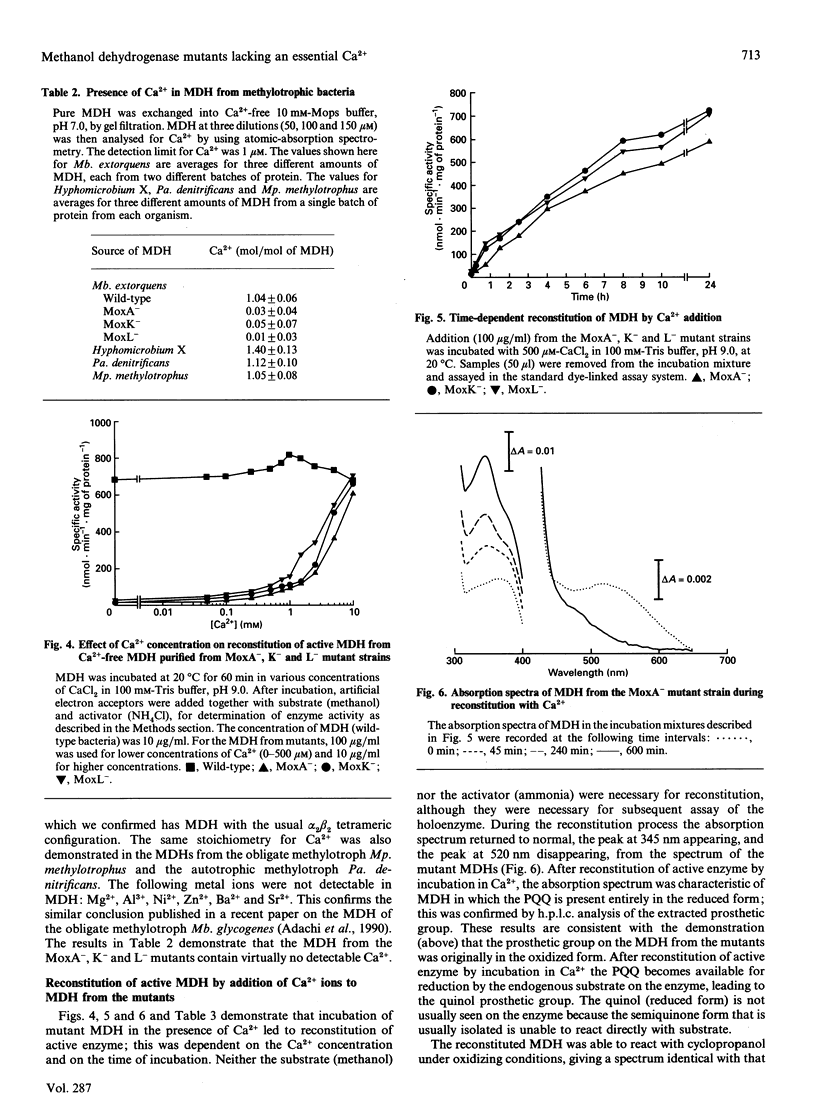
Methanol dehydrogenase (MDH) from Methylobacterium extorquens, Methylophilus methylotrophus, Paracoccus denitrificans and Hyphomicrobium X all contained a single atom of Ca2+ per alpha 2 beta 2 tetramer. The role of Ca2+ was investigated using the MDH from Methylobacterium extorquens. This was shown to be similar to the MDH from Hyphomicrobium X in having 2 mol of prosthetic group (pyrroloquinoline quinine; PQQ) per mol of tetramer, the PQQ being predominantly in the semiquinone form. MDH isolated from the methanol oxidation mutants MoxA-, K- and L- contained no Ca2+. They were identical with the enzyme isolated from wild-type bacteria with respect to molecular size, subunit configuration, pI, N-terminal amino acid sequence and stability under denaturing conditions (low pH, high urea and high guanidinium chloride) and in the nature and content of the prosthetic group (2 mol of PQQ per mol of MDH). They differed in their lack of Ca2+, the oxidation state of the extracted PQQ (fully oxidized), absence of the semiquinone form of PQQ in the enzyme, reactivity with the suicide inhibitor cyclopropanol and absorption spectrum, which indicated that PQQ is bound differently from that in normal MDH. Incubation of MDH from the mutants in calcium salts led to irreversible time-dependent reconstitution of full activity concomitant with restoration of a spectrum corresponding to that of fully reduced normal MDH. It is concluded that Ca2+ in MDH is directly or indirectly involved in binding PQQ in the active site. The MoxA, K and L proteins may be involved in maintaining a high Ca2+ concentration in the periplasm. It is more likely, however, that they fill a 'chaperone' function, stabilizing a configuration of MDH which permits incorporation of low concentrations of Ca2+ into the protein.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. J., Lidstrom M. E. The moxFG region encodes four polypeptides in the methanol-oxidizing bacterium Methylobacterium sp. strain AM1. J Bacteriol. 1988 May;170(5):2254–2262. doi: 10.1128/jb.170.5.2254-2262.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Morris C. J., Nunn D. N., Anthony C., Lidstrom M. E. Nucleotide sequence of the Methylobacterium extorquens AM1 moxF and moxJ genes involved in methanol oxidation. Gene. 1990 May 31;90(1):173–176. doi: 10.1016/0378-1119(90)90457-3. [DOI] [PubMed] [Google Scholar]
- Anthony C. Bacterial oxidation of methane and methanol. Adv Microb Physiol. 1986;27:113–210. doi: 10.1016/s0065-2911(08)60305-7. [DOI] [PubMed] [Google Scholar]
- Anthony C. The c-type cytochromes of methylotrophic bacteria. Biochim Biophys Acta. 1992 Jan 30;1099(1):1–15. [PubMed] [Google Scholar]
