Abstract
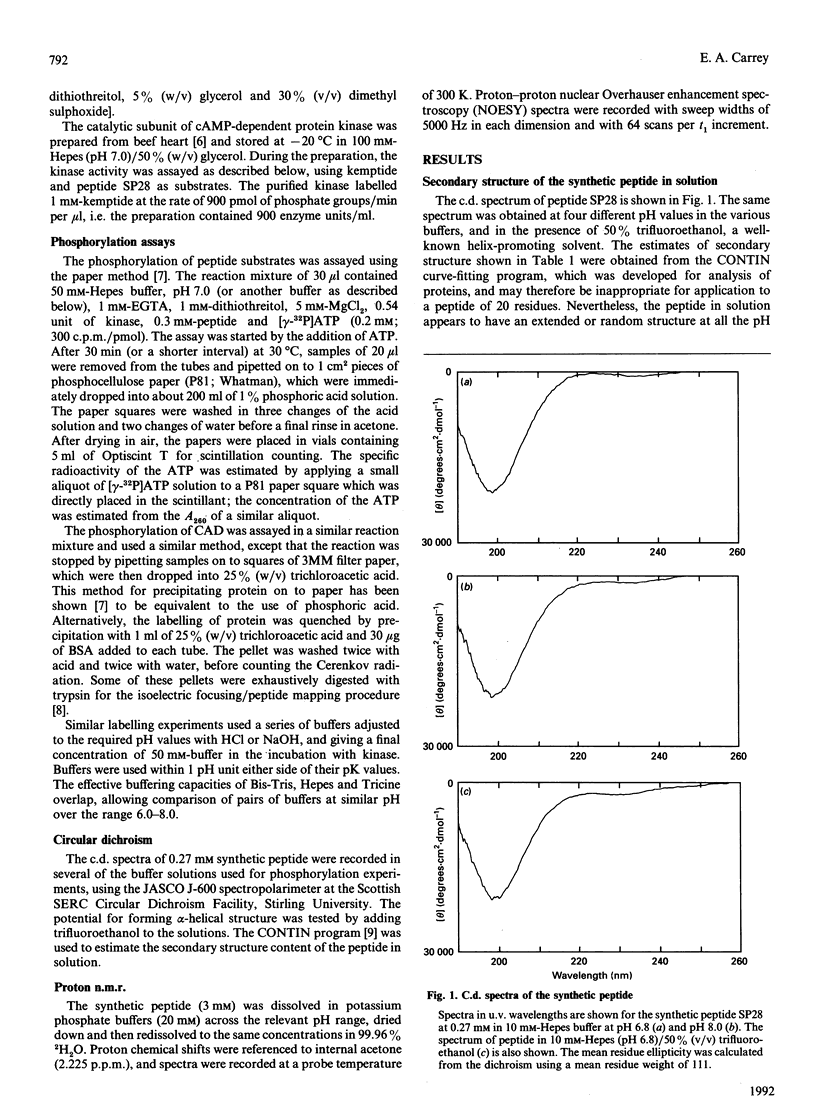
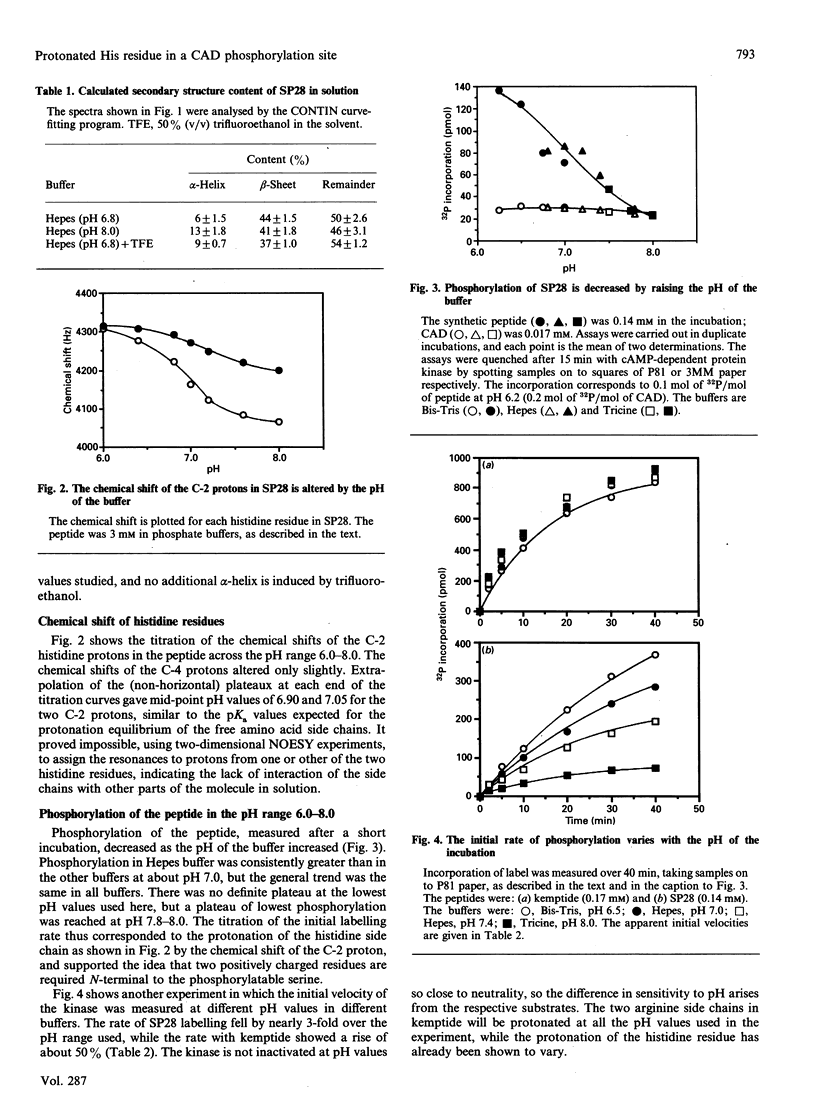
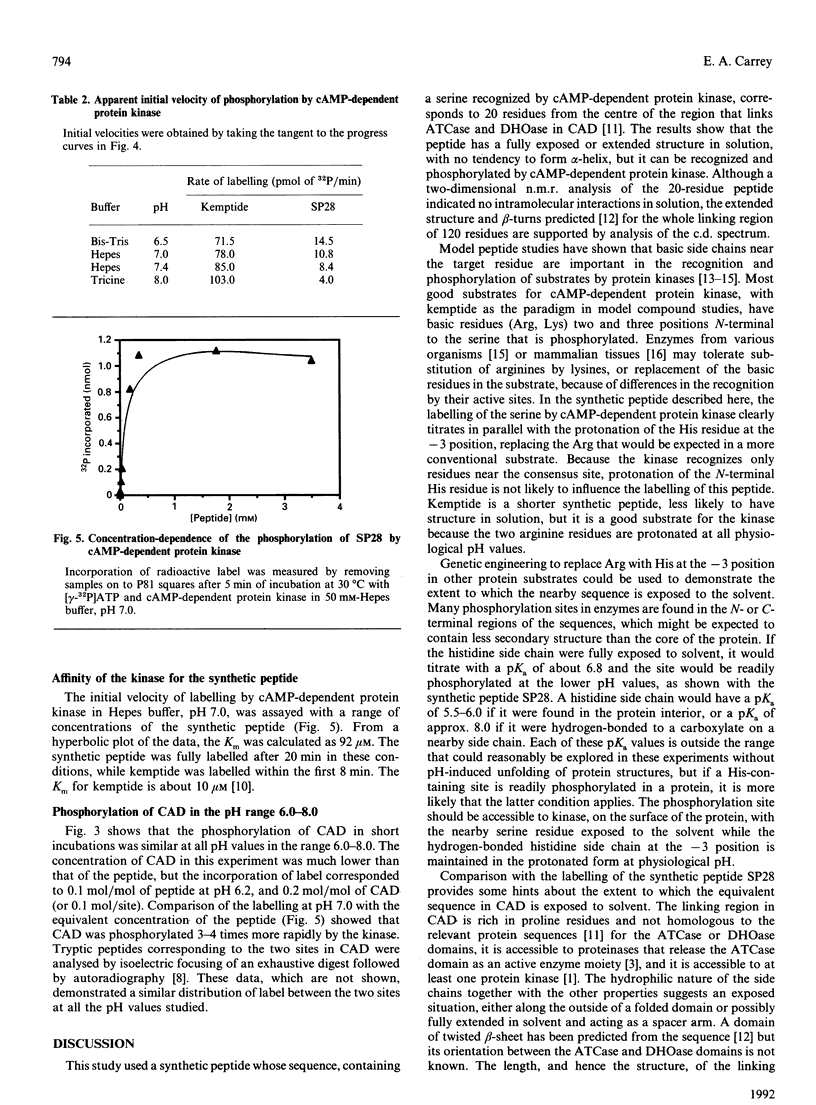
The multienzyme polypeptide CAD is phosphorylated at two sites by cyclic AMP (cAMP)-dependent protein kinase. Site 2 has two interesting features: it is located in a 'linking region' between two discretely folded enzyme domains, and a histidine, instead of the more usual arginine, is found three positions N-terminal to the phosphorylated serine. A synthetic peptide corresponding to the sequence around site 2 has an extended or random structure in solution, and the proton n.m.r. chemical shift of the histidine residues can be titrated against pH in the range 6.0-8.0. The peptide is phosphorylated more rapidly by cAMP-dependent protein kinase at lower pH values, indicating that the protonated histidine side chain corresponds to the arginine in the consensus recognition sequence for the kinase. Kemptide, a specific synthetic substrate for the kinase, was phosphorylated with a higher affinity and at a similar rate at all pH values. CAD was a better substrate than the synthetic peptide, and labelling was not affected by the pH of the incubation conditions. The results indicate that the phosphorylation site in the interdomain linker is sufficiently exposed to the solvent to ensure accessibility to the kinase, but that secondary or tertiary structure in the intact protein allows the histidine residue to remain protonated at physiological pH and enhances recognition of the phosphorylatable serine residue.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carrey E. A., Campbell D. G., Hardie D. G. Phosphorylation and activation of hamster carbamyl phosphate synthetase II by cAMP-dependent protein kinase. A novel mechanism for regulation of pyrimidine nucleotide biosynthesis. EMBO J. 1985 Dec 30;4(13B):3735–3742. doi: 10.1002/j.1460-2075.1985.tb04142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrey E. A., Hardie D. G. Mapping of catalytic domains and phosphorylation sites in the multifunctional pyrimidine-biosynthetic protein CAD. Eur J Biochem. 