Abstract
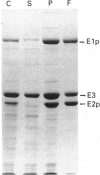
The aceEF-lpd operon of Escherichia coli encodes the pyruvate dehydrogenase (E1p), dihydrolipoamide acetyltransferase (E2p) and dihydrolipoamide dehydrogenase (E3) subunits of the pyruvate dehydrogenase multienzyme complex (PDH complex). An isopropyl beta-D-thiogalactopyranoside-inducible expression system was developed for amplifying fully lipoylated wild-type and mutant PDH complexes to over 30% of soluble protein. The extent of lipoylation was related to the degree of aeration during amplification. The specific activities of the isolated PDH complexes and the E1p component were 50-75% of the values normally observed for the unamplified complex. This could be due to altered stoichiometries of the overproduced complexes (higher E3 and lower E1p contents) or inactivation of E1p. The chaperonin, GroEL, was identified as a contaminant which copurifies with the complex. Site-directed substitutions of an invariant glycine residue (G231A, G231S and G231M) in the putative thiamine pyrophosphate-binding fold of the E1p component had no effect on the production of high-molecular-mass PDH complexes but their E1p and PDH complex activities were very low or undetectable, indicating that G231 is essential for the structural or catalytic integrity of E1p. A minor correction to the nucleotide sequence, which leads to the insertion of an isoleucine residue immediately after residue 273, was made. Substitution of the conserved histidine and arginine residues (H602 and R603) in the putative active-site motif of the E2p subunit confirmed that H602 of the E. coli E2p is essential, whereas R603 could be replaced without inactivating E2p. Deletions affecting putative secondary structural elements at the boundary of the E2p catalytic domain inhibited catalytic activity without affecting the assembly of the E2p core or its ability to bind E1p, indicating that the latter functions are determined elsewhere in the domain. The results further consolidate the view that chloramphenicol acetyltransferase serves as a useful structural and functional model for the catalytic domain of the lipoate acyltransferases.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ali S. T., Guest J. R. Isolation and characterization of lipoylated and unlipoylated domains of the E2p subunit of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1990 Oct 1;271(1):139–145. doi: 10.1042/bj2710139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen A. G., Perham R. N. Two lipoyl domains in the dihydrolipoamide acetyltransferase chain of the pyruvate dehydrogenase multienzyme complex of Streptococcus faecalis. FEBS Lett. 1991 Aug 5;287(1-2):206–210. doi: 10.1016/0014-5793(91)80052-5. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Danson M. J., Hale G., Hooper E. A., Perham R. N. Self-assembly and catalytic activity of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Nature. 1977 Jul 28;268(5618):313–316. doi: 10.1038/268313a0. [DOI] [PubMed] [Google Scholar]
- Borges A., Hawkins C. F., Packman L. C., Perham R. N. Cloning and sequence analysis of the genes encoding the dihydrolipoamide acetyltransferase and dihydrolipoamide dehydrogenase components of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. Eur J Biochem. 1990 Nov 26;194(1):95–102. doi: 10.1111/j.1432-1033.1990.tb19432.x. [DOI] [PubMed] [Google Scholar]
- Brookfield D. E., Green J., Ali S. T., Machado R. S., Guest J. R. Evidence for two protein-lipoylation activities in Escherichia coli. FEBS Lett. 1991 Dec 16;295(1-3):13–16. doi: 10.1016/0014-5793(91)81373-g. [DOI] [PubMed] [Google Scholar]
- Chung C. T., Niemela S. L., Miller R. H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlison M. G., Spencer M. E., Guest J. R. Nucleotide sequence of the sucA gene encoding the 2-oxoglutarate dehydrogenase of Escherichia coli K12. Eur J Biochem. 1984 Jun 1;141(2):351–359. doi: 10.1111/j.1432-1033.1984.tb08199.x. [DOI] [PubMed] [Google Scholar]
- Diefenbach R. J., Candy J. M., Mattick J. S., Duggleby R. G. Effects of substitution of aspartate-440 and tryptophan-487 in the thiamin diphosphate binding region of pyruvate decarboxylase from Zymomonas mobilis. FEBS Lett. 1992 Jan 13;296(1):95–98. doi: 10.1016/0014-5793(92)80411-9. [DOI] [PubMed] [Google Scholar]
