Abstract
Yersinia pestis has been infecting humans since the Late Neolithic (LN). Whether those early infections were isolated zoonoses or initiators of a pandemic remains unclear. We report Y. pestis infections in two individuals (of 133) from the LN necropolis at Warburg (Germany, 5300–4900 cal BP). Our analyses show that the two genomes belong to distinct strains and reflect independent infection events. All LN genomes known today (n = 4) are basal in the phylogeny and represent separate lineages that probably originated in different animal hosts. In the LN, an opening of the landscape resulted in the introduction of new rodent species, which may have acted as Y. pestis reservoirs. Coincidentally, the number of dogs increased, possibly leading to Y. pestis infections in canines. Indeed, we detect Y. pestis in an LN dog. Collectively, our data suggest that Y. pestis frequently entered human settlements at the time without causing significant outbreaks.
Subject terms: Evolutionary genetics, Infectious diseases
Ancient pathogen genomics sheds light on Yersinia pestis evolution and provides evidence of a dog infection during the Late Neolithic, indicating that the pathogen came frequently into contact with humans without causing significant outbreaks.
Introduction
Plague is considered one of the most notorious scourges of humanity and was responsible for at least three pandemics in historical times1. Its causative agent, Yersinia pestis, has been infecting humans for more than 5000 years. The oldest Y. pestis genomes so far have been detected in remains of a hunter-gatherer from Riņņukalns, Latvia (RV2039; 5300–5050 cal BP)2 and a farmer from Gökhem, Sweden (Gökhem_2; 5040–4867 cal BP)3 (Fig. 1A), here referred to as Late Neolithic (LN). The two LN genomes represent strains in the early stages of Y. pestis evolution and were shown to be ancestral to a phylogenetically distinct lineage termed Late Neolithic and Bronze Age (LNBA; 4800–2500 cal BP), which was present throughout Eurasia until at least the third millennium BP4. Both LN strains lacked the virulence factors and mutations observed in much later forms that were required for efficient transmission from rodents to humans via fleas2–5. However, it remains unclear whether the Y. pestis infections detected in the LN skeletal remains were due to isolated zoonoses or marked the beginning of a long-lasting pandemic across Eurasia sustained by human-to-human contact2–4,6. Another question centers on what may have facilitated the transmission of Y. pestis to humans before adaptations to the flea evolved. Various as yet unknown rodent species could have acted as primary hosts for Y. pestis at that time. Human exposure to these rodents and possibly Y. pestis might have been increased through domesticated carnivores, i.e. dogs, which hunted these animals. This could also have led to Y. pestis infections in the canines themselves. In this study, we extend our understanding of Y. pestis evolution during the LN by presenting two new genomes from human individuals and providing evidence of a Y. pestis infection in a dog.
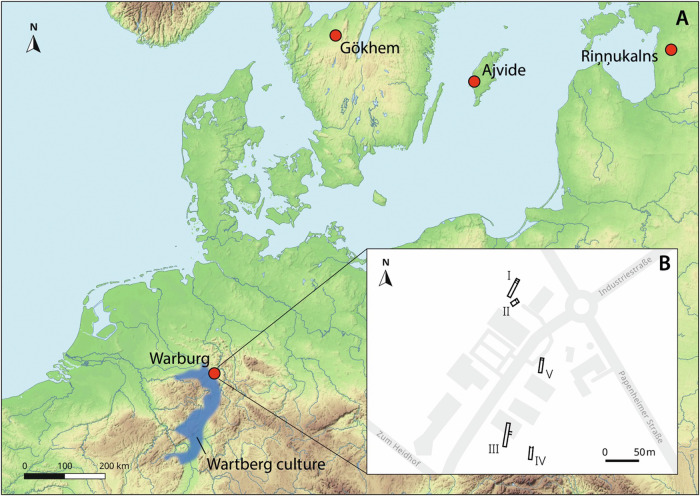
Fig. 1. Map of LN sites.
correctLocation of archaeological sites in which LN Y. pestis was identified in human (Warburg, Riņņukalns, Gökhem) and canine remains (Ajvide) (A). Warburg necropolis with the gallery graves I, III-V and building II (B).
