Abstract
In-depth multiomic phenotyping provides molecular insights into complex physiological processes and their pathologies. Here, we report on integrating 18 diverse deep molecular phenotyping (omics-) technologies applied to urine, blood, and saliva samples from 391 participants of the multiethnic diabetes Qatar Metabolomics Study of Diabetes (QMDiab). Using 6,304 quantitative molecular traits with 1,221,345 genetic variants, methylation at 470,837 DNA CpG sites, and gene expression of 57,000 transcripts, we determine (1) within-platform partial correlations, (2) between-platform mutual best correlations, and (3) genome-, epigenome-, transcriptome-, and phenome-wide associations. Combined into a molecular network of > 34,000 statistically significant trait-trait links in biofluids, our study portrays “The Molecular Human”. We describe the variances explained by each omics in the phenotypes (age, sex, BMI, and diabetes state), platform complementarity, and the inherent correlation structures of multiomics data. Further, we construct multi-molecular network of diabetes subtypes. Finally, we generated an open-access web interface to “The Molecular Human” (http://comics.metabolomix.com), providing interactive data exploration and hypotheses generation possibilities.
Subject terms: Metabolomics, Type 2 diabetes, Glycomics, Lipidomics, Proteomics
Multiomic phenotyping provides molecular insights into complex physiological processes and pathologies. The study uses 18 omics platforms to analyze biofluids from 391 participants. It constructs a comprehensive molecular network based on omics integration, revealing insights into diabetes and other traits.
Introduction
The quote “Learn how to see. Realize that everything connects to everything else” by Leonardo Da Vinci becomes substantive in the context of high-throughput deep molecular phenotyping technologies that enable the measurement of hundreds or even thousands of quantitative readouts of the genome, transcriptome, proteome, metabolome, and glycome as well as related intermediate omics layers, such as the epigenome, and microRNA-ome1. Integrated into a single study, these readouts simultaneously can provide complementary insights into the molecular interactions that define the physiological and pathophysiological processes in the human body.
Indeed, molecular processes have been monitored in human biofluids through the integration of various omics approaches, including genomics, methylation, transcriptomics, proteomics, and metabolomics2–11. However, studies that deploy a broader range of omics techniques tend to have a smaller sample size, typically involving 1–36 individuals. For instance, these studies investigate dynamic changes in diverse molecular components in response to factors such as viral infection10, spaceflight8, as well as extensive exercise9. In contrast, larger cohort studies (≥100 individuals) tend to focus on a more limited spectrum of omics measurements2–4,6. For example, the impact of lifestyle changes was monitored at the molecular level in processes related to obesity, diabetes, liver function, or cardiovascular disease using genomics, proteomics, and metabolomics3. Similarly, proteomics and metabolomics were deployed to determine molecular signatures associated with schizophrenia2, while metabolomics and lipidomics were used for studying HIV infection6. The limited array of omics approaches was also used in a very large population study where the cohort size exceeds 1000 subjects. For instance, genomics, proteomics, and metabolomics, were used to monitor metabolite-protein interactions in over 3,600 healthy subjects5. Additionally, they were also employed to investigate the molecular network related to Alzheimer’s disease based on the molecular alterations measured in over 1200 subjects7.
Thus, deep molecular phenotyping at large-scale using multiple platforms and matrices (“multiomics”) in large cohort studies is becoming increasingly attractive. It is already being driven by the UK Biobank consortia, which genotyped 500,000 participants and are currently acquiring transcriptomics, proteomics, and metabolomics data for a large fraction of them. However, with many different technologies and platforms available, questions arise as to the choice of the platforms to use, their complementarity, and in particular, how to integrate these complex data sets once they have been collected.
Here, we report on what is arguably one of the most deeply phenotyped cohort studies to date. The Qatar Metabolomics Study of Diabetes (QMDiab)12 was originally designed as a diabetes case-control study in the multiethnic population of Qatar. We collected multiple aliquots of blood, urine, and saliva samples from 391 volunteers, with and without diabetes, of predominantly Arab, Filipino, or Indian ethnic backgrounds with the goal of acquiring sufficient material for multiomic analysis (see methods). The collected samples were subsequently characterized on 18 different high-throughput omics platforms. The methods included analyses of blood circulating micro-RNAs, proteins, molecular levels of IgG- and IgA- glycosylations, N-glycosylation of total protein, metabolites in urine, saliva, and plasma measured on targeted and non-targeted Nuclear Magnetic Resonance (NMR)- and mass spectrometry (MS)-based metabolomics platforms, and lipid composition by size-resolved lipo-proteomics as well as complex lipid profiles. Over 6300 individual omics data points were collected for each of the 391 participants. In addition, samples were genotyped for 1.2 million genetic variants, the white blood cell transcriptome was sequenced at a depth of 20 million reads to quantify the expression of 57,000 transcripts, and DNA methylation levels for 450,000 CpG sites were determined.
We previously utilized an individual layer of the generated omics data to answer questions concerning the metabolic signatures of T2D12,13, provide insight into the epigenetic regulation of molecular processes related to smoking, obesity, and T2D14, connect genetic risk to disease endpoints while utilizing proteomics genome-wide association studies (GWAS)15, and proteomics epigenome-wide association studies (EWAS)16, as well as basis for developing various systems biology tools for data analysis17–19. Our data sets have also already served as replication cohorts for multiple large-scale studies16,20,21. The summary of previously published work utilizing QMDiab cohort is outlined in Supplementary Fig. 1.
However, an integrative multiomics analysis of all 18 multiomics layers has not been conducted with this study set.
Here, we combine all data that we ever generated on the QMDiab study with the aim to simultaneously answer technical questions related to omics platform complementarity and data integration. We also asked biological questions related to the interrelationships between these molecular traits and their association with various phenotypes including T2D. Further, we visualized the molecular interactions in the form of interactive network to which we provide free access. The primary goal of our investigation was to utilize these biofluid-based omics’ layers to draw an image of what we call “The Molecular Human”.
To achieve this goal, we connected all multiomics traits using appropriate measures. This included partial correlations to construct Gaussian Graphical Models (GGMs) within individual omics-layers22, mutual best hits (MBH) of between-platform correlations23,24, and genome-wide (GWAS), epigenome-wide (EWAS) and transcriptome-wide (TWAS) associations between the high-dimensional genomics readouts and the other omics layers25–27. Finally, we integrated all obtained connections into a multiomics network with clinical endpoints through phenome wide disease associations and GWAS catalog lookups. We evaluated each omics layer for its potential to explain the inter-individual variability of the study participants’ age, sex, BMI, and diabetes state. We further quantified the between-layer degree of shared mutual information. We subsequently utilized these data to comprehensively characterize the multiomics layers underlying T2D.
To facilitate rapid sharing of our results and also to provide the user with the possibility of testing the interactions of their molecules of interest in the context of other omics layers, we developed a web-based tool called Connecting Omics (COmics) (http://comics.metabolomix.com) together with a blog (http://www.metabolomix.com/comics/) on which we continue to document new case-studies (depicted as Comics take on …). Finally, to show the generalizability of the COmics approach and the capability of utilizing COmics for hypothesis generation we present four distinct use cases by creating molecular network for 5-methyluridine, lactate, LILRA5, and IGFBP6, which extend on their potential involvement in various pathologies.
Results
Deep phenotyping of 391 individuals with 18 omics platforms
Urine, saliva, and blood samples from 391 subjects in the QMDiab study were analyzed on 18 technically distinct platforms (see Table 1 for platform abbreviation) relying on sequencing-, microarray-, mass spectroscopy (MS)-, nuclear magnetic resonance (NMR)-, affinity binding-, chromatography-, and biochemistry assay-based technologies (see Methods, Table 1, Fig. 1, and Supplementary Data 1 for all molecules measured on the non-genomics platforms). The number of quantitative molecular traits determined by the non-genomics platforms ranged from 36 to 1201, and the number of samples shared between every two platforms from 229 to 356 (Table 2). In total, we determined quantitative measures for up to 6304 molecular traits per sample along with genotypes for 1,221,345 autosomal SNPs, expression levels of 57,773 transcripts, and DNA methylation of 470,837 CpG sites.
Table 1.
Overview on applied omics technologies
| Omics | Measurement/Output | Technique/Platform | Matrix | Label |
|---|---|---|---|---|
| GENOMICS | Genotype | Infinium Human Omni 2.5–8 v1.2 BeadChip kit | DNA extracted from buffy coat fraction from whole blood | DNA |
| METHYLOMICS | DNA methylation | Illumina Infinium HumanMethylation450 BeadChip kit | DNA, same as for genomics | MET |
| TRANSCRIPTOMICS | Gene expression | RNA-sequencing based Illumina ~20 M reads | RNA extracted from PaxTube | RNA |
| microRNA expression | microRNA profiling based multiplex qPCR, Exiqon | RNA extracted from EDTA plasma | miRNA | |
| PROTEOMICS | Relative protein abundance | Slow Off-rate Modified Aptamer (SOMAmer), Somalogic 1,1k | EDTA plasma | SOMA |
| Relative protein abundance | Proximity Extension Assay (PEA) based Olink Target 96 Metabolism & Cardiometabolism panels | Heparin plasma | OLINK | |
| GLYCOMICS |
Total plasma N-glycosylation |
Hydrophilic interaction ultra-performance liquid chromatography (HILIC-UPLC) based Genos pipeline | EDTA plasma | PGP |
| IgG glycosylation | Liquid chromatography mass spectrometry (LC-MS) based Genos pipeline | EDTA plasma | IgG | |
| IgA & IgG glycosylation | LC-MS based124 pipeline | EDTA plasma | IgA | |
| LIPOPROTEOMICS | Lipoproteins | Proton nuclear magnetic resonance (1H NMR) based Nightingale technology | EDTA plasma | BRAIN |
| LIPIDOMICS | Absolute lipid concentration | LC-MS based on Lipidyzer technology at Metabolon | EDTA plasma | LD |
| Lipids and other metabolite concentration | Flow injection analysis (FIA)- MS based Biocrates technology | EDTA plasma | BM | |
| METABOLOMICS | Metabolite level | (HILIC-MS) & (UPLC-MS) based HD4 Metabolon | EDTA plasma | HDF |
| Metabolite level | Gas chromatography (GC)-MS (UPLC-MS) based HD2 Metabolon | EDTA plasma | PM | |
| Metabolite level | (GC-MS) & (UPLC-MS) based HD2 Metabolon | Saliva | SM | |
| Metabolite level | GC-MS & UPLC-MS based HD2 Metabolon | Urine | UM | |
| Metabolite level | 1H NMR deploying Chenomx for annotation, based on131 pipeline | Urine | CM | |
| CLINICAL | Clinical biochemistry and blood counts | Cobas 6000; Roche Diagnostics | Blood/Urine | CLIN |
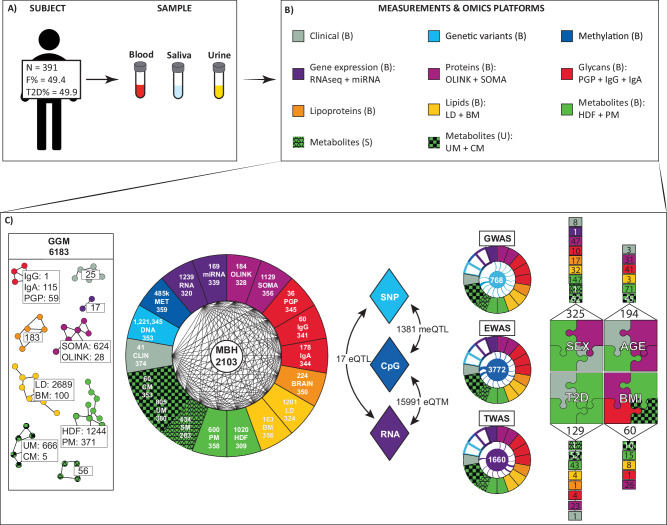
Fig. 1. Overview of the subject and data sets.