- Griffin T. A., Chuang D. T. Genetic reconstruction and characterization of the recombinant transacylase (E2b) component of bovine branched-chain alpha-keto acid dehydrogenase complex. Implication of histidine 391 as an active site residue. J Biol Chem. 1990 Aug 5;265(22):13174–13180. [PubMed] [Google Scholar]
- Guest J. R., Angier S. J., Russell G. C. Structure, expression, and protein engineering of the pyruvate dehydrogenase complex of Escherichia coli. Ann N Y Acad Sci. 1989;573:76–99. doi: 10.1111/j.1749-6632.1989.tb14988.x. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Lewis H. M., Graham L. D., Packman L. C., Perham R. N. Genetic reconstruction and functional analysis of the repeating lipoyl domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Mol Biol. 1985 Oct 20;185(4):743–754. doi: 10.1016/0022-2836(85)90059-2. [DOI] [PubMed] [Google Scholar]
- Hawkins C. F., Borges A., Perham R. N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989 Sep 11;255(1):77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- Hemilä H., Palva A., Paulin L., Arvidson S., Palva I. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J Bacteriol. 1990 Sep;172(9):5052–5063. doi: 10.1128/jb.172.9.5052-5063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmingsen S. M., Woolford C., van der Vies S. M., Tilly K., Dennis D. T., Georgopoulos C. P., Hendrix R. W., Ellis R. J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988 May 26;333(6171):330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landt O., Grunert H. P., Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990 Nov 30;96(1):125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Leslie A. G., Moody P. C., Shaw W. V. Structure of chloramphenicol acetyltransferase at 1.75-A resolution. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4133–4137. doi: 10.1073/pnas.85.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G. Refined crystal structure of type III chloramphenicol acetyltransferase at 1.75 A resolution. J Mol Biol. 1990 May 5;213(1):167–186. doi: 10.1016/S0022-2836(05)80129-9. [DOI] [PubMed] [Google Scholar]
- Lewendon A., Murray I. A., Shaw W. V., Gibbs M. R., Leslie A. G. Evidence for transition-state stabilization by serine-148 in the catalytic mechanism of chloramphenicol acetyltransferase. Biochemistry. 1990 Feb 27;29(8):2075–2080. doi: 10.1021/bi00460a016. [DOI] [PubMed] [Google Scholar]
- Marsh P. Ptac-85, an E. coli vector for expression of non-fusion proteins. Nucleic Acids Res. 1986 Apr 25;14(8):3603–3603. doi: 10.1093/nar/14.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattevi A., Obmolova G., Schulze E., Kalk K. H., Westphal A. H., de Kok A., Hol W. G. Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex. Science. 1992 Mar 20;255(5051):1544–1550. doi: 10.1126/science.1549782. [DOI] [PubMed] [Google Scholar]
- Matuda S., Nakano K., Ohta S., Saheki T., Kawanishi Y., Miyata T. The alpha-ketoacid dehydrogenase complexes. Sequence similarity of rat pyruvate dehydrogenase with Escherichia coli and Azotobacter vinelandii alpha-ketoglutarate dehydrogenase. Biochim Biophys Acta. 1991 May 2;1089(1):1–7. doi: 10.1016/0167-4781(91)90076-x. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R., Radford S. E., Perham R. N. Investigation of the mechanism of active site coupling in the pyruvate dehydrogenase multienzyme complex of Escherichia coli by protein engineering. J Mol Biol. 1988 Jul 5;202(1):97–106. doi: 10.1016/0022-2836(88)90522-0. [DOI] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R. Subgenes expressing single lipoyl domains of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1987 Aug 1;245(3):869–874. doi: 10.1042/bj2450869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X. D., Stoops J. K., Reed L. J. Overexpression and mutagenesis of the catalytic domain of dihydrolipoamide acetyltransferase from Saccharomyces cerevisiae. Biochemistry. 1990 Sep 18;29(37):8614–8619. doi: 10.1021/bi00489a017. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Packman L. C., Green B., Perham R. N. Lipoylation of the E2 components of the 2-oxo acid dehydrogenase multienzyme complexes of Escherichia coli. Biochem J. 1991 Jul 1;277(Pt 1):153–158. doi: 10.1042/bj2770153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packman L. C., Perham R. N. Chain folding in the dihydrolipoyl acyltransferase components of the 2-oxo-acid dehydrogenase complexes from Escherichia coli. Identification of a segment involved in binding the E3 subunit. FEBS Lett. 1986 Oct 6;206(2):193–198. doi: 10.1016/0014-5793(86)80979-6. [DOI] [PubMed] [Google Scholar]