Results
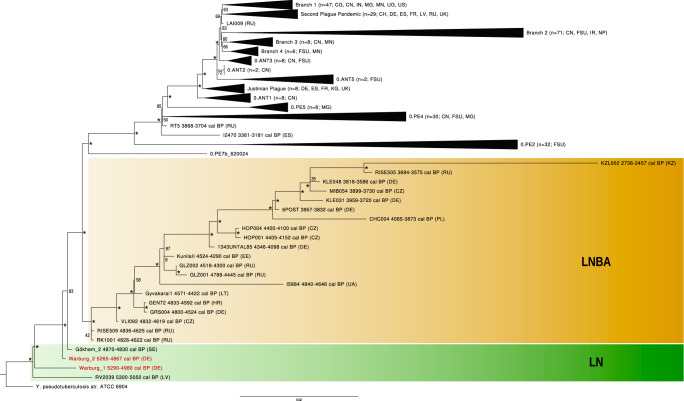
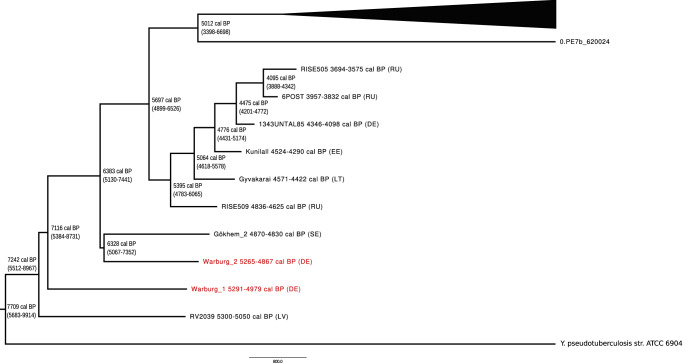
Here, we report on two new LN Y. pestis genomes that were identified in human remains from the Warburg necropolis (5300–4900 cal BP), located in present-day Germany (Fig. 1A). The site is archaeologically attributed to the LN Wartberg Culture (WBC)7 and consists of five gallery graves (I-V)7 with commingled skeletal remains of at least 198 individuals8 (Fig. 1B). For 133 individuals from gallery graves I, III and IV, we generated genome-wide shotgun sequences. Metagenomic screening of the datasets for Y. pestis DNA revealed two positive individuals, hereafter labelled Warburg_1 (5291-4979 cal BP; from gallery grave I) and Warburg_2 (5265-4857 cal BP; from gallery grave III). Their remains were discovered in two separate gallery graves and the radiocarbon dates likely placed them in different times. With a likelihood of 88%, Warburg_1 is older than Warburg_2 and the probability distributions indicate a difference of about 200 years (OxCal v4.4.4., Supplementary Data 1, Supplementary Fig. 1). We found that the two individuals had typical WBC ancestry components9 and were not related to each other (Supplementary Data 2 and 3, Supplementary Figs. 2 and 3). Regarding Y. pestis reads, both samples showed a high mean coverage of up to 28 x for the chromosome and 117 x for the three plasmids (Table 1, Supplementary Data 1, Supplementary Figs. 4 and 5). The genomes had virulence factors described for modern Y. pestis, apart from the ymt gene, the filamentous prophage YpfΦ and YPMT1.66c that are all known to be absent in LN strains4,5,10 (Supplementary Fig. 6). Consistent with this finding, all four LN genomes had a basal position in the phylogenetic tree (Figs. 2 and 3). Warburg_1 branched off shortly after RV2039 and Warburg_2 diverged shortly before Gökhem_2. To assess the potential influence of single-nucleotide polymorphisms (SNPs) characteristic of the LN lineage (Warburg_1, Warburg_2, RV2039, Gökhem_2), we carried out a SNP effect analysis. We focused only on non-synonymous SNPs which lead to nonsense or missense mutations due to the loss/gain of a start/stop codon. In doing so, we discovered six SNPs in two members of the LN branch that were not shared by later-dating strains (Supplementary Data 4). These six SNPs either introduce or remove stop codons. The introduction of a stop codon in these cases creates short N-terminal protein fragments, unlikely to retain the function of the protein. Removal of a stop codon leads to C-terminal extensions by translation of intergenic regions or potential fusion proteins including sequences from the neighbouring gene. Based on the sequences, a functional assessment is not possible, and additional experiments are required to distinguish between the two possibilities. The accumulation of pseudogenes, which is a hallmark of the LNBA clade4, was not visible in the LN genomes. This may indicate that adaptation to one specific host had not yet occurred.
Table 1.