A Study cohort and collected samples; B Data and omics platforms used for data generation; C Calculation strategies used to define: Within platform significant associations GGM—(Gaussian Graphical Model); Between platform significant associations MBH—(Mutual best hit); Multiomics GWAS—(Genome-wide association studies); Multiomics EWAS—(Epigenome-wide association studies); and Multiomics TWAS—(Transcriptome-wide association studies); as well as statistical associations between each platform and the phenotype such as SEX, AGE, type 2 diabetes (T2D) and body mass index (BMI). CLIN clinical chemistry parameters, DNA genotype data, MET DNA methylation sites, RNA RNA transcripts determined with RNA-sequencing, miRNA microRNA profiles, SOMA blood circulating proteins measured with aptamer-based technology (SomaLogic), OLINK blood circulating proteins measured using high-multiplex immunoassays (Olink), PGP glycan traits N-glycosylation, IgG IgG-glycopepdides, IgA IgA and IgG-glycopeptides BRAIN plasma lipoproteins, LD plasma lipids quantified using Lipidyzer, BM plasma lipids quantified with Biocrates p150 kit, HDF plasma metabolic traits profiled on HD4 platform (Metabolon), PM plasma metabolic traits profiled on HD2 platform (Metabolon), SM saliva metabolic traits profiled on HD2 platform (Metabolon), UM urine metabolic traits profiled on HD2 platform (Metabolon), CM urine metabolites quantified with 1H NMR deploying Chenomx. N number of subjects, F female, B blood, U urine, S saliva. The source data for (C) is available in the Supplementary Data (SD) 2−9 and 11−14.
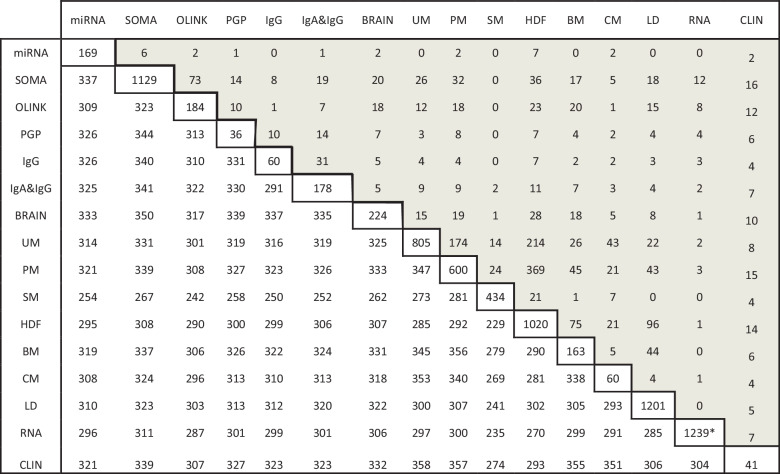
Table 2.
Mutual best hits (MBH) between platforms
*Note Genotype (DNA) and methylation (MET) data were not included in the MBH computation. Transcriptome (RNA) was limited to 1239 transcripts that are also covered by the two proteomics platforms (SOMA, OLINK).
The upper triangle of this matrix indicates the number of mutual best hits identified between the respective platforms, the diagonal contains the number of traits evaluated for that platform, and the lower triangle reports the number of samples for which data was available for both platforms in parallel. Platform abbreviations are explained in Table 1.
Complex correlation structures within and between platforms pose major challenges to integrating these datasets. For example, correlations between complex lipid species may be driven by the abundance of common precursor fatty acids, but also by factors determining interconversion between different classes. To cope with these challenges, and based on prior experience15,22, we adopted a strategy using partial correlations within platforms by deploying GGM’s, MBHs correlation between platforms, and linear model associations for genomics traits (GWAS, EWAS, and TWAS hits). In total, we identified 6183 partial correlations GGM’s (Supplementary Data 2), 2103 unique MBHs between platforms (Table 2 and Supplementary Data 3), 1381 associations of SNPs with methylation levels (meQTL’s) (Supplementary Data 4), 15,991 association of methylation levels with mRNA expression levels (eQTM’s) (Supplementary Data 5), 17 association of SNPs with mRNA expression (eQTL’s) (Supplementary Data 6), 768 GWAS with multiomics (moQTLs) at 586 independent genetic loci (Supplementary Data 7), 3772 EWAS with multiomics (moEWAS) (Supplementary Data 8), and 1660 TWAS with multiomics (moTWAS) (Supplementary Data 9). All the included associations were limited to statistically significant once at stringent Bonferroni correction levels (see methods) and serve as the foundation to portray “The Molecular Human”.
Mutual best hits deployed for between platforms assessment
In the biological system, homology reflects on molecular, structural, or physiological similarity in different species28. Genetic elements inherited in two species by a common ancestor are defined as homologs29, and are further classified as orthologs if they diverged through speciation or as paralogs if they evolved through duplication30,31. The gene orthologs are typically the most similar genes in the respective species in terms of sequence, structure, and function32. Among different approaches, deployed for identification of orthology, the most used is bidirectional best hit (BBH), which defines as orthologs all pairs of genes between two species that are reciprocally similar to one another than to any other gene in a sequence similarity search23,24. Inspired by this concept, we hypothesized that BBH, which is hypothesis-free approach, could be utilized beyond genomics to identify molecular orthologs between platforms. Here we define the BBH applied for multiomics as MBHs and use this approach to identify ortholog readouts between two platforms. This can be challenging when the platforms capture related features using different techniques and at varying depth. Examples are the analytical resolution to differentiate between lipid side chains or protein glycosylation. Examining these individual MBHs could assist in revealing potential issues with molecule annotations and help define the general overlap between platforms.
The number of MBHs between every two platforms (⇔) is presented in Table 2, and the correlation levels of all statistically significant MBHs are provided in Supplementary Data 3. The number of traits determined by each platform varies, so the relative information content provided by one platform compared to another is also different. For example, 60 urine metabolites were measured using the NMR-based platform (CM; see Table 1 for platform abbreviations), and 805 molecules were quantified on the MS-based platform (UM). We identified 43 significant MBHs between the two platforms, accounting for 72% of the traits determined by the CM platform but only 5% by the UM platform. This exemplifies the substantial difference in the extent of trait determination and, thus, information content provided by each platform.
While analyzing MBH between platforms capturing overlapping set of molecules (PM ⇔ HDF, OLINK ⇔ SOMA, and IgG ⇔ IgA) but utilizing different detection strategies (e.g., GC vs. LC; aptamer vs. antibody) we found that those display between 72% and 93% of concordance in respect of detected molecules, which underscores good quality of the selected methods applied in different laboratories (Supplementary Note 1 and Supplementary Data 10).
Evaluation of platform performance through GWAS hits
Even though GWAS studies are preferably conducted in large sample cohorts to ensure that the study is sufficiently powered to identify hits, the strength of a genetic association also depends on the effect size and the technical and biological variability of the phenotype. Replication of genetic signals across platforms provides an additional independent assessment of the utility of that platform. This is especially so when sample aliquots from the same study are evaluated and where technical variability is the only factor that differs between platforms. Thus, the calculated association p values for each QTLs with different omics phenotypes, conducted on an identical genetic variant, could serve as an objective measure, enabling the comparison of readouts from two platforms.
Exploiting this property, we monitored p values of GWAS associations with identical molecules, measured across different platforms (see Supplementary Note 2). We found none of the platforms to generally outcompete its alternative when considering strength of genetic association. Instead, we observed that individual platforms exhibited superior performance for certain subsets of molecules.
Platform-defined variance in age, sex, BMI, and diabetes state
The molecular composition of the body at different omics layers is usable to explore effects of sex33–35, measure biological age36–42, or study diabetes progression43–45. Here we investigated which of the molecular traits and platforms most accurately characterize phenotypes such as age, sex, BMI, and type 2 diabetes (T2D). First, we determined molecules associated with the phenotypes age, sex, BMI, and T2D and identified 194, 325, 60, and 129 associated molecules, respectively (Supplementary Data 11–14). Next, we examined the percentage of age, sex, BMI, and T2D variance, which may be explained by data from each individual platform. We trained a random forest model for two continuous (age and BMI) and two dichotomous traits (sex and diabetes state) on each platform. We estimated the variation explained by the respective omics phenotype (Table 3). We found that both metabolomics and proteomics accurately describe the variation of these investigated phenotypic traits. For instance, the variations in sex (95%) and T2D (86%) were most precisely captured by the HDF platform, age (54% and 52%) by the OLINK and SOMA platforms respectively, and BMI (42%) by the SOMA platform. The molecules measured on clinical chemistry data (CLIN) were accurate towards age (55%), sex (93%), and T2D (92%).
Table 3.
Percentage of the variance explained in age, sex, BMI, and diabetes state by platform
| AGE [%] | SEX [%] | BMI [%] | DIAB [%] | |
|---|---|---|---|---|
| CLIN | 54.9 | 92.5 | 17.7 | 92.0 |
| RNA | 24.6 | 70.6 | 9.4 | 67.2 |
| miRNA | 9.7 | 61.4 | 3.3 | 58.4 |
| OLINK | 53.8 | 85.4 | 22.1 | 79.6 |
| SOMA | 52.5 | 93.3 | 41.7 | 82.3 |
| PGP | 44.3 | 73.3 | 26.6 | 72.5 |
| IgG | 46.3 | 63.9 | 4.3 | 71.9 |
| IgA | 45.1 | 73.0 | 7.0 | 73.0 |
| BRAIN | 28.7 | 77.4 | 16.5 | 76.6 |
| LD | 26.5 | 76.9 | 18.9 | 68.2 |
| BM | 22.4 | 77.5 | 23.9 | 83.2 |
| HDF | 50.9 | 95.2 | 28.6 | 86.4 |
| PM | 51.7 | 91.6 | 24.7 | 86.3 |
| SM | 20.7 | 64.3 | 5.1 | 71.0 |
| UM | 50.3 | 89.4 | 27.3 | 81.9 |
| CM | 37.0 | 83.9 | 16.3 | 81.9 |
This data identifies individual platform capability to explain variance in age, sex, BMI, and T2D phenotypes.
Crosstalk between metabolites of urine, saliva, and plasma
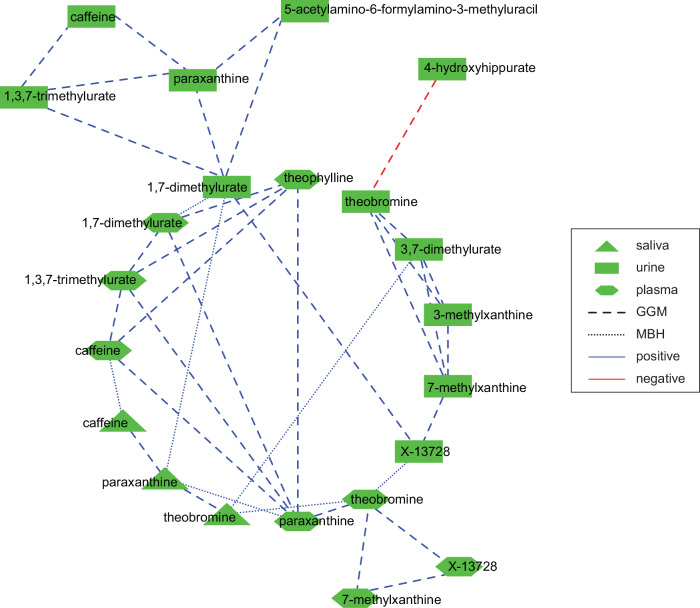
MBH between urine, saliva, and plasma metabolites, measured on MS-based platform, is capturing dependencies between those matrices, and thus informs about the interactions between them. We found 174, 24, and 14 MBHs between urine and plasma metabolites, plasma and saliva metabolites, and urine and saliva metabolites, respectively. Most MBHs connected identical molecules, reflecting on homeostasis between saliva and plasma and the detoxification processes that occur in the kidney. Yet, MBHs found between metabolites from different matrices, could potentially be used to inform on physiological processes as well as disease-related pathologies. For instance, caffeine metabolism may serve as an example showcasing organ molecular diffusion between saliva, blood, and urine. The caffeine is metabolized in ~80% to paraxanthine, ~12% into theobromine, and ~ 4% theophylline which are all further metabolized whereas ~4% of caffeine is excreted without transformation46. We found MBH between paraxanthine [SM] ⇔ 1,7-dimethylurate [UM] and theobromine [SM] ⇔ 3,7-dimethylurate [UM]. Identified MBH are depicting substrate/product relation between those molecules47.
