Abstract
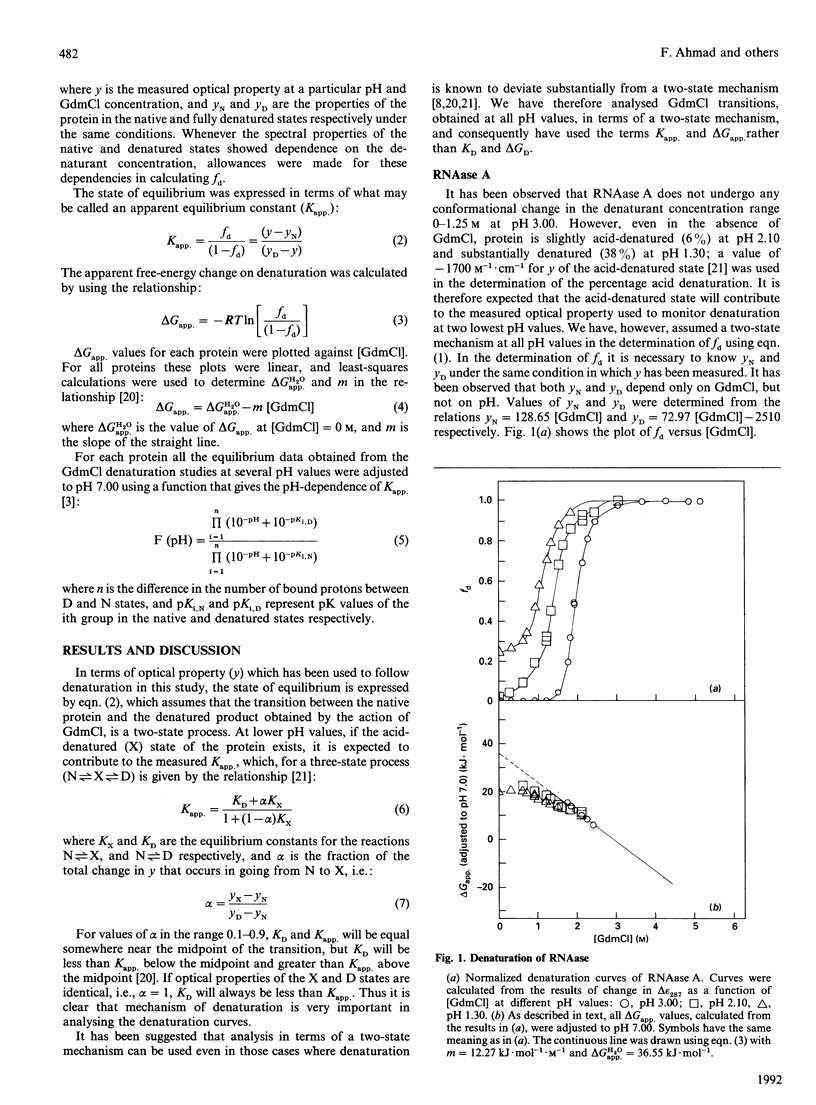
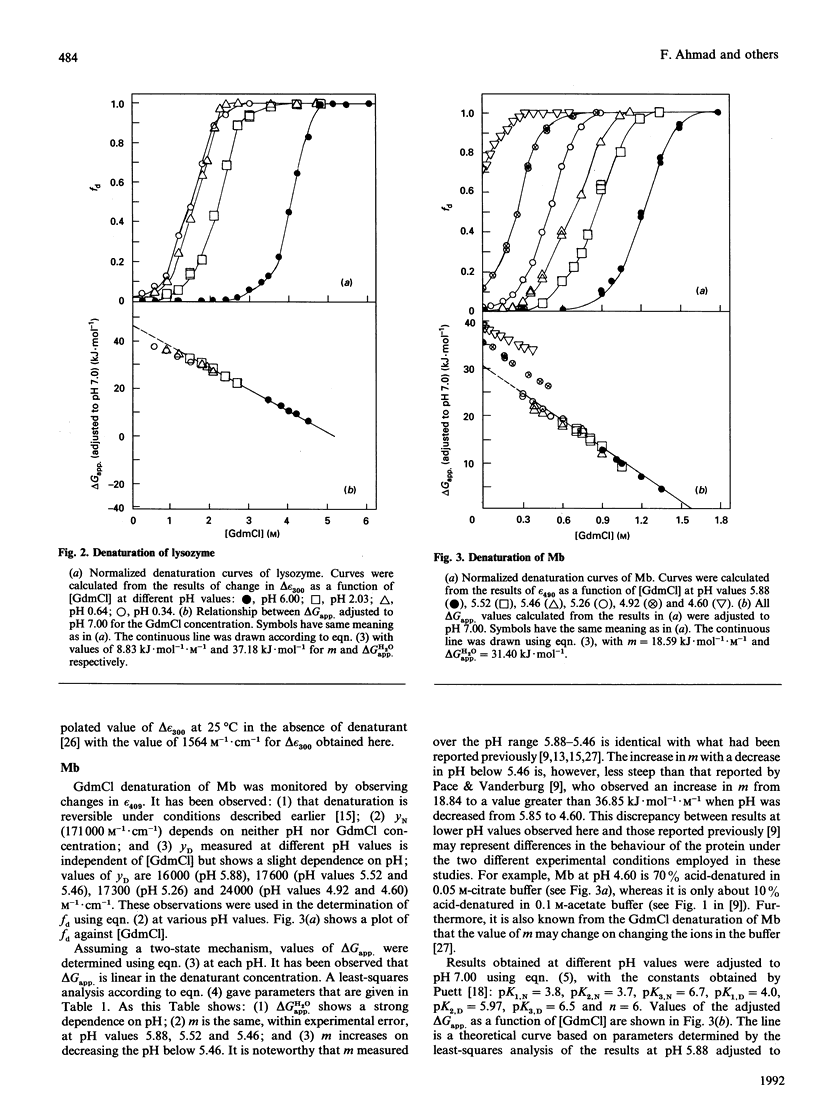
The guanidinium chloride (GdmCl) denaturation of RNAase A, lysozyme and metmyoglobin was investigated at several pH values by using absorbance measurements at 287, 300 and 409 nm respectively. From these measurements the free-energy change on denaturation, delta Gapp., was calculated, assuming a two-state mechanism, and values of delta Gapp. at zero concentration of the denaturant were measured. For each protein all delta Gapp. values were adjusted to pH 7.00 by using the appropriate relationship between delta Gapp. and pH. Dependence of the adjusted delta Gapp. value on GdmCl concentration increases for metmyoglobin and decreases for the other two proteins as the denaturant concentration decreases. It has been shown that these are expected results if the presence of the acid-denatured state during the GdmCl denaturation of proteins is considered.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad F., Bigelow C. C. Estimation of the free energy of stabilization of ribonuclease A, lysozyme, alpha-lactalbumin, and myoglobin. J Biol Chem. 1982 Nov 10;257(21):12935–12938. [PubMed] [Google Scholar]
- Ahmad F. Complexities in the denaturation of horse metmyoglobin by guanidine hydrochloride. J Biol Chem. 1985 Sep 5;260(19):10458–10460. [PubMed] [Google Scholar]
- Ahmad F., Contaxis C. C., Bigelow C. C. Free energy changes in lysozyme denaturation. J Biol Chem. 1983 Jul 10;258(13):7960–7963. [PubMed] [Google Scholar]
- Ahmad F. Protein stability from denaturation transition curves. Indian J Biochem Biophys. 1991 Jun;28(3):168–173. [PubMed] [Google Scholar]
- Aune K. C., Tanford C. Thermodynamics of the denaturation of lysozyme by guanidine hydrochloride. II. Dependence on denaturant concentration at 25 degrees. Biochemistry. 1969 Nov;8(11):4586–4590. doi: 10.1021/bi00839a053. [DOI] [PubMed] [Google Scholar]
- BIGELOW C. C. Difference spectra of ribonuclease and two ribonuclease derivatives. C R Trav Lab Carlsberg. 1960;31:305–324. [PubMed] [Google Scholar]
- Bolen D. W., Santoro M. M. Unfolding free energy changes determined by the linear extrapolation method. 2. Incorporation of delta G degrees N-U values in a thermodynamic cycle. Biochemistry. 1988 Oct 18;27(21):8069–8074. doi: 10.1021/bi00421a015. [DOI] [PubMed] [Google Scholar]