Mapping statistics for the Y. pestis-positive samples
| Sample | Mapping reads | Cov 1x (%) | Cov 2x (%) | Cov 3x (%) | Mean coverage |
|---|---|---|---|---|---|
| chromosome NC_003143.1 | |||||
| Warburg_1 | 521,157 | 89.45 | 76.32 | 58.09 | 3.22 x |
| Warburg_2 | 3,315,529 | 94.84 | 94.68 | 94.58 | 28.89 x |
| C90 | 31,549 | 8.42 | 0.31 x | ||
| pCD1 NC_003131.1 | |||||
| Warburg_1 | 16,491 | 92.14 | 90.62 | 88.44 | 8.38 x |
| Warburg_2 | 94,944 | 92.86 | 92.33 | 92.09 | 43.96 x |
| C90 | 263 | 12.45 | 0.14 x | ||
| pMT1 NC_003134.1 | |||||
| Warburg_1 | 7,857 | 57.52 | 33.57 | 16.49 | 1.5 x |
| Warburg_2 | 75,513 | 74.55 | 73.04 | 72.67 | 17.03 x |
| C90 | 187 | 6.04 | 0.07 x | ||
| pPCP1 NC_003132.1 | |||||
| Warburg_1 | 4,386 | 79.85 | 79.80 | 79.67 | 11.34 x |
| Warburg_2 | 42,311 | 80.28 | 80.20 | 80.19 | 117.31 x |
| C90 | 63 | 20.92 | 0.26 x | ||
Overview of statistics after mapping to the chromosome and plasmids of the Y. pestis reference genome. The abbreviation “cov“ refers to the percentage of the genome with at least 1 x, 2 x or 3 x coverage, respectively.
Fig. 2. Phylogeny of ancient and modern Y. pestis genomes.
Maximum likelihood phylogenetic tree based on the SNP alignment (10,743 positions) of 226 modern genomes, 62 published ancient genomes, the novel genomes Warburg_1 and Warburg_2 (red) and the outgroup Y. pseudotuberculosis. LN strains are highlighted in green and LNBA strains in yellow. Dating of the ancient strains is given as calibrated years before the present (cal BP). Country abbreviation is given in brackets (CH = Switzerland, CG = Congo, CN = China, CZ = Czech Republic, DE = Germany, EE = Estonia, ES = Spain, FR = France, FSU = Former Soviet Union, HR = Croatia, IN = India, IR = Iran, KG = Kyrgyzstan, KZ = Kazakhstan, LT = Lithuania, LV = Latvia, MG = Madagascar, MM = Myanmar, MN = Mongolia, NP = Nepal, PL = Poland, RU = Russia, SE = Sweden, UA = Ukraine, UG = Uganda, UK = United Kingdom, US = United States). Unique positions to the outgroup were excluded to facilitate the visualisation. Bootstrap values were calculated for 1000 replicates and nodes with a support above 90 are marked with an asterisk. The scale corresponds to substitutions per site. Genomes included in the phylogeny are listed in Supplementary Data 5.
Fig. 3. Molecular dating of early Y. pestis.
Maximum clade credibility tree based on 42 modern and ancient Y. pestis genomes and the two genomes from Warburg (red). All dates are given as calibrated years before the present (cal BP). Country abbreviation is provided in brackets as described in Fig. 2. Genomes included in the molecular dating analysis are listed in Supplementary Data 6.
To test the hypothesis that the prehistoric dog could have been infected with Y. pestis, we screened publicly available shotgun datasets from Eurasian Neolithic (n = 15) and Bronze Age canines (n = 6)11–15 for pathogens in general and Y. pestis in particular. In one canine (labelled C90), we detected Y. pestis-specific reads which had not been reported in the original publication12. Specimen C90 consisted of a mandible found in the cultural layers of the settlement site Ajvide on the island of Gotland in present-day Sweden and dated to 4900–4500 cal BP (Pitted Ware Culture; Fig. 1A)12. The reads in C90 showed the typical damage profiles for ancient DNA. Although the data was previously only generated for canine population genetic analyses12, we could reconstruct 8% of the bacterial genome. However, due to the low coverage, we did not explore its placement in the phylogenetic tree.
Discussion
In this study, we identified Y. pestis in two individuals from the LN necropolis at Warburg in Germany. Our results suggest that the two Y. pestis genomes (Warburg_1 and Warburg_2) belonged to distinct strains. They differed in 82 positions (Supplementary Fig. 7). Due to this genetic distance, it can be assumed that the two genomes did not directly evolve from each other. This finding is also reflected in the typology of the phylogenetic tree indicating a divergence of the genomes long before the infections occurred (Fig. 3). In addition, the genomes were detected in unrelated individuals who were buried in different gallery graves. All evidence suggests that both infections represent independent events and thus appear not epidemiologically linked.