The components of caffeine metabolism identified with MBH can be further substantiated with the GGM’s associations22 which we calculated separately for each platform (Supplementary Note 3). We find multiple substrate/product associations from caffeine metabolism (e.g., PM: caffeine/paraxantine, caffeine/theophylline or UM: paraxanthine/5-acetylamino-6-formylamino-3-methyluracil, theobromine/3,7-dimethylurate), which shows how GGMs provide a simplified overview of the actual biological processes.
The caffeine network reassembled by using MBH and GGM’s reflects on caffeine metabolism across body biofluids, in which all three pathways of caffeine metabolism can be found (Fig. 2).
Fig. 2. The cross-talk between human body fluids captured by Mutual Best Hit (MBH) and Gaussian Graphical Model (GGM) reassembles caffeine metabolism.
Green indicates measurements conducted with metabolomics. The Supplementary Data (SD) 2 and 3 serves as data source for this figure.
Thus, we demonstrated that MBH’s and GGM’s capture biological processes accurately. We further showed that data between different sample matrices could be integrated and interpreted to provide further insights into molecular processes and thereby inform about system-rich biology.
Omics associations reflect on biological process
We further investigated the relevance of MBH for capturing biologically relevant process. For example, HbA1C [CLIN], known marker for diagnosis and monitoring of Type 2 Diabetes (T2D)48,49, was showing association with the elevated blood glucose level measured on different platforms as well as other molecules previously described in the context of diabetes including betaine50, mannose51 and X-1433113 (Supplementary Fig. 2A). We also found MBH between thyroxine and SERPINA7, a thyroxine-binding globulin, which in the bloodstream carries thyroxine and triiodothyronine into thyroid gland52; the MBH was found independently of used technical platform (thyroxine (HDF) ⇔ SERPINA7 (OLINK) (p = 1.4 × 10−28; r = 0.62); thyroxine (HDF) ⇔ SERPINA7 (SOMA) (p = 2.8 × 10−18; r = 0.51)) (Supplementary Fig. 2B). The MBHs detected between Apolipoprotein E (APOE), involved in lipid metabolism, and different lipid molecules across various platforms e.g., APOE (SOMA) ⇔ Total cholesterol in VLDL (BRAIN) (p = 1.9 × 10−38; r = 0.65); APOE (SOMA) ⇔ Total [FA16:0] (LD) (p = 4.2 × 10−37; r = 0.62); APOE (SOMA) ⇔ palmitoyl-linoleoyl-glycerol (16:0/18:2) (HDF) (p = 5.5 × 10−20; r = 0.58); APOE (SOMA) ⇔ 1-palmitoylglycerol (1-monopalmitin) (PM) (p = 1.9 × 10−19; r = 0.49); APOE (SOMA) ⇔ PC.aa.C34.2 (BM) (p = 1.0 × 10−11; r = 0.34), further suggest that actual biological processes can be captured by the MBH (Supplementary Fig. 2C). The majority of MBH identified between proteomics and glycomics replicated the associations reported by us previously53. The MBH’s between proteomics and transcriptomics showed frequently the gene transcripts and corresponding proteins SIGLEC14 (RNA) ⇔ SIGLEC14 (SOMA) (p = 1.1 × 10−37; r = 0.60); GNLY (RNA)⇔ GNLY (SOMA) (p = 9.6 × 10−22; r = 0.49); LILRA5 (RNA)⇔ LILRA5 (OLINK) (p = 2.0 × 10−9; r = 0.33). This data indicates that associations depicted by the MBH reflect on the actual biological processes. Yet, the MBH could be also used in different capacities. In Supplementary Note 4 we showed that MBH linking lipidomics with metabolomics can provide further insight into the structure of complex lipids.
Multiomics GWAS, EWAS, and TWAS—relevant for human health
GWAS with intermediate phenotypes such as metabolomics, proteomics, or epigenomics have already been numerously conducted across different populations providing insight into human physiology and various pathophysiological processes54–58. Similarly, our previous EWAS were conducted using data from 10 out of 18 platforms (Supplementary Fig. 2) leading to identification of associations between epigenetic variations and different biological traits14,16.
Here, we showed that our multiomics GWAS and EWAS replicate multiple previous findings Supplementary Note 5 and resulted in identification of new hits (Supplementary Note 6 for GWAS and Supplementary Note 7 for EWAS). We also briefly describe the identified interplay between SNP genotype, DNA methylation, and gene expression (Supplementary Note 8).
Previous TWAS studies focus mainly on gene–trait associations from GWAS datasets27,59,60. Yet, to the best of our knowledge, the TWAS with multiomics phenotype which we report on here represents the first of its kind conducted to date.
Below, we present examples of findings from multiomics GWAS, EWAS, and TWAS, that hold potential relevance for human health and were not reported previously.
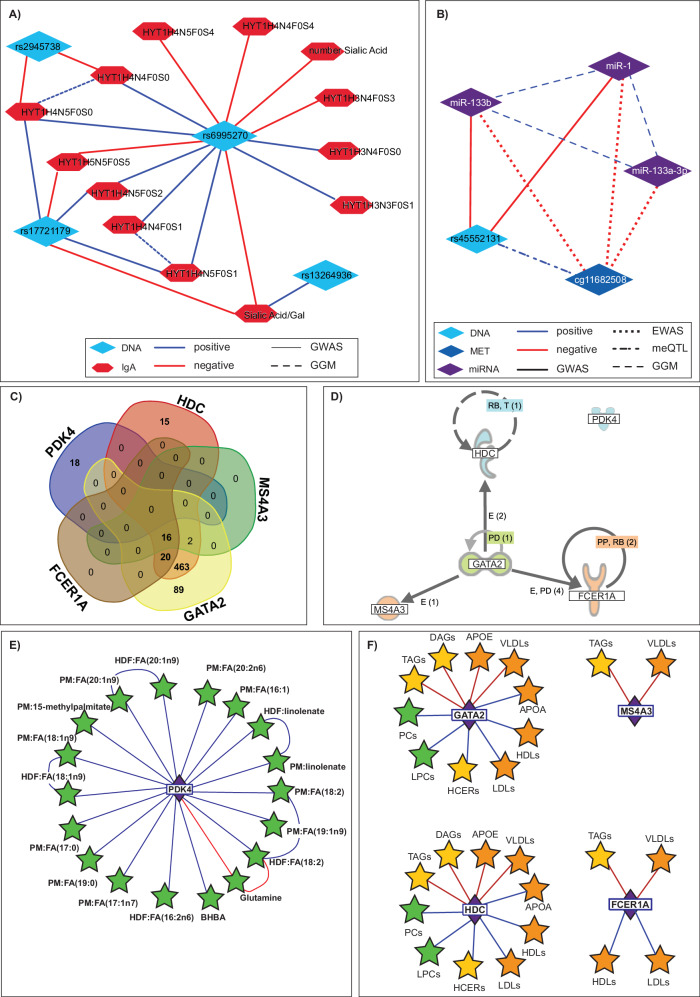
While analyzing our GWAS hits, we found previously unreported GWAS associations between multiple variants near ST3GAL1 (rs6995270, rs17721179, rs2945738, rs13264936) and 14 molecules including 2 sialic acid molecules and 12 different glycans, which all were IgA glycans and included N-acetylneuraminic acid (sialic acid). ST3GAL1 is glycosyltransferase, an enzyme involved in carbohydrate metabolism, that catalyzes the transfer of sialic acid from Cytidine-5′-monophospho-N-acetylneuraminic acid (CMP-sialic acid) to galactosyl β(1,3)-N-acetylgalactosamine] Galβ1-3GalNAc61,62. The molecular network generated from those 4 variants (Fig. 3A) shows different association directionality between genetic variants and IgA glycans. Our finding suggests that IgA glycan composition is genetically driven, which is of significant relevance for autoimmune diseases, given that IgA effector functions were shown to depend on sialylation, where loss of sialic acid increases pro-inflammatory effects63.
Fig. 3. Examples of findings from multiomics GWAS, EWAS, and TWAS associations.
A Glycome GWAS revealed associations between ST3GAL1 variants and IgA1 glycosylation. (Referee to SD2 and SD7 as the data source); B miRNA regulation throughout genetic and epigenetic changes as determined with GWAS and EWAS. (Referee to SD2, SD4, and SD8 as the data source); C Venn diagram showing an overlap between molecules associated with gene transcripts of GATA2, HDC, MS4A3, and FCER1A but not PDK4. (Referee to SD9 as the data source); D Ingenuity pathway analysis (IPA) revealed potential interaction between GATA2, HDC, MS4A3, and FCER1A but not PDK4; E The molecules associated with PDK4. (Referee to ST9 as the data source); F Associations between lipids structures and GATA2, HDC, MS4A3 and FCER1A. (Referee to SD9 as the data source). Molecules measured across platforms are depicted by different colors: DNA (light blue), Methylation (dark blue), RNA (violet), IgA (red), BRAIN (orange), BM (yellow), HDF, PM, UM, CM (green) form the multiomics network.
Genetic and epigenetic changes directly affecting miRNA transcription may provide further insight into regulatory mechanisms associated with the pivotal role of microRNAs in complex human diseases64,65. We identified three miR’s (miR-133b, miR-1, and miR-133a-3p), showing association with cg11682508 (C20orf166) out of which two (miR-133b and miR-1) associated with rs45552131 (near C20orf166-AS1) (Fig. 3B). The observed association between cg11682508 and rs45552131 replicates previous independent findings66. The expression of miR-1 and miR-133a is modulated by insulin and may be involved in insulin signaling. Given that both miRNA’s are derived from introns of protein-coding transcripts (C20orf166)67, it may be reasoned that cg11682508 is also involved in insulin modulation and signaling. Interestingly, cg11682508 was previously described as one of the methylation sites being dysregulated in pancreatic islets of T2D subjects68. By investigating the interplay between genetic variants, methylation, and miRNA we identified a novel CPG—miRNA axis beyond replication of previous findings.
