Abstract
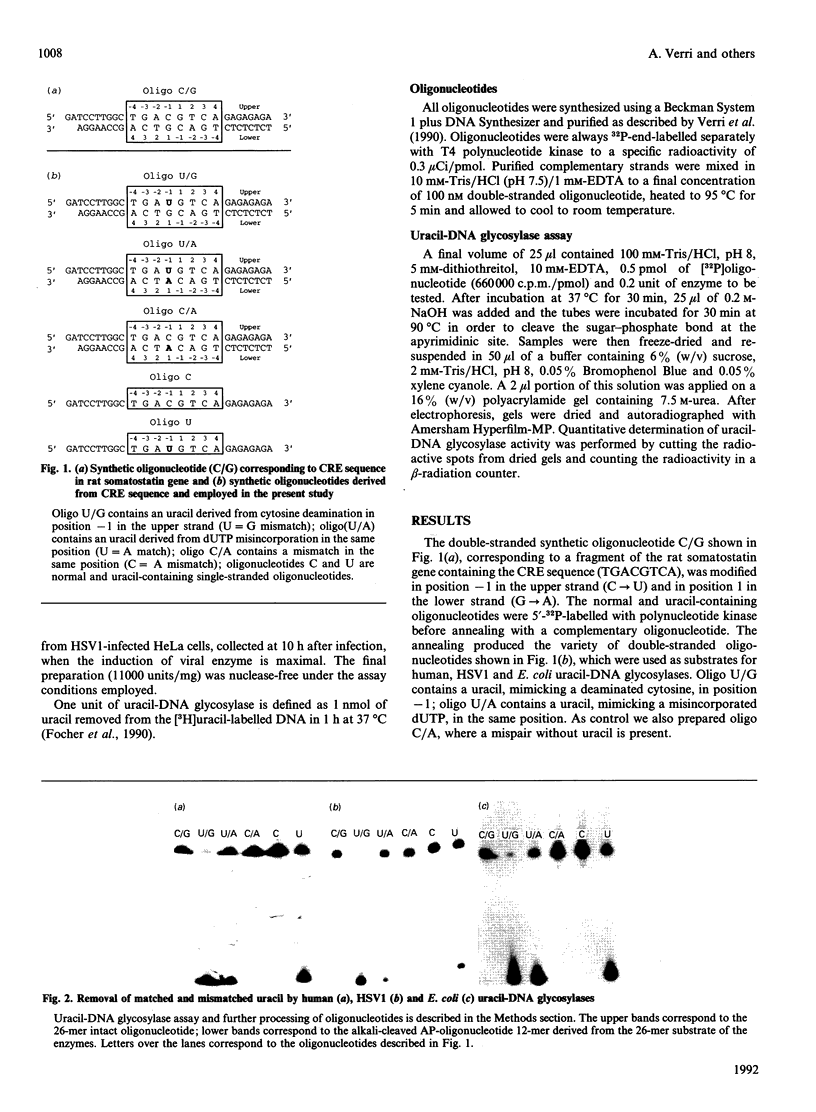
We have investigated the substrate specificity of human, viral and bacterial uracil-DNA glycosylases employing as substrate double-stranded oligonucleotides containing in the same position of the 5'-32P-labelled strand an uracil residue facing, on the complementary strand, guanine (mimicking cytosine deamination) or adenine (mimicking dUTP misincorporation). The enzyme removal of uracil was monitored and quantified by the generation of alkali-sensitive apyrimidinic sites. All three uracil-DNA glycosylases excise uracil from mispaired oligonucleotides (U/G) more efficiently than from paired oligonucleotides (U/A). The enzymes also remove uracil from single-stranded oligonucleotide with an efficiency similar to that observed with U/A paired oligonucleotide. The efficient recognition of U/G mispair by uracil-DNA glycosylase is important in minimizing miscoding transcripts and C/G-->T/A transitions in proliferating cells.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blaisdell P., Warner H. Partial purification and characterization of a uracil-DNA glycosylase from wheat germ. J Biol Chem. 1983 Feb 10;258(3):1603–1609. [PubMed] [Google Scholar]
- Caradonna S. J., Cheng Y. C. Uracil DNA-glycosylase. Purification and properties of this enzyme isolated from blast cells of acute myelocytic leukemia patients. J Biol Chem. 1980 Mar 25;255(6):2293–2300. [PubMed] [Google Scholar]
- Dianov G., Lindahl T. Preferential recognition of I.T base-pairs in the initiation of excision-repair by hypoxanthine-DNA glycosylase. Nucleic Acids Res. 1991 Jul 25;19(14):3829–3833. doi: 10.1093/nar/19.14.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., Rockstroh P. A., Warner H. R. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978 Jun;134(3):1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982 Aug;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focher F., Mazzarello P., Verri A., Hübscher U., Spadari S. Activity profiles of enzymes that control the uracil incorporation into DNA during neuronal development. Mutat Res. 1990 Mar;237(2):65–73. doi: 10.1016/0921-8734(90)90012-g. [DOI] [PubMed] [Google Scholar]
- Impellizzeri K. J., Anderson B., Burgers P. M. The spectrum of spontaneous mutations in a Saccharomyces cerevisiae uracil-DNA-glycosylase mutant limits the function of this enzyme to cytosine deamination repair. J Bacteriol. 1991 Nov;173(21):6807–6810. doi: 10.1128/jb.173.21.6807-6810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan H., Wittwer C. U. Uracil DNa-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981 Jun 11;9(11):2599–2613. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Mullaney J., Moss H. W., McGeoch D. J. Gene UL2 of herpes simplex virus type 1 encodes a uracil-DNA glycosylase. J Gen Virol. 1989 Feb;70(Pt 2):449–454. doi: 10.1099/0022-1317-70-2-449. [DOI] [PubMed] [Google Scholar]
- Sirover M. A. Induction of the DNA repair enzyme uracil-DNA glycosylase in stimulated human lymphocytes. Cancer Res. 1979 Jun;39(6 Pt 1):2090–2095. [PubMed] [Google Scholar]
- Talpaert-Borlé M., Campagnari F., Creissen D. M. Properties of purified uracil-DNA glycosylase from calf thymus. An in vitro study using synthetic DNA-like substrates. J Biol Chem. 1982 Feb 10;257(3):1208–1214. [PubMed] [Google Scholar]
- Talpaert-Borlé M., Clerici L., Campagnari F. Isolation and characterization of a uracil-DNA glycosylase from calf thymus. J Biol Chem. 1979 Jul 25;254(14):6387–6391. [PubMed] [Google Scholar]
- Tye B. K., Chien J., Lehman I. R., Duncan B. K., Warner H. R. Uracil incorporation: a source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1978 Jan;75(1):233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verri A., Mazzarello P., Biamonti G., Spadari S., Focher F. The specific binding of nuclear protein(s) to the cAMP responsive element (CRE) sequence (TGACGTCA) is reduced by the misincorporation of U and increased by the deamination of C. Nucleic Acids Res. 1990 Oct 11;18(19):5775–5780. doi: 10.1093/nar/18.19.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrad D. M., Caradonna S. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J Virol. 1988 Dec;62(12):4774–4777. doi: 10.1128/jvi.62.12.4774-4777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]