Interestingly, despite the screening of numerous specimens from each gallery grave (total n = 133), we found no further infections with Y. pestis or any other pathogen. In another collective WBC grave at the site of Niedertiefenbach (n = 42; present-day Germany), no signs of pathogens were detected either9. It must be acknowledged that pathogen-negative results do not necessarily mean absence of infection, as taphonomic processes may have degraded any microbial traces. In addition, both the Warburg and Niedertiefenbach samples consisted of petrous bones as well as teeth, the latter being the better source material for the detection of blood-borne viruses and bacteria16. These limitations notwithstanding, the arguments presented above (i.e., independent infection events at Warburg) and the overall small number of 2 positives among the 175 tested WBC individuals suggest that the collective graves were not used for the burial of victims of a plague outbreak or other epidemics, as previously suggested for the same period3. The few Y. pestis cases in the WBC are consistent with the results of other large-scale pathogen screenings that have so far revealed only single infections with human pathogens (Y. pestis, Salmonella enterica) or endemically occurring infections (hepatitis B virus, parvovirus B19, Helicobacter pylori) in Neolithic remains17. Also, the mortuary practice of single and multiple inhumations during that period does not indicate mass mortality, as would be expected in an epidemic. The findings from the Neolithic are thus in marked contrast to the short-term mass burials and the high pathogen load seen in the Middle Ages18,19.
The two Warburg genomes increase the number of Y. pestis genomes from the LN to four. All LN genomes were distinct from each other and represent lineages separated by an extended period of independent evolution (Figs. 2 and 3). The high diversity and the basal position of the LN Y. pestis lineages may reflect a low level of specialization at this early evolutionary stage. Such reduced specialization potentially facilitated their survival across diverse environments and a wide host range. According to the current phylogeny, the LN strains gave rise to two lineages, one from which the pathogens of the deadly Justinianic and medieval plagues emerged and another that led to the LNBAs (Fig. 2). The LNBA clade went extinct sometime in the third millennium BP4 (Figs. 2 and 3). For more than 2000 years, the LNBA strains were the dominant Y. pestis lineage in humans across Eurasia4. They may represent an adaptation to a very specialized Y. pestis ecology (e.g., host(s)), as reflected by the increasing pseudogenization of bacterial genes over time4. This process could have led to the evolutionary dead-end of the LNBA lineage and to a less severe, perhaps even chronic, manifestation of plague in humans that resembled an endemic rather than a pandemic disease2,4.
During the LN, woodland clearance increasingly created open landscapes in central and northern Europe20–23 that attracted a variety of new rodent species (e.g., European hamster Cricetus cricetus24) originally native to the steppe further east or south. Some of these species could have been natural reservoirs of Y. pestis1 and an infection in humans would have been feasible through close contact with a Y. pestis-positive wild animal or carcass25,26. However, we do not know how frequent such encounters were, especially when the animals in question were not normally hunted or touched by humans. Therefore, we propose the dog as facilitator which could have increased exposure of humans to Y. pestis from various wild animals, especially those with which humans did not come into regular contact. The archaeological record during the LN shows large numbers of dog remains, for instance, as material for ornamentation (pendants and jewellery made from dog teeth, e.g., 356 teeth [canines] in the grave Wewelsburg 118 in Altendorf)27. In addition, the animals were likely used for hunting and herding28,29. If dogs preyed on infected animals, this could have increased the probability of Y. pestis transmission from rodents to humans. Since modern dogs can develop pneumonic plague and infect humans directly without the need for flea adaptation1,26, the question arises whether this was also the case during the LN. Given instances of dog-to-human transmissions today1, it is possible that dogs themselves acted as a Y. pestis reservoir for humans at the time, or vice versa.
To the best of our knowledge, C90 is so far the only case of Y. pestis infection in a Neolithic dog. This small number may be explained by the innate resistance of dogs to Y. pestis which leads to rapid pathogen clearance and a low fatality rate1,30. If Neolithic dogs recovered from plague as frequently and quickly as their modern counterparts30, most that were ever infected would be Y. pestis-negative at the time of their death (and in the palaeogenomic pathogen screening). Thus, we might underestimate the actual number of infected dogs.