To the best of our knowledge, this is the first, conducted to date, multiomics TWAS, that includes proteomics, metabolomics, lipidomics, lipoproteomics and glycomics in addition to methylation and miRNA. To our surprise, we discovered that the majority 67% (1114 out of 1660) of the identified TWAS associations were with lipids and lipoproteins, while fewer (Supplementary Note 9 and Supplementary Data 15) were with proteins (300) and metabolites (157). Those gene expressions—lipid/lipoprotein associations were found to be grouped predominantly around five gene transcripts including GATA Binding Protein 2 (GATA2), Histidine Decarboxylase (HDC), Fc Epsilon Receptor 1 alpha (FCER1A), Pyruvate Dehydrogenase Lipoamide Kinase 4 (PDK4), and Membrane Spanning 4-Domains A3 (MS4A3), showing association with 590, 516, 36, 18, and 18 lipids/lipoproteins respectively. An overlap between molecules associated with gene transcripts of GATA2, HDC, MS4A3, and FCER1A but not with PDK4 (Fig. 3C) reproduces ingenuity pathway analysis (IPA) that suggests potential interaction between GATA2, HDC, MS4A3, and FCER1A but not PDK4 (Fig. 3D). Lipids associated with PDK4 were largely fatty acids with various chain lengths (Fig. 3E). Changes in PDK4 expression were shown to play a role in lipid-related metabolic adaptation by stimulating fatty acids oxidation69, which explains the findings. For GATA2, HDC, MS4A3, and FCER1A we observed associations, exhibiting diverse directionalities, with a various high-density lipoproteins (HDL), low-density lipoproteins (LDL), and very-low-density lipoproteins (VLDL), as well as triacylglycerols (TAG) containing different fatty acid chains. Additionally, apolipoprotein A (APOA) and APOE were associated only with GATA2 and HDC (Fig. 3F). This observation could be of relevance for cardiovascular and autoimmune (systemic lupus, psoriasis, or rheumatoid arthritis) disease where lipids and lipoproteins are strongly altered. Importantly, we monitored gene expression from blood, which constitutes of immune cells. GATA2, HDC, MS4A3, and FCER1A but not PDK4 are predominantly expressed by basophils (70, Human Protein Atlas proteinatlas.org), further suggesting the interplay between basophils and circulating lipoproteins. Thus, we potentially identified an array of lipids/lipoproteins with immunostimulatory properties throughout our lipidomics TWAS, which is of importance for cardiovascular and autoimmune disease.
The molecular human—insight into T2D via multiomics
Diseases such as diabetes, cardiovascular, and autoimmune disorders are multifactorial71–73. Thus, molecular relationships, as defined by the correlation across and between different omics and reported here, may substantiate previous discoveries related to a disease as well as any single molecule association (e.g., gene, protein, metabolite) or the interactions between them, defined by e.g., pQTLs or mQTLs.
To address diabetes heterogeneity, Ahlqvist et al. used clinical parameters and defined new diabetes subgroups including four defining T2D: (1) mild age related (MARD) characterized by low insulin resistance, but a much lower age of onset of T2D; (2) mild obesity related (MOD) characterized by high BMI with low insulin resistance; (3) severe insulin resistant (SIRD) characterized by highest level of insulin resistance (HOMA2-IR) and high BMI; and (4) severe insulin deficient (SID) characterized by young age at onset, low BMI, low insulin secretion (HOMA2-B) and poor glycemic control (high HbA1c)74. In our latest study, we deployed metabolomics and proteomics to further characterize those diabetes subgroups in Qatar Biobank (QBB) cohort75. However, when focusing solely on the protein and metabolite signatures, without a context of their molecular milieu, the understanding of pathologies associated with these diabetes subgroups remains limited.
Here, to contextualize the molecular milieu of metabolic and protein signatures of T2D subgroups (MARD, MOD, SIDD, and SIRD), we utilized all our calculated multiomics associations and generated subgroup-specific multiomic networks. The list of metabolic and protein signatures defining T2D subgroups, detected in our previous study75, and overlapping with our multiomics dataset, is presented in Supplementary Data 16.
We found differences in the number of molecules forming networks for MARD (Fig. 4, and Supplementary Data 17), MOD (Supplementary Fig. 3, and Supplementary Data 18), SIRD (Supplementary Note Fig. 4, and Supplementary Data 19), and SIDD (Supplementary Fig. 5, and Supplementary Data 20), which was expected given various number of proteins and metabolic signatures characterized each subgroup75. The molecular network generated around the protein and metabolic signatures of MARD subcluster, detailed below, as well as MOD, SIDD, and SIRD in Supplementary Note 10 offer additional valuable insights pertinent to the pathological process in which those proteins and metabolites are involved.
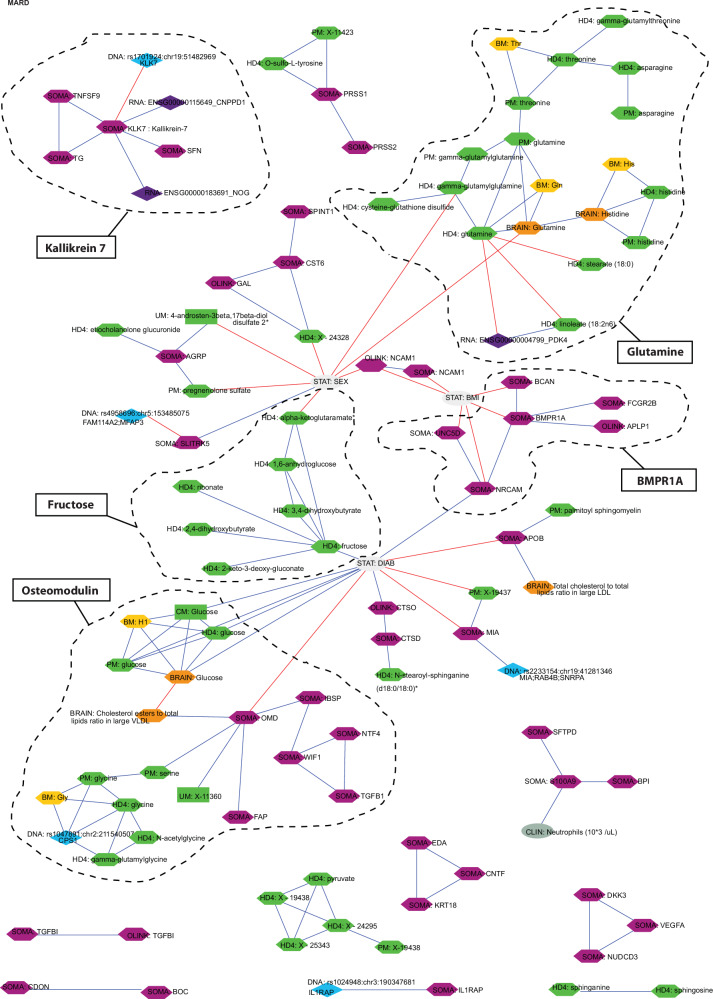
Fig. 4. Multiomics interactions of molecular (proteins and metabolites) signatures of MARD cluster.
Molecules measured across 11 platforms CLIN (gray), DNA (blue), RNA (violet), SOMA & OLINK (purple), BRAIN (orange), BM (yellow), HDF, PM, UM, CM (green) form the multiomics network. The Supplementary Data (SD) 2, 3, 7, 9, and 17 serves as data source for this figure.
MARD network indicates risk to cardiovascular calcification
The MARD network comprises 108 molecules detected on 11 platforms (Supplementary Data 19 and Fig. 4) forming five molecular subclusters around osteomodulin, glutamine, bone morphogenetic protein receptor type 1A (BMPR1A), kallikrein 7 (KLK7), and fructose. Those subclusters can reflect on specific molecular processes. For instance, the molecules clustered around osteomodulin consist of bone sialoprotein 2 (IBSP), the noncollagenous bone matrix protein, indicated as a component actively regulating aortic valve calcification76, a circulating Wnt inhibitory factor 1, which is associated with IBSP, was previously shown to be involved in the development of cardiovascular disease and atherosclerotic plaque formation77, and FAP propyl endopeptidase directly associated with osteomodulin was shown as a negative regulator of cardiac repair78. The glycine, also found in osteomodulin cluster, was reported as a major quantitative and structural constituent of collagen molecule79, which was found to promote cardiovascular calcification80. We also identified and replicated81 glycine association with rs1047891 near CPS1, carbamoyl-phosphate synthase 1. Thus, the molecular network around the osteomodulin indicates mechanisms underlying cardiovascular complications, particularly calcification, which might be relevant for the MARD subgroup. The comprehensive characterization of processes related to osteomodulin was feasible after the deployment of multiomics data.
Overall, monitoring of complex diseases such as diabetes through multiomics layers results in a more accurate description of the pathological processes as we showed by defining molecular network of diabetes subgroups. Now, the molecular interactions established across omics layers by our study can provide further insight into various pathological processes beyond T2D. We provide the full network in digital format (Cytoscape82) for free download.
COmics—server to explore multiomics interactions
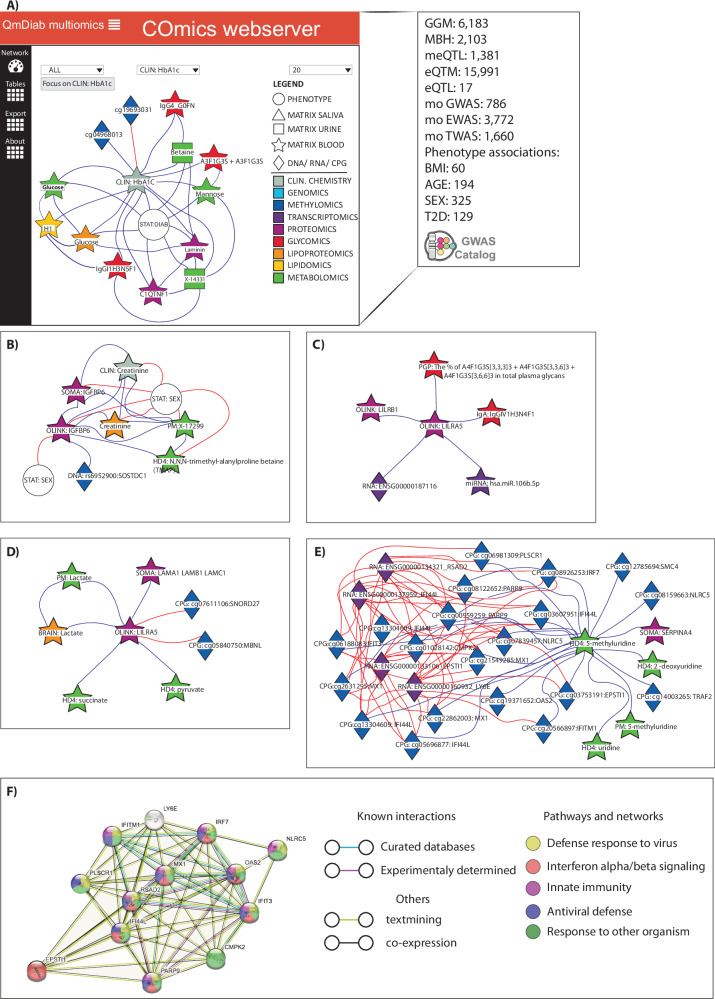
Although Cytoscape82 is a powerful tool that could be deployed for the exploration of molecular networks, it requires specialized knowledge and software familiarity. To simplify data access and result visualization, we integrated all associations along with GWAS catalog information. We constructed a molecular network consisting of over 34,000 edges and 6304 nodes, which we made available as on an open-access COmics server (http://comics.metabolomix.com) (Fig. 5A). This server is intuitive to use, requiring minimal skills, resources to explore molecular interactions related to physiological and pathophysiological processes, and can also serve as a tool for hypothesis generation. We present examples defining molecular milieu of IGFBP6 (“COmics takes on IGFBP6: ”, Fig. 5B), LILRA5 (“COmics takes on LILRA5: ”, Fig. 5C), lactate (“COmics takes on lactate: ”, Figs. 5D), and 5-methyluridine (“COmics takes on 5-methyluridine (ribothymidine):”, Fig. 5E) which extend on our understanding related to their potential role in human health, including T2D and autoimmune diseases.
Fig. 5. Overview on COmics functionality.
A COmics webpage layout and the information on integrated data; B COmics generated molecular network of IGFBP6. (Referee to SD2, SD3, SD8 as the data source); C COmics generated molecular network of LILRA5. (Referee to SD3 and SD9 as the data source); D COmics generated molecular network of lactate. (Referee to SD2, SD3 and SD8 as the data source); E COmics generated molecular network of 5-methyluridine; (Referee to SD3, SD5, and SD8 as the data source); F STRING generated clusters of molecules associated with 5-methyluridine showed their involvement in immune responses. The color code for the network representation (B-E) represents following omics: Dark blue diamond—Methylomics, Vialet diamond—Transcriptomics, Purple star—Proteomics, Red star—Glycomics, Orange star—Lipoproteomics, Green star—Metabolomics, Gray star—clinical chemistry.