- Dill K. A. Theory for the folding and stability of globular proteins. Biochemistry. 1985 Mar 12;24(6):1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- Greene R. F., Jr, Pace C. N. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, alpha-chymotrypsin, and beta-lactoglobulin. J Biol Chem. 1974 Sep 10;249(17):5388–5393. [PubMed] [Google Scholar]
- HAMAGUCHI K., KURONO A. STRUCTURE OF MURAMIDASE (LYSOZYME). I. THE EFFECT OF GUANIDINE HYDROCHLORIDE ON MURAMIDASE. J Biochem. 1963 Aug;54:111–122. [PubMed] [Google Scholar]
- McPhie P. pH dependence of the thermal unfolding of ribonuclease A. Biochemistry. 1972 Feb 29;11(5):879–883. doi: 10.1021/bi00755a029. [DOI] [PubMed] [Google Scholar]
- Nozaki Y. The preparation of guanidine hydrochloride. Methods Enzymol. 1972;26:43–50. doi: 10.1016/s0076-6879(72)26005-0. [DOI] [PubMed] [Google Scholar]
- Pace C. N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Laurents D. V. A new method for determining the heat capacity change for protein folding. Biochemistry. 1989 Mar 21;28(6):2520–2525. doi: 10.1021/bi00432a026. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Laurents D. V., Thomson J. A. pH dependence of the urea and guanidine hydrochloride denaturation of ribonuclease A and ribonuclease T1. Biochemistry. 1990 Mar 13;29(10):2564–2572. doi: 10.1021/bi00462a019. [DOI] [PubMed] [Google Scholar]
- Pace C. N. The stability of globular proteins. CRC Crit Rev Biochem. 1975 May;3(1):1–43. doi: 10.3109/10409237509102551. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Vanderburg K. E. Determining globular protein stability: guanidine hydrochloride denaturation of myoglobin. Biochemistry. 1979 Jan 23;18(2):288–292. doi: 10.1021/bi00569a008. [DOI] [PubMed] [Google Scholar]
- Puett D. The equilibrium unfolding parameters of horse and sperm whale myoglobin. Effects of guanidine hydrochloride, urea, and acid. J Biol Chem. 1973 Jul 10;248(13):4623–4634. [PubMed] [Google Scholar]
- Salahuddin A., Tanford C. Thermodynamics of the denaturation of ribonuclease by guanidine hydrochloride. Biochemistry. 1970 Mar 17;9(6):1342–1347. doi: 10.1021/bi00808a007. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. The thermodynamic stability of proteins. Annu Rev Biophys Biophys Chem. 1987;16:115–137. doi: 10.1146/annurev.bb.16.060187.000555. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]