1988 Feb 1;171(3):583–588. doi: 10.1111/j.1432-1033.1988.tb13828.x. [DOI] [PubMed] [Google Scholar]
- Carrey E. A., Hardie D. G. Mapping of radiolabeled peptides derived from proteolysis of polypeptides bound to nitrocellulose after "Western" blotting. Anal Biochem. 1986 Nov 1;158(2):431–435. doi: 10.1016/0003-2697(86)90571-3. [DOI] [PubMed] [Google Scholar]
- Carrey E. A. Nucleotide ligands protect the inter-domain regions of the multifunctional polypeptide CAD against limited proteolysis, and also stabilize the thermolabile part-reactions of the carbamoyl-phosphate synthase II domains within the CAD polypeptide. Biochem J. 1986 Jun 1;236(2):327–335. doi: 10.1042/bj2360327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The role of protein phosphorylation in the hormonal control of enzyme activity. Eur J Biochem. 1985 Sep 16;151(3):439–448. doi: 10.1111/j.1432-1033.1985.tb09121.x. [DOI] [PubMed] [Google Scholar]
- Coleman P. F., Suttle D. P., Stark G. R. Purification from hamster cells of the multifunctional protein that initiates de novo synthesis of pyrimidine nucleotides. J Biol Chem. 1977 Sep 25;252(18):6379–6385. [PubMed] [Google Scholar]
- Denis C. L., Kemp B. E., Zoller M. J. Substrate specificities for yeast and mammalian cAMP-dependent protein kinases are similar but not identical. J Biol Chem. 1991 Sep 25;266(27):17932–17935. [PubMed] [Google Scholar]
- House C., Wettenhall R. E., Kemp B. E. The influence of basic residues on the substrate specificity of protein kinase C. J Biol Chem. 1987 Jan 15;262(2):772–777. [PubMed] [Google Scholar]