The presence of Y. pestis in a dog from present-day Sweden fits well with the geographical distribution pattern of the four infected human individuals (Fig. 1A). Surprisingly, all five Y. pestis findings from the LN occupy a relatively small geographical area in northwestern Europe. Overall, the results suggest a significant Y. pestis exposure in and around human settlements at the time, most likely leading to isolated infections rather than large-scale disease outbreaks.
Methods
Human skeletal samples
Archaeologically, the Warburg necropolis belongs to the Late Neolithic Wartberg culture (WBC) and is dated to 5300–4900 cal BP. The burial complex is located 1.6 km northwest of the town Warburg (Germany). It consisted of four stone gallery graves (I, III, IV, V) and one wooden burial chamber or ritual building (II)7. The gallery graves contained commingled skeletal remains of at least 198 individuals8. In total, we included bone and tooth samples from 133 individuals in the pathogen screening for which we could generate at least 2.5 million sequencing reads each.
DNA isolation, sequencing and processing
All lab work was carried out in the Kiel Ancient DNA Laboratory, following established guidelines31. Tooth and bone samples were cleaned with pure bleach, rinsed with purified water, dried overnight at 37 °C, and subsequently ground in a ball mill homogenizer for 45 sec at maximum speed. Between 80–120 mg of powder was used for extraction based on a published protocol32 as outlined in a previous study33. The double-stranded half-UDG libraries with unique index combinations for each sample were shotgun sequenced on an Illumina HiSeq platform (2x100bp) and the generated data was pre-processed by removing sequencing adaptor remnants and merging overlapping reads with ClipAndMerge v1.7.834.
Metagenomic screening
For the metagenomic screening, a custom database was created with the “MALT-build” function of the software MALT35, following the manual’s instructions. For this purpose, all bacterial and viral genomes that included the description “complete genome” were downloaded from NCBI36 (as of 12.04.2021). This final database comprised 38,273 complete bacterial and viral genomes. Subsequently, metagenomic screening on both the pre-processed human (n = 133) and published canine datasets (n = 21)11–15 was performed using the “MALT-run” function in semi-global alignment mode with a 90% sequence identity threshold. The output files were visualized in MEGAN637.
Yersinia pestis alignment
For the two human Y. pestis-positive samples Warburg_1 and Warburg_2, more shotgun data was generated from the initial libraries without additional enrichment. Subsequently, the reads of the three Y. pestis-positive samples (human: Warburg_1 and Warburg_2; canine: C9012) were mapped against the reference genome of the Y. pestis CO92 chromosome (NC_003143.1) and the three plasmids pCD1 (NC_003131.1), pMT1 (NC_003134.1) and pPCP1 (NC_003132.1). The ancient origin of the samples was authenticated with DamageProfiler v1.1 (Supplementary Data 1, Supplementary Figs. 4 and 5)38.
SNP effect analysis
VCF files from Warburg_1 and Warburg_2 were filtered for SNPs with a coverage of at least 3 x, a calling quality of 30 and a 90% majority call. This set of filtered SNPs was provided to SNPEff v4.339 to analyze the effects of SNPs in the ancient genomes. The output file was filtered for high-impact SNPs and evaluated. A SNP effect analysis was not possible on the data from the dog genome (C90) due to insufficient coverage.
Analysis of virulence factors
The Y. pestis reads from Warburg_1 and Warburg_2 were screened for the presence or absence of 115 chromosomal and 44 plasmid-associated virulence genes compiling a bed file based on supplementary information of a previous study4. To asses the coverage of these virulence genes for each Y. pestis genome, the percentage of the coverage was determined using bedtools v2.25.040 with the “genomecov” and “coverage” functions. A short awk script (awk ‘$5! = 0 {print $1 “\t” $4 “\t” $8}’ [sampleID].histogram.bed > [sampleID].coverage_table.bed) was used to extract columns for chromosome/plasmid name ($1), gene name ($4) and gene coverage ($8) from the bed file. The values were all collected in an excel table that was used as input for R v3.6.341. Heatmaps were created with the R pheatmap package.