COmics takes on IGFBP6
http://comics.metabolomix.com/?focus=OLINK:IGFBP6_P24592&maxnodes=1
Insulin-like growth factor-binding protein-6 (IGFBP6) is a high-affinity IGFBP shown to play a role in multiple processes, including tissue remodeling and repair, fibrosis, and immunological responses83. Nevertheless, its molecular interactome was not previously described and could shed light on the pathophysiology related to IGFBP6. The network (Fig. 5B) consists of eight direct associations with IGFBP6 including six molecular associations (IGFBP6 [SOMA], creatinine [CLIN], creatinine [BRAIN], N,N,N-trimethyl-alanylproline betaine [HDF], PM: X-17299 [PM], rs6952900 near SOSTDC1 [GWAS]) and two phenotypic associations (SEX and DIAB). Given that creatinine and N,N,N-trimethyl-alanylproline betaine were recognized as markers defining kidney function84 it could be reasoned that IGFBP6 is implicated in physiological or pathophysiological kidney processes. Indeed, elevated levels of IGFBP6 were identified in children with chronic renal failure85. Additionally, IGFBP6 was associated with the diabetes phenotype, for which a compromised kidney function was identified in ~40% of T2D patients86. The identified network further indicates the involvement of IGFBP6 in kidney pathology, which could be relevant for T2D.
COmics takes on LILRA5
http://comics.metabolomix.com/?focus=OLINK:LILRA5_A6NI73&maxnodes=1
Leukocyte immunoglobulin-like receptor 5 (LILRA5) was shown to be expressed by monocytes as well as neutrophils1, and recent study reported on its expression by macrophages87. LILRA5, expressed by macrophages of synovial tissue, was shown to trigger selectively pro-inflammatory cytokines and IL-10 in rheumatoid arthritis patients88. We created the molecular network of LILRA5 (Fig. 5C) to further understand the molecular process related to LILRA5. We identified five molecules directly associated with LILRA5 (ENSG00000187116_ LILRA5 [TWAS], miR-106b-5p [miRNA], LILRB1 [OLINK], PGP32 [PGP] and IgGIV1H3N4F1 [IgA]). The identified TWAS associations between LILRA5 [OLINK] and ENSG00000187116 indicate translational processes, whereas LILRA5/miR-106b-5p [miRNA] suggest that miRNA might be involved in the regulation of LILRA5 levels or function. Recently, miR-106b-5p was reported as a molecule released by macrophages involved in inflammation and communication between macrophages and renal juxtaglomerular cells89. The association identified here extends the miR-106b-5p-related knowledge. Interestingly, we also found an association between LILRA5 and another protein from the same family, LILRB1, known to be involved in the immune response modulation90, suggesting a potential interaction between those molecules. Indeed, a recent study reported and validated protein-protein interactions between LILRA5 and LILRB191, further confirming our finding. Identification of glycans (total N-glycans and IgG) in the LILRA5 clusters is not surprising, given their extensively described involvement in inflammation and rheumatoid arthritis92, but informatively pointing towards glycan-protein axis in this regard.
COmics takes on lactate
http://comics.metabolomix.com/?focus=HD4:lactate&maxnodes=1
Lactate is a critical metabolite for proper physiology, and alterations in lactate metabolism are involved in various diseases, including cancer, cardiovascular diseases, inflammation, and many others93. Here we describe molecular interaction with lactate in blood using COmics (Fig. 5D). This molecular network identified expected metabolite-metabolite interactions as the one observed between lactate and metabolites of TCA cycle (succinate and private) as well as less expected lactate association such as the one observed with laminin (LAMA) [SOMA], and as well as cg07611106 and cg05840750 (detected by EWAS). LAMA is one of the largest non-collagenous glycoproteins in the basement membrane and is an essential component of the extracellular matrix (ECM). A previous study described that high glucose and insulin levels trigger increased LAMA production by renal cells, further suggesting its relevance in diabetes94. We also found a significant association between diabetes phenotype and LAMA (Supplementary Data 15). Thus, it could be reasoned that the association identified between lactate and LAMA might relate to glucose and insulin metabolism. This hypothesis could be supported by our previous study showing elevated lactate levels in diabetes patients with acute disease onset13, thus suggesting the lactate-LAMA association as relevant for disrupted glucose-insulin axis in diabetic subjects. Yet, it would be important to understand the mechanistic nature of lactate-LAMA association.
COmics takes on 5-methyluridine (ribothymidine)
http://comics.metabolomix.com/?focus=HD4:5-methyluridine(ribothymidine)&maxnodes=1
5-methyluridine, is endogenous methylated nucleoside metabolized from uridine in the reaction catalyzed by methyltransferase in which S-adenosyl methionine (SAM) serves as methyl donor95. Although 5-methyluridine was reported in context of various conditions including respiratory process in asthma96, major adverse cardiovascular events97, as well as COVID-1998, the actual impact of alterations in levels of plasma 5-methyluridine is unclear. Using COmics we found 28 molecules associated with 5-methyluridine (Fig. 5E). The associations between metabolites (uridine and 2-deoxyuridine) were expected and reflected in substrate-product relation in uridine metabolism. The associations identified between 5-methyluridine and 20 cpg sides near 13 different genes (SMC4, TRAF2, NLRC5, IFITM1, OAS2, IRF7, PLSCR1, PARP9, IFI44L, MX1, EPSTI1, IFIT3, CMPK2), as well as 4 gene expressions (IFI44L, RSAD2, LY6E, EPSTI1) were more intriguing, mainly because we also identified eQTM between the gene transcripts and methylation sides suggesting interplay between those molecules. To assess whether the network identified with COmics reflects on protein-protein interactions involved in biological processes, we used STRING database99. Indeed, supportive evidence for the interactions between molecules identified in the cluster genes was found. Moreover, majority of those molecules were involved in immune responses predominantly innate immunity and interferon signaling (Fig. 5F), which we further assessed and confirmed with Interferome database100. Thus, the negative association, which we observed between 5-methyluridine and all 4 gene transcripts, and the positive association between 20 methylation sides, suggests immunosuppressive properties of 5-methyluridine. With COmics we also identified a protein SERPINA4 (Kallistatin) to be associated with 5-methyluridine. Interestingly, kallisterin was reported in context of rheumatoid arthritis as a molecule with anti-inflamatory properties, inhibiting accumulation of immune cells101. Thus, the molecules directly associate with 5-methyluridine reveal multiomics axis extending on the mechanisms with regulatory effect on the immune system.
These examples demonstrate the utility of COmics (http://comics.metabolomix.com) as a resource to explore highly complex molecular relationships related to physiological processes and disease phenotypes.
Discussion
The Molecular Human could be considered as holistic description of molecular interactions in the human body, which we achieved here by integrating molecules detected across 18 platforms and 8 omics. Although, to date this is the largest effort in terms of the number of measurements conducted in the relatively big human cohort (391 subjects), we are aware that future attempts might extend the molecular interactome towards process concerning in greater detail secretome by focusing on sweat and tears, exhalome focused on the molecular composition of the breath as well as microbiome aiming to provide comprehensive description of the gut and skin microbiota and their interactions with the host. Thus, we see our approach as an overture into future large-scale multiomics study for which we are setting a stage.
Understanding inter-molecular relationships and platform complementarity is central to working with large genetic and epidemiological meta-analyses, evaluating data integration options, and extracting additional information. Deployment of MBH across omics platforms covering different and overlapping molecular traits, which we investigated here, can indeed be used to define molecular orthology bridging different platforms and omics. While investigating MBH between platforms containing overlapping molecular traits (e.g., SOMA ⇔ OLINK; HDF ⇔ PM; IgG ⇔ IgA) and identifying association between them we showed good platform performance regarding components identification. On the other hand, MBH applied to molecular traits between different omics (e.g., thyroxine (HDF) ⇔ SERPINA7 (OLINK); APOE (SOMA) ⇔ Total cholesterol in VLDL (BRAIN)), reveal biologically relevant molecular interactions. Those examples underscore the value of the measurements and suggests the utility of suggested here data integration.
However, ~28% of common protein targets were not detected by the MBH for the two affinity-based platforms used for proteomics. This suggests that integrating the measurement approaches can be challenging and may require special attention for the molecules that MBH did not identify. Many of our observations are in line with previous studies assessing proteomics methods in multiple cohorts102. They could be linked to differences in their analytical performance for common protein targets.
Complex and multifactorial conditions such as diabetes, cardiovascular, and autoimmune diseases require comprehensive characterization for proper diagnostics and treatment. This is particularly relevant when the disease progression is not well defined, as well as when various comorbidities occur. For instance, treatment of T2D diabetes patients depends on multiple factors including their blood glucose level (tested with HbA1C or 1,5-AG), insulin resistance status (tested with hyperinsulinemia-euglycemic clamp or HOMA-IR), capability to produce insulin (e.g., fasting insulin or C-peptide test) as well as presence of other diseases (e.g., cardiovascular disease, neuropathy, kidney disease, and retinopathy)103. The identification of five subgroups among diabetes patients, stratifying individuals with respect to disease progression and diabetic complication risks, adds to the complexity but could further navigate more personalized treatment options74. The multiomics analysis offers a powerful framework that could be utilized to better phenotype patients with complex diseases by defining molecular interactions across omics layers with functional relevance for disease endpoints. GWAS or EWAS with intermediate phenotypes e.g., miRNA, protein, glycan, or metabolite, has shown a potential to provide insight into human physiology and complex diseases in the past4,10,104–107. The integration of additional multiomics layer resulted in identifying processes relevant to human biology, including biochemical reactions and metabolism of the components involved, as well as molecular interactions previously identified by multiomics GWAS, EWAS, and TWAS. In our cohort study, we reproduced a plethora of literature-reported hits which proves the robustness of our approach. Moreover, we identified previously unreported associations shedding a light on a range of biological processes relevant for diabetes, autoimmune, and cardiovascular disease.
We further utilized the integrated multiomics data to describe molecular milieu of proteins and metabolites, recognized as signatures of T2D subgroups75, which enabled us to provide further insight into potential pathologies, relevant for those subtypes which would require further investigation. For instance, based on the integrated analysis, we suggest cardiovascular complications such as calcification could be a risk factor for the MRAD subgroup. We also describe other molecular events relevant to MOD, SIDD, and SIRD subgroups which were not suggested before as the multiomics component was missing in the previous analysis. For instance, the multiomics network applied to MOD subgroup resulted in the identification of known as well as previously unreported interactions as the one found between leptin and CXCL5, cytokine implicated in the chemotaxis of inflammatory cells (Supplementary Note 10). Given that CXCL5 was recently implicated in the browning of WAT108 and leptin was determined as a molecule enabling browning of white adipose tissue (WAT)109,110, our identified association might suggest interplay between CXCL5 and leptin in the processes of WAT remodeling. This further extends our understanding of metabolically healthy obesity, characterized, among others, by high BMI and low insulin resistance111, which to some extent is characteristic of MOD subgroup. Despite the valuable insights provided by this multiomics integration, it is essential to note that the associations observed are hypothesis-generating in nature. Thus, further study would be required to provide definitive biological conclusions. Nevertheless, the perspective obtained through utilizing multiomics layers in understanding human biology in this study is relevant and can serve as a foundational framework for future multiomics initiatives.