- Packman L. C., Perham R. N. Limited proteolysis and sequence analysis of the 2-oxo acid dehydrogenase complexes from Escherichia coli. Cleavage sites and domains in the dihydrolipoamide acyltransferase components. Biochem J. 1987 Mar 1;242(2):531–538. doi: 10.1042/bj2420531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perham R. N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991 Sep 3;30(35):8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Laue E. D., Perham R. N., Miles J. S., Guest J. R. Segmental structure and protein domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Genetic reconstruction in vitro and 1H-n.m.r. spectroscopy. Biochem J. 1987 Nov 1;247(3):641–649. doi: 10.1042/bj2470641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto B., Tzagoloff A. Structure and regulation of KGD2, the structural gene for yeast dihydrolipoyl transsuccinylase. Mol Cell Biol. 1990 Aug;10(8):4221–4232. doi: 10.1128/mcb.10.8.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell G. C., Guest J. R. Overexpression of restructured pyruvate dehydrogenase complexes and site-directed mutagenesis of a potential active-site histidine residue. Biochem J. 1990 Jul 15;269(2):443–450. doi: 10.1042/bj2690443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell G. C., Guest J. R. Sequence similarities within the family of dihydrolipoamide acyltransferases and discovery of a previously unidentified fungal enzyme. Biochim Biophys Acta. 1991 Jan 29;1076(2):225–232. doi: 10.1016/0167-4838(91)90271-z. [DOI] [PubMed] [Google Scholar]
- Russell G. C., Guest J. R. Site-directed mutagenesis of the lipoate acetyltransferase of Escherichia coli. Proc Biol Sci. 1991 Feb 22;243(1307):155–160. doi: 10.1098/rspb.1991.0025. [DOI] [PubMed] [Google Scholar]
- Schulze E., Westphal A. H., Obmolova G., Mattevi A., Hol W. G., de Kok A. The catalytic domain of the dihydrolipoyl transacetylase component of the pyruvate dehydrogenase complex from Azotobacter vinelandii and Escherichia coli. Expression, purification, properties and preliminary X-ray analysis. Eur J Biochem. 1991 Nov 1;201(3):561–568. doi: 10.1111/j.1432-1033.1991.tb16315.x. [DOI] [PubMed] [Google Scholar]
- Spencer M. E., Guest J. R. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol Gen Genet. 1985;200(1):145–154. doi: 10.1007/BF00383328. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983 Jul 1;133(3):481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Lewis H. M., Darlison M. G., Guest J. R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983 Oct 3;135(3):519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- Texter F. L., Radford S. E., Laue E. D., Perham R. N., Miles J. S., Guest J. R. Site-directed mutagenesis and 1H NMR spectroscopy of an interdomain segment in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry. 1988 Jan 12;27(1):289–296. doi: 10.1021/bi00401a044. [DOI] [PubMed] [Google Scholar]
- Watson N., Dunyak D. S., Rosey E. L., Slonczewski J. L., Olson E. R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992 Jan;174(2):530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler I. D., Hemalatha S. G., Patel M. S. Sequence conservation in the alpha and beta subunits of pyruvate dehydrogenase and its similarity to branched-chain alpha-keto acid dehydrogenase. FEBS Lett. 1991 Apr 22;282(1):209–213. doi: 10.1016/0014-5793(91)80479-m. [DOI] [PubMed] [Google Scholar]
- Zhang H., Scholl R., Browse J., Somerville C. Double stranded DNA sequencing as a choice for DNA sequencing. Nucleic Acids Res. 1988 Feb 11;16(3):1220–1220. doi: 10.1093/nar/16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]