- Musmanno L. A., Maley J. A., Davidson J. N. Synthesis of the nonconserved dihydroorotase domain of the multifunctional hamster CAD protein in Escherichia coli. Gene. 1991 Mar 15;99(2):211–216. doi: 10.1016/0378-1119(91)90129-y. [DOI] [PubMed] [Google Scholar]
- Pearson R. B., Woodgett J. R., Cohen P., Kemp B. E. Substrate specificity of a multifunctional calmodulin-dependent protein kinase. J Biol Chem. 1985 Nov 25;260(27):14471–14476. [PubMed] [Google Scholar]
- Provencher S. W., Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981 Jan 6;20(1):33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- Reimann E. M., Beham R. A. Catalytic subunit of cAMP-dependent protein kinase. Methods Enzymol. 1983;99:51–55. doi: 10.1016/0076-6879(83)99039-0. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr Assays of protein kinase. Methods Enzymol. 1983;99:3–6. doi: 10.1016/0076-6879(83)99034-1. [DOI] [PubMed] [Google Scholar]
- Simmer J. P., Kelly R. E., Scully J. L., Grayson D. R., Rinker A. G., Jr, Bergh S. T., Evans D. R. Mammalian aspartate transcarbamylase (ATCase): sequence of the ATCase domain and interdomain linker in the CAD multifunctional polypeptide and properties of the isolated domain. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4382–4386. doi: 10.1073/pnas.86.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhar S., Munday M. R. Evidence for different isozymic forms of catalytic subunit of cyclic AMP-dependent protein kinase in heart and lactating mammary gland of the rat. Biochem Soc Trans. 1992 Aug;20(3):307S–307S. doi: 10.1042/bst020307s. [DOI] [PubMed] [Google Scholar]
- Williams N. K., Simpson R. J., Moritz R. L., Peide Y., Crofts L., Minasian E., Leach S. J., Wake R. G., Christopherson R. I. Location of the dihydroorotase domain within trifunctional hamster dihydroorotate synthetase. Gene. 1990 Oct 15;94(2):283–288. doi: 10.1016/0378-1119(90)90399-c. [DOI] [PubMed] [Google Scholar]