Yersinia pestis phylogenetic analysis
Generation of VCF files and phylogenetic analyses were based on 226 modern and 62 ancient strains, including Warburg_1, Warburg_2 (Supplementary Data 5), and carried out as described in Susat et al.42. To ensure the authenticity of the SNPs used, we took the following precautions. For a SNP to be called, it must be supported by 90% of the reads and a minimum coverage of 4 reads. SNPs located in previously identified homoplastic regions4 were excluded (based on a list provided by https://github.com/aidaanva/LNBAplague/blob/main/multiVCFAnalyzer/SNPstoExclude). For single-stranded libraries generated from samples of the LNBA lineage (downloaded from the open repository ENA, project no. PRJEB51099; libraries KLE031, KLE048), we implemented the publicly available genoSL.R script (https://github.com/aidaanva/GenoSL) to exclude SNPs that are based on the C → T deaminations only visible in single-stranded libraries4. Lastly, after generation of the multi-vcf alignment, we excluded Y. pseudotuberculosis-specific SNPs to allow easier visualization of the tree, with an intentionally shortened branch for the outgroup Y. pseudotuberculosis. Due to insufficient Y. pestis coverage in sample C90, a reliable placement of the bacterial genome in the phylogeny was not possible.
Molecular dating
A SNP-based phylogenetic tree was generated on a subset of 42 representative Y. pestis genomes, including Warburg_1 and Warburg_2 (Supplementary Data 6), as described in the previous paragraph. The obtained tree was used as a starting tree for BEAST2 v2.643. Dates were provided in BP (before the present) format, with 1950 AD corresponding to age 0 and the mean age calculated for each dating range. The following parameters were used: uncorrelated relaxed clock, lognormal distribution, GTR + G4 substitution model and a coalescent constant population, as applied in previous studies2,3,43. Two different runs were performed: one run based on a provided phylogenetic tree that can be altered by the tool, and one run without a starting tree. The multiple output files from both independent runs were combined using the LogCombiner v2.6.744 and evaluated with Tracer v1.7.245 with effective sample size (ESS) exceeding a minimum value of 250. Trees were combined with TreeAnnotator v2.6.044.
Human population genetic analyses
Reads from Warburg_1 and Warburg_2 were aligned to the human genome reference (hg19). The ancient origin of the DNA was authenticated with DamageProfiler v1.1 (Supplementary Data 1)38. Mapping of reads, contamination estimation, genetic sex determination as well as mitochondrial DNA and Y chromosome haplogroup determination were performed as described in Immel et al.9. For the two individuals, pseudo-haploid genotypes on ~1240 K informative SNPs46 were generated and subsequently merged with previously published genotypes from various ancient and modern West Eurasian populations (Allen Ancient DNA Resource)47. For the principal component analysis (PCA) with smartpca48, the genotypes of all ancient populations were projected onto the principal components calculated from the modern populations. An unsupervised ADMIXTURE v1.3.049 analysis was performed using 3–12 components (K) with 10 bootstraps each. Additionally, two-way admixture models were tested with qpAdm from Admixtools50. Kinship between the two Warburg individuals was assessed with READ51 using the normalization value of 0.2493 derived from the median mismatch rate observed in the Warburg dataset, which comprised 8256 pairwise comparisons from 129 samples.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through the CRC 1266 – project number 290391021.
Author contributions
A.N. and B.K.-K. conceived and designed the study. K.S. and M.M.R. provided the human skeletal material. J.S., M.H.-C., J.H.B and N.A.d.S. generated and analyzed aDNA data. K.S., M.M.R., U.S., W.K., C.R. and J.M. provided environmental, archaeological and archaeozoological context and interpretation. H.S. interpreted the results of the SNP effect analysis. A.N. and B.K.-K. interpreted the results and wrote the manuscript with input from all the other authors. J.H.B edited the final manuscript.
Peer review
Peer review information
Communications Biology thanks Sébastien Calvignac-Spencer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Tobias Goris. A peer review file is available.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The generated datasets are available in the European Nucleotide Archive under the accession number PRJEB49648.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Materials availability:
This study did not generate new unique reagents.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06676-7.
References
- 1.Barbieri, R. et al. Yersinia pestis: the natural history of plague. Clin. Microbiol. Rev.34, e00044–19 (2020). 10.1128/CMR.00044-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Susat, J. et al. A 5,000 year-old hunter-gatherer already plagued by Yersinia pestis. Cell Rep.35, 109278 (2021). 10.1016/j.celrep.2021.109278 [DOI] [PubMed] [Google Scholar]
- 3.Rascovan, N. et al. Emergence and spread of basal lineages of Yersinia pestis during the Neolithic decline. Cell176, 295–305.e10 (2019). 10.1016/j.cell.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 4.Andrades Valtueña, A. et al. Stone Age Yersinia pestis genomes shed light on the early evolution, diversity, and ecology of plague. Proc. Natl Acad. Sci. USA119, e2116722119 (2022). 10.1073/pnas.2116722119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun, Y.-C., Jarrett, C. O., Bosio, C. F. & Hinnebusch, B. J. Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe15, 578–586 (2014). 10.1016/j.chom.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swali, P. et al. Yersinia pestis genomes reveal plague in Britain 4000 years ago. Nat. Commun.14, 2930 (2023). 10.1038/s41467-023-38393-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raetzel-Fabian, D. Absolute Chronologie. In Die Kollektivgäber-Nekropole Warburg I-V, Bodenaltertümer Westfalens (ed. Günther, Klaus) 165–178 (Mainz, 1997).