Additionally, our multiomics network, created based on molecular interactions across 18 platforms, is giving possibilities beyond molecular characterization of diabetes subtypes. It can be utilized in more generalizable approach to better understand molecular milieu (both direct and distant) of each measured molecule and consequently, for hypothesis generation as we outlined in Fig. 5 and under vignette “COmics takes on”. For instance, while analyzing the molecular network of 5-methyluridine we pointed out its potential immunosuppressive properties and suggested involvement of methylation and alteration in expression of e.g., IFI44L, EPSTI1, and LY6E genes, relevant in context of autoimmune diseases such as systemic lupus and rheumatoid arthritis112,113. We extend the effort of documenting new molecular case studies where a multiomics approach provides further insight into molecule function and their potential involvement in various pathologies through identified omics associations. These case studies are presented in the form of a blog (http://www.metabolomix.com/comics/), depicted as ‘Comics take on …’. Finally, we provide the scientific community with access to this multiomics network via the developed web server COmics (http://comics.metabolomix.com) to facilitate global testing of the interactions of molecules of interest in the context of other omics layers. This can contribute to more rapid hypothesis generation, followed by its testing, and thus progress in the field.
Yet, it is crucial to bear in mind that the implementation of such a broad array of platforms is frequently not feasible and not always necessary. The selection of specific omics and platforms should be driven by the scientific question, as well as the process or phenotype requiring investigation. As demonstrated in this study, phenotypes such as age, sex, or diabetes necessitate omics that closely recapitulate the specific phenotype. For example, metabolomics, glycomics, or proteomics were identified as the main molecular hubs enabling the construction of networks for diabetes subtypes, which was not feasible with transcriptomics or methylation alone, even though they are also components of the network. In contrast, associations revealed by TWAS, reflecting on immunostimulatory processes, were predominantly captured by transcriptomics and lipidomics/lipoproteomics, with other omics not contributing significantly to this discovery. This underscores the importance of 2-way comparisons rather than fully multiomics approaches in capturing certain processes. Now, with access to this data, each investigator has the freedom to monitor the molecular milieu of the molecule or phenotype of interest, allowing them to decide on the most suitable omics/platform approach for their study. Our study has strengths and weaknesses. The diversity of the QMDiab participants provides access to a wide range of individuals from various ethnicities including Arabs, South Asians, and Filipinos Given that the majority of the study focusses on Caucasian population, multiethnic nature of our work especially in multiomics context is truly unique and is adding to the previously conducted omics research on Asian and Middle eastern population114–116. Yet, mixed ethnicity in the QMDiab might result in population-specific stratification and thus in inflated p values. Indeed, our previous study showed that the first three principal components (PC’s) of the genotype variants capture self-reported ethnicity15. Therefore, we added the first three principal components of the genotyping data (genoPCs) to represent accurately the ethnicity. Additionally, participants were enrolled continuously on an availability basis (i.e., without selection for diabetes state, age, sex, BMI or ethnicity) at the dermatology department of the major public hospital in Doha, Qatar, using identical collection kits and protocols, to avoid batch effects between cases and controls, which could occur during the initial phase of patient enrollment and sample collection. The fact that participants were from diverse ethnic backgrounds introduces variations on multiple levels, including lifestyle, dietary habits, physical activity levels, and health behaviors, among other factors. This diversity may be advantageous when investigating correlations between omics layers, as it could potentially increase the signal-to-noise ratio. Similarly, the fact that study participants were not fasting implies further biological variation in the data, which may strengthen correlation signals related to processes confounded by fasting when case-control studies are conducted. Noteworthy, while the average time between meals for individuals with or without T2D was not assessed, and the fasting status of the participants was not defined, our previous study demonstrated that the increased variability is random and does not tend to bias the associations13. Because our cohort consists of healthy and T2D subjects some of the observed associations could be driven by the molecular alterations which are known features of T2D (e.g., elevated carbohydrates, lipids, and branch chain amino acid levels). For instance, identified TWAS associations, dominated by 5 genes linked to various lipids could reflect on the enrolled participants’ characteristics, which might be recognized as study limitation. Nevertheless, such an experimental setting enabled us to uncover a range of lipids/lipoproteins with immunostimulatory properties in our lipidomics TWAS, which holds significance for cardiovascular disease.
Taken together, we have drawn a multiomic image of the Molecular Human by providing a comprehensive description of biological processes based on the integrated data generated by 18 technologically diverse platforms in human samples obtained from 391 subjects. We provide open access to this resource via the COmics web server and Github. Our study describes the complementarity of various omics layers and demonstrates the capacity for integrated omics data to mirror biological processes. It sets the stage for future studies that utilize such resources to understand the molecular networks surrounding molecules of interest that link them to the disease endpoints.
Methods
Ethics
The study was approved by the Institutional Review Boards of HMC and Weill Cornell Medicine-Qatar (WCM-Q) under research protocol #11131/11 and complies with all relevant ethical regulations. For ongoing work related to this study, a non-human subjects research determination was obtained. The study design and conduct adhered to all relevant regulations regarding the use of human material and data and was conducted in accordance with the criteria set by the Declaration of Helsinki.
Cohort characteristics
The subjects were enrolled in the framework of the Qatar Metabolomics Study on Diabetes (QMDiab), a cross-sectional diabetes case-control study at the Dermatology Department of HMC in Doha, Qatar as previously described12. Written informed consent was obtained from all participants. No compensation was given to the participants. The study enrolled 391 participants with at least one omics phenotype and includes 17 additional subjects that were not a part of Mook-Kanamori et al. The cohort consists of 193 females and 198 males. The average participants age was 46.5 years (s.d. = 12.9) and the average BMI was 29.7 kg/m2 (s.d. = 6.0). This cohort includes 195 participants with T2D and 196 without T2D.
Sample collection
Non-fasting blood, saliva and urine were collected according with standard protocols as previously described12. Blood was collected using EDTA, Heparin, citrate and PAXgene Blood RNA tubes. Blood collected in EDTA and Heparin was centrifuged at 2500 g for 10 min, plasma was collected aliquoted and stored at −80 °C until analysis. The blood collected into PAXgene Blood RNA tubes was centrifuged for 10 min at 4000 g. The supernatant was removed, and the pellet was used for the RNA extraction. The saliva was collected using Salivette system (Salivette®, SARSTEDT AG & Co. KG) according with manufacturer’s protocol. Collected saliva samples were centrifuged at 2000 × g for 2 min, aliquoted and stored at −80 °C until analysis. The urine was collected into the URINE CAPS mixed transferred into the falcon tube, centrifuged at 2500 g for 10 min, aliquoted and stored at −80 °C.
Deep molecular phenotyping
The obtained samples were submitted for deep molecular phenotyping which utilized clinical chemistry parameters along with omics measurements across 18 technically diverse platforms. All the cases and controls were measured simultaneously on each analytical platform to minimize measurements biases. We determined: 41 clinical chemistry parameters (CLIN); genotype data of 1,221,345 variants (DNA); 450k DNA methylation sites (MET); (4) 57,942 transcriptomic traits, including 57,773 RNA transcripts (RNA) using RNA-sequencing (Illumina, 20M reads), and 169 microRNA profiles (miRNA) with multiplex qPCR (Exicon); 1313 blood circulating proteins using two different technologies 1129 proteins (SOMA) from aptamer-based technology (SomaLogic) and 184 proteins (OLINK) from high-multiplex immunoassays (Olink); 274 glycan traits including 36 total plasma N-glycosylation (PGP) using HILIC-UPLC and 60 IgG-glycopepdides (IgG) deploying LC-MS, both profiled at Genos Ltd. as well as 178 IgA and IgG-glycopeptides (IgA) measured with LC-MS in Wuhrer lab; 225 plasma lipoproteins (BRAIN) quantified with 1H NMR (Nightingale), 1494 lipids including 1,331 plasma lipids (LD) quantified using Lipidyzer deploying LC-MS system (Metabolon), and 163 plasma lipids and other metabolites (BM) quantified with FIA-MS (Biocrates p150 kit); 3415 metabolic traits profiled with different approaches and matrixes including 1104 plasma metabolites (HDF) determined with HILIC-MS and UPLC-MS on HD4 platform (Metabolon), 2251 metabolites (758 in plasma (PM), 602 in saliva (SM) 891 in urine (UM),) measured using GC-MS and UPLC-MS on HD2 platform (Metabolon), and 60 urine lipids (CM) quantified with 1H NMR deploying Chenomx (University Greifswald). For the cross-platform analyses we limited the RNA profiles to 1239 transcripts, which were also assayed by SOMA and OLINK platforms.
Clinical chemistry data
The obtained blood samples were analyzed within 4 h of blood collection at the Department of Laboratory Medicine and Pathology of HMC with the Cobas® 6000 (Roche Diagnostics, Basel, Switzerland).
Genotyping
The genotyping was conducted by the Genomics Core at WCM-Q as we previously reported15. Briefly, the Illumina Omni 2.5 array (version 8) was used. Out of 359 genotyped samples, high-quality genotype data (2,338,671 variants) was obtained for 353 samples, and six samples, which displayed overall low call rate (<90%), were excluded. After duplicate variants removal, 2,327,362 variants left. Variant removal due to (1) The missing genotype data(in all 134,830 variants) (PLINK option -geno 0.02) resulting in 2,192,532 variants; (2) minor allele threshold (in all, 941,058 variants) (PLINK option -maf 0.05), resulting in 1,251,474 variants; (3) Violation of Hardy-Weinberg equilibrium (in all, 28,175 variants) (PLINK option -hwe 1E-6), leaving 1,223,299 variants out of which 1,221,345 were autosomal variants. The total genotyping rate of these remaining variants was 99.7%.
Methylation
The methylation analysis was conducted by Genomic Core at WCM-Q as previously described14. Samples were probed for genome-wide DNA methylation profiling of over 485,000 methylation site using the Illumina Infinium HumanMethylation450 (450 K) BeadChip array. The assay performance was assessed with implemented in the Genome Studio software and all the samples passed the quality control. The obtained data was further normalized using the Lumi: BMIQ pipeline, which includes color bias adjustment, quantile normalization (QN), and beta mixture quantile dilation normalization (BMIQ).
Transcriptomics (RNA-seq)
The obtained pellets from the PAXgene Blood RNA tubes were used for the isolation of total RNA with PAXgene Blood miRNA Kit (Qiagene). In brief, the obtained pellets were mixed with RNaze-free water, and vortexed until the pellets dissolved. The samples were centrifuged for 10 min at 4000 × g and pellet was formed. The supernatant was removed and 350 µL of BM1 buffer provided with the kit was added into the pellet. The samples were vortexed until the pellet dissolved, and mixed with 300 µL of BM2 buffer as well as 40 µL of proteinase K, provided with the kit. The samples were incubated for 10 min. at 55 °C under constant shaking followed by transfer onto the PAXgene shredder spin column placed in a processing tube. The samples were centrifuge for 3 min at 15,000 × g and the supernatant was placed into the fresh tube, mixed with 700 µL of 100% isopropanol and transferred onto PAXgene RNA spin column. The samples were centrifuged for 1 min at 15,000 × g the flow-throw was removed, and 350 µL of BM3 buffer was placed onto PAXgene RNA spin column. The samples were centrifuged for 15 s. at 15,000 × g, the PAXgene RNA spin column was placed in the fresh collection tube and 80 µL of RDD buffer containing DNase-I was placed onto PAXgene RNA spin column followed by 15 min. incubation at room temperature. 350 µL of BM3 buffer was placed onto PAXgene RNA, the samples were centrifuged for 15 s at 15,000 × g, the flow-throw was removed, and 500 µL of BM4 buffer was added. The samples were centrifuged for 15 s at 15,000 × g, the flow-throw was removed, and additional 500 µL of BM4 buffer was added. After centrifugation for 2 min. at 15,000 × g, the PAXgene RNA spin column was placed into the fresh collection tube, and the samples were eluted from the column with 80uL of BR5 buffer. The obtained eluent was incubated for 5 min at 60 °C, and afterwards chilled on ice. The integrity and quantity of the isolated RNA was measured using Qubit RNA HS Assay Kit (high sensitivity, 5 to 100 ng quantification range) Assay Kit and Qubit 3.0 fluorometer (Life Technologies) according to the manufacturer’s protocol. The samples were kept at -80 °C until measurements.