- Anthony C. The structure of bacterial quinoprotein dehydrogenases. Int J Biochem. 1992;24(1):29–39. doi: 10.1016/0020-711x(92)90226-q. [DOI] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochem J. 1967 Sep;104(3):960–969. doi: 10.1042/bj1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. T., Anthony C. The interaction of methanol dehydrogenase and cytochrome cL in the acidophilic methylotroph Acetobacter methanolicus. Biochem J. 1991 Nov 15;280(Pt 1):139–146. doi: 10.1042/bj2800139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleton-Jansen A. M., Goosen N., Fayet O., van de Putte P. Cloning, mapping, and sequencing of the gene encoding Escherichia coli quinoprotein glucose dehydrogenase. J Bacteriol. 1990 Nov;172(11):6308–6315. doi: 10.1128/jb.172.11.6308-6315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. M., Day D. J., Anthony C. The interaction of methanol dehydrogenase and its electron acceptor, cytochrome cL in methylotrophic bacteria. Biochim Biophys Acta. 1992 Feb 13;1119(1):97–106. doi: 10.1016/0167-4838(92)90240-e. [DOI] [PubMed] [Google Scholar]
- Cross A. R., Anthony C. The purification and properties of the soluble cytochromes c of the obligate methylotroph Methylophilus methylotrophus. Biochem J. 1980 Nov 15;192(2):421–427. doi: 10.1042/bj1920421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day D. J., Anthony C. Soluble cytochromes c of methanol-utilizing bacteria. Methods Enzymol. 1990;188:298–303. doi: 10.1016/0076-6879(90)88046-d. [DOI] [PubMed] [Google Scholar]
- Dijkstra M., Frank J., Jongejan J. A., Duine J. A. Inactivation of quinoprotein alcohol dehydrogenases with cyclopropane-derived suicide substrates. . Eur J Biochem. 1984 Apr 16;140(2):369–373. doi: 10.1111/j.1432-1033.1984.tb08111.x. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Jr Studies on methanol dehydrogenase from Hyphomicrobium X. Isolation of an oxidized form of the enzyme. Biochem J. 1980 Apr 1;187(1):213–219. doi: 10.1042/bj1870213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. J. Molecular chaperones: the plant connection. Science. 1990 Nov 16;250(4983):954–959. doi: 10.1126/science.250.4983.954. [DOI] [PubMed] [Google Scholar]
- Geiger O., Görisch H. Enzymatic determination of pyrroloquinoline quinone using crude membranes from Escherichia coli. Anal Biochem. 1987 Aug 1;164(2):418–423. doi: 10.1016/0003-2697(87)90513-6. [DOI] [PubMed] [Google Scholar]
- Geiger O., Görisch H. Reversible thermal inactivation of the quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus. Ca2+ ions are necessary for re-activation. Biochem J. 1989 Jul 15;261(2):415–421. doi: 10.1042/bj2610415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarek Z., Gergely J. Zero-length crosslinking procedure with the use of active esters. Anal Biochem. 1990 Feb 15;185(1):131–135. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- Groen B. W., Duine J. A. Quinoprotein alcohol dehydrogenase from Pseudomonas aeruginosa and quinohemoprotein alcohol dehydrogenase from Pseudomonas testosteroni. Methods Enzymol. 1990;188:33–39. doi: 10.1016/0076-6879(90)88009-y. [DOI] [PubMed] [Google Scholar]