- 8.Löwen, H. Menschenreste. In Die Kollektivgäber-Nekropole Warburg I-V, Bodenaltertümer Westfalens (ed. Günther, Klaus) 39−47; 81−91; 112−128; 191−194 (Mainz, 1997).
- 9.Immel, A. et al. Genome-wide study of a Neolithic Wartberg grave community reveals distinct HLA variation and hunter-gatherer ancestry. Commun. Biol.4, 113 (2021). 10.1038/s42003-020-01627-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonczarowska, J. H. et al. Ancient Yersinia pestis genomes lack the virulence-associated YpfΦ prophage present in modern pandemic strains. Proc. Biol. Sci.290, 20230622 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergström, A. et al. Grey wolf genomic history reveals a dual ancestry of dogs. Nature607, 313–320 (2022). 10.1038/s41586-022-04824-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergström, A. et al. Origins and genetic legacy of prehistoric dogs. Science370, 557–564 (2020). 10.1126/science.aba9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botigué, L. R. et al. Ancient European dog genomes reveal continuity since the early Neolithic. Nat. Commun.8, 16082 (2017). 10.1038/ncomms16082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frantz, L. A. F. et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science352, 1228–1231 (2016). 10.1126/science.aaf3161 [DOI] [PubMed] [Google Scholar]
- 15.Ollivier, M. et al. Dogs accompanied humans during the Neolithic expansion into Europe. Biol. Lett.14, 20180286 (2018). 10.1098/rsbl.2018.0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margaryan, A. et al. Ancient pathogen DNA in human teeth and petrous bones. Ecol. Evol.8, 3534–3542 (2018). 10.1002/ece3.3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spyrou, M. A., Bos, K. I., Herbig, A. & Krause, J. Ancient pathogen genomics as an emerging tool for infectious disease research. Nat. Rev. Genet.20, 323–340 (2019). 10.1038/s41576-019-0119-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonczarowska, J. H. et al. Pathogen genomics study of an early medieval community in Germany reveals extensive co-infections. Genome Biol.23, 250 (2022). 10.1186/s13059-022-02806-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haller, M. et al. Mass burial genomics reveals outbreak of enteric paratyphoid fever in the late medieval trade city Lübeck. iScience24, 102419 (2021). 10.1016/j.isci.2021.102419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trondman, A. K. et al. Pollen-based quantitative reconstructions of Holocene regional vegetation cover (plant-functional types and land-cover types) in Europe suitable for climate modelling. Glob. Change Biol.21, 676–697 (2015). 10.1111/gcb.12737 [DOI] [PubMed] [Google Scholar]
- 21.Fyfe, R. M., Woodbridge, J. & Roberts, N. From forest to farmland: pollen‐inferred land cover change across Europe using the pseudobiomization approach. Glob. Change Biol.21, 1197–1212 (2015). 10.1111/gcb.12776 [DOI] [PubMed] [Google Scholar]
- 22.Zanon, M., Davis, B. A. S., Marquer, L., Brewer, S. & Kaplan, J. O. European forest cover during the past 12,000 years: a palynological reconstruction based on modern analogs and remote sensing. Front. Plant Sci.9, 253 (2018). 10.3389/fpls.2018.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Githumbi, E. et al. European pollen-based REVEALS land-cover reconstructions for the Holocene: methodology, mapping and potentials. Earth Syst. Sci. Data14, 1581–1619 (2022). 10.5194/essd-14-1581-2022 [DOI] [Google Scholar]
- 24.Schmölcke, U. Nutztierhaltung, Jagd und Fischfang. Zur Nahrungsmittelwirtschaft des frühgeschichtlichen Handelsplatzes von Groß Strömkendorf, Landkreis Nordwestmecklenburg. Beiträge zur Ur- und Frühgeschichte Mecklenburg-Vorpommerns43 (Lübstorf, 2004).