The samples containing total RNA (400 ng) were submitted to the Genomics Core at WCMQ for the RNA-sequencing. The total RNA was depleted of rRNA and Globin using the NEBNext rRNA & Globin Depletion Kit for Human/Mouse/Rat (New England BioLabs, Ipswich, MA). The depleted RNA was used to generate strand-specific libraries with BIOO NEXTflex Rapid Directional RNA-Seq Kit (Bioo-Scientific, Austin, TX). Library quality and quantity were analyzed with the Bioanalyzer 2100 (Agilent, Santa Clara, CA) on a High Sensitivity DNA chip. 10 libraries were then pooled in equimolar ratios and paired-end sequenced at 75 bp on one lane of an Illumina HiSeq 4000 (Illumina, San Diego, CA). Total of 57,773 RNA transcripts were measured in 320 subjects.
microRNA quantification
RNA extraction
The miRNAs were isolated from 200 µL EDTA-plasma sample using the miRNeasy serum/plasma kit (Qiagen) following the manufacturer’s instructions. Briefly, the samples were lyzed using QIAzol Lysis Reagent and spiked with 3.5 μl miRNeasy Serum/Plasma Spike-In Control included in the kit. The chloroform was added, samples were mixed and the centrifuged. The obtained after centrifugation upper aqueous phase was transferred into the fresh tube, mixed with 1.5 volume of 100% ethanol, and transferred into an RNeasy MinElute spin column in a 2 ml collection tube, provided in the kit. The samples were centrifuged, the flow-throw was removed, and RWT buffer provided with the kit was added onto the RNeasy MinElute spin column. The samples were centrifuged, the flow-throw was discarded, RPE buffer, provided with the kit, was added onto the RNeasy MinElute spin column. The samples were centrifuged and flow-throw was removed. The 80% ethanol prepared in RNaze-free water was placed onto the MinElute spin column, the samples were centrifuged until the spin column membrane dried. The MinElute spin column was placed in fresh collection tube and the total RNA including miRNA was eluted with 14 μl RNase-free water.
miRNA profiling
Prior the profiling, the isolated RNA samples were reverse transcribed to cDNA using the Exiqon Universal cDNA Synthesis Kit II (Exiqon Inc., MA, USA) according with the manufacturer instruction. Briefly, 2 μL of total RNA (5 ng/μL) were used for cDNA synthesis. All processes were conducted in 384 well plate format. The quality and integrity of the synthesized cDNA was assessed using the miRNA QC PCR Panel (V4.M; Exiqon Inc.). Obtained cDNA was 50-fold diluted and mixed with 2x Exilent SYBR Green master mix (Exiqon Inc.), and ROX reference dye (4 μl/2 ml) (Thermo Fisher Scientific, MA, USA). The samples were loaded onto human serum/plasma focus miRNA PCR panels, and quantitative real-time PCR was performed using the QuantStudio 12 K Flex real-time PCR System (Applied Biosystems, CA, USA). The PCR data were processed using Exiqon GenEx qPCR analysis software (version 6). The inter-plate calibration was performed using the mean value of UniSp3 interplate calibrator. The samples with a high degree of hemolysis were identified after monitoring of calculated ΔCt between hsa-miR-23a-3p and hsa-miR-451a. The samples with ΔCt > 7 were removed from the analysis. Only microRNA assays with Ct ≤ 35, expressed in at least 60% of the samples were counted and the remaining samples were removed from the analysis. The global average of all expressed microRNAs with Ct < 35 was used to normalize individual assays. Total of 169 miRNAs were profiled in 339 subjects.
Proteomics measurements using SOMAscan technology
The EDTA-plasma samples were used for proteomics analysis based on SOMAscan assay (version 1.1) technology, which was conducted at the WCM-Q Proteomics Core15. The method employed protein-capture by Slow Offrate Modified Aptamers (SOMAmer)117. Briefly, undepleated EDTA-plasma was diluted and the following assay steps were performed: (1) Binding: analytes and SOMAmers, carrying a biotin moiety via a photocleavable linker were equilibrated; (2) Catch I: analyte/SOMAmer complexes were immobilized on streptavidin‐support, followed by washing steps to remove proteins not stably interacting with SOMAmers; (3) Cleave: release of analyte/SOMAmer complexes from streptavidin beads through exposure to long‐wave ultraviolet light resulting in linker cleavage; (4) Catch II: biotinylation of proteins in analyte/SOMAmer complexes and subsequent repeated immobilization on streptavidin support followed by washing steps to select against non‐specific analyte/SOMAmer complexes; (5) Elution: denaturation of analyte/SOMAmer complexes and SOMAmer release; (6) Quantification: hybridization to custom arrays of SOMAmer‐complementary oligonucleotides. The primary data were submitted to Somalogic for normalization of raw intensities, across-batch calibration and steps of quality control. In total 1129 molecules were quantified in 356 samples.
Proteomics measurements using Olink technology
Heparin-plasma samples were used for the proteomics measurements based on the Olink® technology (Olink Proteomics AB, Uppsala, Sweden) at the WCM-Q Proteomics Core. The technology is based on a proximity extension assay (PEA)118, and enables for simultaneous analysis of 92 analytes in 1 µL of sample. We used two different Olink® panels, namely Cardiometabolic and Metabolism, for measurements of 184 unique proteins. The samples were processed along with 8 control samples according to the manufacturer’s protocol using the following steps: (1) Immunoassay: the sample was mixed and incubated with 92 supplier-provided optimized antibody pairs labeled individuallywith oligonucletotides (PEA probes). Pair coupled oligonucleotides carry unique annealing sites that allows specific hybridization of matching probes; (2) Extension: Target binding by antibody pairs brings the corresponding probe oligonucleotides in close proximity and allows for hybridization. Hybridized templates are extended by DNA polymerase, which generates a DNA template for amplification; (3) Preamplification: Universal primers enable parallel preamplification of all 92 DNA templates by PCR; (4) Detection: The resulting DNA sequence is subsequently detected and quantified using a microfluidic real-time PCR instrument (Biomark HD, Fluidigm, South San Francisco, CA, USA). The data obtained were normalized using an internal extension control and an inter-plate control, to adjust for intra- and inter-run variation. In total 184 proteins were quantified in 328 samples.
Total plasma N-glycosylation (Genos platform)
Sample processing
he EDTA-plasma samples were analyzed by Genos Ltd. (Zagreb, Croatia) using ultra-performance liquid chromatography (UPLC) glycoprofiling as previously described53,119. Briefly, the sample processing for total plasma N-glycosylation measurements was conducted in 96-well plate format out of 10 μL of plasma sample in following steps: (1) Release of N-glycans from plasma proteins: The plasma proteins were denaturated with 20 µl of sodium dodecyl sulfate (SDS) 2% (w/v) (Invitrogen, USA) for 10 min at 65 °C, followed by cooling to room temperature for 30 min, and mixing with 10 µl of 4% (v/v) Igepal-CA630 (Sigma-Aldrich, USA) under constant shaking for 15 min. N-glycans were released after incubation of samples with enzyme, N-glycosidase-F (1.2 U of PNGase F (Promega, USA)) overnight at 37 °C; (2) Fluorescent labeling of released plasma glycans: The obtained N-glycans were mixed with freshly prepared labeling mixture containing (70: 30 v/v) 2-aminobenzamide and 2-picoline borane in dimethylsulfoxide (Sigma-Aldrich) and glacial acetic acid (Merck, Germany) for 15 min followed by 2 h incubation at 65 °C; (3) Cleaning and elution of labeled N-glycans: The excess free label and reducing agent were removed from the samples using hydrophilic interaction liquid chromatography solid-phase extraction (HILIC-SPE). The samples were loaded into the wells of 0.2 µm 96-well GHP filter-plate (Pall Corporation, USA), which was used as stationary phase, and were washed 5 times with cold 96% acetonitrile (ACN). Glycans were eluted with 2 × 90 μL of ultrapure water under constant shaking for 15 min at room temperature. The eluates were combined and stored at −20 °C until use.
Sample measurements
Total plasma N-glycans were measured using HILIC-UPLC as previously described120. Briefly, the labeled N-glycans were gradient eluted from Waters BEH Glycan chromatography column (Waters UPLC BEH particles 2.1 × 150 mm, 1.7 μm) using 100 mM ammonium formate at pH 4.4, and ACN. The flow rate was 0.56 ml/min in a 23 min of the analytical run. The fluorescence was measured at 420 nm with excitation at 330 nm using Waters Acquity UPLC H-class system consisting of a fluorescence (FLR) detector set with 250 nm excitation and 428 nm emission wavelengths.
The data processing was performed using an automatic processing method enabling to obtain chromatograms separated into 39 peaks. The data was further quantified and annotated into 36 primary glycan traits120. All N-glycans have core sugar sequence consisting of two N-acetylglucosamines (GlcNAc) and three mannose residues; F indicates a core fucose α1–6 linked to the inner GlcNAc; Ax indicates the number of antennas (GlcNAc) on trimannosyl core; Gx indicates the number of β1–4 linked galactoses on antenna; G1 indicates that the galactose is on the antenna of the α1–6 mannose; Sx indicates the number (x) of sialic acids linked to galactose. In total 36 total plasma N-glycans were measured in 345 subjects.
IgG glycosylation (Genos platform)
Sample processing
The IgG isolation and measurements were conducted by Genos, Ltd as previously described21,121. Briefly, the sample processing for plasma IgG-glycosylation measurements was conducted in the following steps: (1) Preparation of protein G monolithic plates: the 96-well protein G monolithic plate (BIA Separations, Ajdovščina, Slovenia) was washed with 10 column volumes of ultrapure water, 10 column volumes of binding buffer (1 x PBS), and 5 column volumes of 0.1 M formic acid (pH 2.5). The protein G plate was equilibrated with 10 column volumes of 10 x binding buffer and 20 column volumes of 1 x binding buffer; (2) Isolation of IgG from human plasma: For the IgG isolation the protein G monolithic plate was used. The IgG were obtained from 70 to 100 µl of plasma. The samples were diluted 10 times with binding buffer and filtered through GHP AcroPrep 96-well filter plates. The samples were applied onto the protein G monolithic plates and instantly washed three times with PBS to remove the unbound proteins; (3) Elution of IgGs: The IgG were eluted from the protein G monoliths into 96-well plate with 5 column volumes of 0.1 M formic acid (Merck, Germany) followed by immediate neutralization with 1 M ammonium bicarbonate (Merck, Germany)122; (4) IgG digestion and purification: Aliquots of 40 µl from the obtained samples, containing isolated IgG, were used for further processing. The samples were incubated with 2% SDS [20 µL (w/v)] for 10 min at 60 °C, and followed by overnight incubation with 200 ng trypsin at 37 °C. The obtained IgG tryptic glycopeptides samples were purified by reverse phase solid phase extraction using Chromabond C18 beads applied to each well of an OF1100 96-well polypropylene filter plate. The beads were activated with 80% ACN containing 0.1% trifluoroacetic acid (TFA); (5) IgG elution: The tryptic digests were diluted 10 times in 0.1% TFA, loaded onto the C18 beads in vacuum manifold and washed 3 times with 0.1% TFA. IgG glycopeptides were eluted into an PCR 96 well plate with 120 µl of 20% ACN containing 0.1% TFA by 5 min centrifugation at 105 × g. Eluates containing glycopeptides were dried by vacuum centrifugation and −20 °C until analysis by MS.