- Groen B. W., van Kleef M. A., Duine J. A. Quinohaemoprotein alcohol dehydrogenase apoenzyme from Pseudomonas testosteroni. Biochem J. 1986 Mar 15;234(3):611–615. doi: 10.1042/bj2340611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heptinstall J., Quayle J. R. Pathways leading to and from serine during growth of Pseudomonas AM1 on C1 compounds or succinate. Biochem J. 1970 Apr;117(3):563–572. doi: 10.1042/bj1170563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom M. E. Genetics of carbon metabolism in methylotrophic bacteria. FEMS Microbiol Rev. 1990 Dec;7(3-4):431–436. doi: 10.1111/j.1574-6968.1990.tb04949.x. [DOI] [PubMed] [Google Scholar]
- Nunn D. N., Anthony C. The nucleotide sequence and deduced amino acid sequence of the cytochrome cL gene of Methylobacterium extorquens AM1, a novel class of c-type cytochrome. Biochem J. 1988 Dec 1;256(2):673–676. doi: 10.1042/bj2560673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Day D., Anthony C. The second subunit of methanol dehydrogenase of Methylobacterium extorquens AM1. Biochem J. 1989 Jun 15;260(3):857–862. doi: 10.1042/bj2600857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):581–590. doi: 10.1128/jb.166.2.581-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Phenotypic characterization of 10 methanol oxidation mutant classes in Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):591–597. doi: 10.1128/jb.166.2.591-597.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oefner C., Suck D. Crystallographic refinement and structure of DNase I at 2 A resolution. J Mol Biol. 1986 Dec 5;192(3):605–632. doi: 10.1016/0022-2836(86)90280-9. [DOI] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Salisbury S. A., Forrest H. S., Cruse W. B., Kennard O. A novel coenzyme from bacterial primary alcohol dehydrogenases. Nature. 1979 Aug 30;280(5725):843–844. doi: 10.1038/280843a0. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Verwiel P. E., Frank J., Verwiel E. J. Characterization of the second prosthetic group in methanol dehydrogenase from hyphomicrobium X. Eur J Biochem. 1981 Aug;118(2):395–399. doi: 10.1111/j.1432-1033.1981.tb06415.x. [DOI] [PubMed] [Google Scholar]
- van der Meer R. A., Groen B. W., van Kleef M. A., Frank J., Jongejan J. A., Duine J. A. Isolation, preparation, and assay of pyrroloquinoline quinone. Methods Enzymol. 1990;188:260–283. doi: 10.1016/0076-6879(90)88043-a. [DOI] [PubMed] [Google Scholar]