- 25.Slavin, P. & Sebbane, F. Emergence and spread of ancestral Yersinia pestis in Late Neolithic and Bronze Age Eurasia, ca. 5,000 to 2,500y B.P. Proc. Natl Acad. Sci. USA119, e2204044119 (2022). 10.1073/pnas.2204044119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell, S. B., Nelson, C. A., Hinckley, A. F. & Kugeler, K. J. Animal exposure and human plague, United States, 1970–2017. Emerg. Infect. Dis.25, 2270–2273 (2019). 10.3201/eid2512.191081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schierhold, K. Studien zur Hessisch-Westfälischen Megalithik: Forschungsstand und -perspektiven im europäischen Kontext. Münstersche Beiträge zur ur- und frühgeschichtlichen Archäologie6 (Leidorf, 2012).
- 28.Ewersen, J., Ziegler, S., Ramminger, B. & Schmölcke, U. Stable isotopic ratios from Mesolithic and Neolithic canids as an indicator of human economic and ritual activity. J. Archaeol. Sci. Rep.17, 346–357 (2018). [Google Scholar]
- 29.Benecke, N. Archäozoologische Studien zur Entwicklung der Haustierhaltung in Mitteleuropa und Südskandinavien von den Anfängen bis zum ausgehenden Mittelalter (De Gruyter Akademie Forschung, 1994, Reprint 2015 Edn).
- 30.Nichols, M. C. et al. Yersinia pestis infection in dogs: 62 cases (2003-2011). J. Am. Vet. Med. Assoc.244, 1176–1180 (2014). 10.2460/javma.244.10.1176 [DOI] [PubMed] [Google Scholar]
- 31.Cooper, A. & Poinar, H. N. Ancient DNA: do it right or not at all. Science289, 1139 (2000). 10.1126/science.289.5482.1139b [DOI] [PubMed] [Google Scholar]
- 32.Rohland, N., Harney, E., Mallick, S., Nordenfelt, S. & Reich, D. Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B. Biol. Sci.370, 20130624 (2015). 10.1098/rstb.2013.0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krause-Kyora, B. et al. Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans. Nat. Commun.9, 1569 (2018). 10.1038/s41467-018-03857-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peltzer, A. et al. EAGER: efficient ancient genome reconstruction. Genome Biol.17, 60 (2016). 10.1186/s13059-016-0918-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vågene, Å. J. et al. Salmonella enterica genomes from victims of a major sixteenth-century epidemic in Mexico. Nat. Ecol. Evol.2, 520–528 (2018). 10.1038/s41559-017-0446-6 [DOI] [PubMed] [Google Scholar]
- 36.O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res.44, D733–D745 (2016). 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huson, D. H., Auch, A. F., Qi, J. & Schuster, S. C. MEGAN analysis of metagenomic data. Genome Res.17, 377–386 (2007). 10.1101/gr.5969107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neukamm, J., Peltzer, A. & Nieselt, K. DamageProfiler: fast damage pattern calculation for ancient DNA. Bioinformatics37, 3652–3653 (2021). 10.1093/bioinformatics/btab190 [DOI] [PubMed] [Google Scholar]
- 39.Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (Austin)6, 80–92 (2012). 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.RStudio Team. RStudio: Integrated Development Environment for R.http://www.rstudio.com (2021).
- 42.Susat, J. et al. Yersinia pestis strains from Latvia show depletion of the pla virulence gene at the end of the second plague pandemic. Sci. Rep.10, 14628 (2020). 10.1038/s41598-020-71530-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouckaert, R. et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol.15, e1006650 (2019). 10.1371/journal.pcbi.1006650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol.7, 214 (2007). 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol.67, 901–904 (2018). 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathieson, I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature528, 499–503 (2015). 10.1038/nature16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mallick, S. et al. The Allen Ancient DNA Resource (AADR): a curated compendium of ancient human genomes. bioRxiv10.1101/2023.04.06.535797 (2023). [DOI] [PMC free article] [PubMed]
- 48.Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet.2, e190 (2006). 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res.19, 1655–1664 (2009). 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patterson, N. et al. Ancient admixture in human history. Genetics192, 1065–1093 (2012). 10.1534/genetics.112.145037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monroy Kuhn, J. M., Jakobsson, M. & Günther, T. Estimating genetic kin relationships in prehistoric populations. PLoS ONE13, e0195491 (2018). 10.1371/journal.pone.0195491 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The generated datasets are available in the European Nucleotide Archive under the accession number PRJEB49648.