Sample measurement
Purified tryptic IgG glycopeptides were analyzed as previously described121. For the separation and measurements nanoACQUITY UPLC system (Waters, Milford Massachusetts, USA), consisting of binary pump, auxillary pump, autosampler maintained at 10 °C and column oven compartment set at 30 °C coupled to and the Bruker Compact Q-TOF-MS were used. 9 μL of purified IgG glycopeptides sample was applied to a Thermo Scientific PepMap 100 C8 (5 mm × 300 μmi.d., 5 μm) SPE trap column. After sample loading the trap column was switched in-line with the gradient and C18 nano-LC column (150 mm × 100 μm i.d., 2.7 μm HALO fused core particles; Advanced Materials Technology, Wilmington, Delaware, USA) for 9.5 min while sample elution took place. IgG glycopeptides were reconstituted in 20 µl MQ water before nano-LC-ESI-MS analysis. Separation was achieved at 1 ml/min using the following gradient of mobile phase A and mobile phase B (80% ACN and 20% 0.1% TFA): 0.5 min 12% B, 0.5–4 min 12% B - 17% B, 4–5 min 17% B. The column outlet tubing was directly applied as sprayer needle. Quadrupole and collision energy was set at 4 eV. Spectra were recorded from m/z 600 to 1900 with 2 averaged scans at a frequency of 0.5 Hz. Per sample the total analysis time was 15 min.
The nanoACQUITY UPLC system and the Bruker Compact Q-TOF-MS were operated under HyStar software version 3.2.
Glycan data was first normalized (total area normalization) and then batch corrected using Combat. Batch correction was performed on the log-transformed normalized data. After batch correction, the data was inverse transformed so all values were between 0 and 100. Finally, the data was z-scored. Glycan structural features are given in terms of number of galactoses (G0, G1, and G2), fucose (F), bisecting N-acetylglucosamine (N) and N-acetylneuraminic acid (S). Total of 60 IgGs were measured in 341 samples.
IgA and IgG glycosylation (Univ. Leiden platform)
Sample processing
The purification, separation, and measurements of IgA and IgG was conducted at Leiden University Medical Center as previously described123,124. Briefly, 2 μL and 5 μL of plasma was used for IgG and IgA analysis, respectively. The samples were diluted with PBS to obtain final volume of 200 μL. The samples purification was conducted in duplicate on separate plates using affinity bead chromatography. The samples designated for IgG analysis were purified using 15 μL/well of Protein G Sepharose 4 Fast Flow beads (GE Healthcare) on an Orochem filter plate, followed by three washing steps with PBS. The samples designated for IgA analysis were purified using 2 μL/well of CaptureSelect IgA Affinity Matrix beads (Thermo Fisher Scientific). The plates were incubated for 1 h under constant shaking.
The samples were washed three times with PBS followed by three additional washes with purified water using vacuum manifold. The IgGs and IgAs were eluted from the beads using 100 mM formic acid under constant shaking for 10 min, followed by 1 min centrifugation at 100 × g. The obtained eluates were dried for 2 h at 60 °C in a vacuum centrifuge.
The samples designated for IgG analysis, were resolubilized by addition of ammonium bicarbonate (50 mM) under constant shaking for 5 min. The samples were digested by overnight incubation with tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin at 37 °C.
The samples designated for IgA analysis were reduced and alkylated prior to digestion to obtain peptides covering all glycosylation sites. The samples resolubilization was conducted with ammonium bicarbonate (30 mM) containing 12.5% of acetonitrile under constant shaking for 5 min. The samples were mixed with dithiothreitol (35 mM) and incubated for 5 min at room temperature followed by additional incubation for 30 min at 60 °C. The samples were cooled to room temperature, mixed with iodoacetamide (125 mM), incubated in the dark under shaking for 30 min and mixed with dithiothreitol (100 mM) to quench the iodoacetamide. The samples were digested with TPCK-treated trypsin by the incubation over night at 37 °C.
Sample measurement
The samples designated for IgG and IgA were measured at different days. The sample separation and measurements were conducted on Ultimate 3000 RSLCnano system (Dionex/Thermo Scientific) equipped with an Acclaim PepMap 100 trap column (particle size 5 μm, pore size 100 Å, 100 μm × 20 mm) and an Acclaim PepMap C18 nano analytical column (particle size 2 μm, pore size 100 Å, 75 μm × 150 mm) coupled to a quadrupole-TOF-MS (Impact HD; Bruker Daltonics). 250 μL of sample was injected into the flow (25 μL/min) of aqueous solvent and was trapped on the trap column (Dionex Acclaim PepMap100 C18, 5 mm × 300 μm; Thermo Fisher Scientific, Breda, The Netherlands). The analytes were eluted on the analytical column (Ascentis Express C18 nanoLC column, 50 mm × 75 μm, 2.7 μm fused core particles; Supelco, Bellefonte, PA) under flow rate of 0.9 μL/min and separated in linear gradient from 3% to 30% solvent containing 95% (v/v) ACN. The samples were measured in positive-ion mode using a CaptiveSprayer (Bruker Daltonics) electrospray source at 1300 V. The mass spectra were acquired with a frequency of 1 Hz and the MS ion detection window was set at mass-to-charge ratio (m/z) 550–1800. Fragmentation spectra were recorded with a detection window of m/z 50–2800.
Obtained LC-MS data were examined according with pipeline developed by Manfred Wuhrer lab as previously described123,124. In total 178 molecules including IgGs and IgAs were measured in 344 samples.
Untargeted metabolomics—Metabolon HD2 platform
The EDTA-plasma, saliva, and urine samples were used for untargeted metabolic profiling as we previously described12,13. The measurements were conducted at Metabolon Inc, deploying HD2 platform based on ultra-high-performance liquid chromatography-mass spectrometry (UPLC-MS) and gas chromatography-mass spectrometry (GC-MS) technology125. In brief, the sample was mixed with the recovery standards prior to the extraction for quality control (QC) purposes. The resulting sample extract was divided into aliquots designated for the analysis using the following: (1) UPLC-MS/MS with positive ion mode electrospray ionization (ESI); (2) UPLC-MS/MS with negative ion mode ESI; (3) hydrophilic interaction chromatography (HILIC)/UPLC-MS/MS; (4) GC-MS. The sample extract was dried under nitrogen flow and reconstituted in solvents compatible with each of the four analytical methods.
Three out of the four sample aliquots were designated for LC-MS measurements and were reconstituted in acidic or basic solvents. The first sample aliquot was reconstituted in acidic conditions, and gradient eluted from a C18 column (Waters UPLC BEH C18-2.1 × 100 mm, 1.7 µm) with water and methanol containing 0.1% formic acid (FA). The second sample aliquot was reconstituted in basic solvent, and gradient eluted from C18 column (Waters UPLC BEH C18-2.1 × 100 mm, 1.7 µm) with water and methanol containing 6.5 mM ammonium bicarbonate. The third aliquot was gradient eluted from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 µm) using water and acetonitrile with 10 mM ammonium formate. The flow rate was 350 μL/min, and the sample injection volume was 5 μL.
The separation and measurements of the sample aliquots designated for LC-MS were performed on Waters ACQUITY UPLC in-line to Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and an Orbitrap mass analyzer. In the MS analysis, the scan range varied between methods but fell within the range of 70–1000 m/z.
The remaining fourth sample aliquot was designated for GC-MS measurements. The sample aliquot was derivatized with N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA) followed by drying under nitrogen flow. Separation was conducted under temperature ramp from 60 to 340 °C over a period of 17.5 min, using a 5% diphenyl/95% dimethyl polysiloxane fused silica column (20 m × 0.18 mm ID; 0.18 μm film thickness) and helium at flow rate of 1 ml/min as the carrier gas. The measurements were performed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization (EI), and the MS scan range was from 50 to 750 m/z. The number of measured metabolites in given sample matrix was following: 758 metabolites in 358 EDTA-plasma samples, 602 metabolites in 283 saliva samples, and 891 metabolites in 360 urine samples.
Untargeted metabolomics—Metabolon HD4 platform
The EDTA-plasma samples were used to conduct metabolic profiling at Metabolon Inc on technologically advanced, in comparison with HD2, HD4 platform enabling for increased sensitivity and accurate detection of more metabolites. The main technical difference between HD2 and HD4 platforms was replacement of GC-MS with hydrophilic interaction chromatography (HILIC) method. The method was described in great detail previously126. In brief, sample processing was conducted as we described in “Untargeted profiling - HD2 platform: LC-MS and GC-MS” section, except of the sample dedicated for GC-MS measurement. This sample aliquot instead was gradient eluted from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 μm) using water and acetonitrile with 10 mM ammonium formate at pH 10.8. The measurements were conducted using Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high-resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer126. Total of 1104 metabolites were measured in 309 plasma samples.
Targeted metabolomics—Biocrates p150 platform
The EDTA-plasma samples were used for targeted metabolomics analysis. The samples were measured at the Metabolomics Platform of the Helmholtz Center Munich using AbsoluteIDQTM kit p150 (Biocrates Life Science AG, Innsbruck, Austria) as previously described127,128. For the lipid molecules including PC, lysoPC, SM, and AC, measured with AbsoluteIDQTM kit the information on the sum of the carbons of the fatty acid chains is provided but not the fatty acid chain actual composition. Total of 10 µL of plasma was used to conduct the assay. The samples were applied on the assay kit 96-well plate consisting of filters with internal standards and were dried under a nitrogen stream at room temperature (RT). The samples were derivatized with a reagent containing 5% phenylisothiocyanate (PITC), dryed under a nitrogen stream at RT, and extracted with 300 μL of 5 mM ammonium acetate in methanol. Next, the samples were filtered by centrifugation, the resulting flow-through was diluted 1:6 with running solvent and placed into fresh deep-well plate. The plate was covered with the silicone mat, and mixed. Sample handling was performed by a Hamilton Microlab STARTM robot (Hamilton Bonaduz AG, Bonaduz, Switzerland) and a Ultravap nitrogen evaporator (Porvair Sciences, Leatherhead, U.K.), beside standard laboratory equipment. Metabolites were measured in positive and negative multiple reaction monitoring (MRM) scan mode by direct infusion to an API 4000 triple quadrupole system (SCIEX Deutschland GmbH, Darmstadt, Germany) equipped with a 1200 Series HPLC (Agilent Technologies Deutschland GmbH, Böblingen, Germany) and a HTC PAL autosampler (CTC Analytics, Zwingen, Switzerland) controlled by the software Analyst 1.6.2. The metabolite concentrations were calculated using internal standards and the MetIDQ software provided with AbsoluteIDQTM kit, and are reported in μmol/L. For the lipid molecules including PC, lysoPC, SM, and AC, measured with AbsoluteIDQTM kit the information on the sum of the carbons of the fatty acid chains is provided but not the fatty acid chain actual composition. For example, PC.aa.36.1 describes phosphatidylcholine (PC) where two glycerol residues are bound in diacyl (aa) binding into the fatty acid moiety; the sum of carbons of both fatty acid chains is 36, and there is one double bound (.1). Total of 163 metabolites were quantified in 356 samples.
Lipidomics—Lipidyzer platform
The EDTA-plasma samples were used for in depth profiling of lipids, which was conducted at Metabolon Inc. deploying LipidyzerTM platform of AB Sciex Pte technology as previously described129,130. In brief, the samples were extracted in the presence of internal standards using butanol:methanol (BUME) mixture (3:1) followed by two-phase extraction into 300 µl heptane:ethyl acetate (3:1) using 300 µl 1% acetic acid as buffer. The obtained extracts were dried under nitrogen flow and reconstituted in ammonium acetate dichloromethane:methanol. The samples were analyzed in both positive and negative mode electrospray using Sciex SelexIon-5500 QTRAP. The molecules were detected in MRM mode with a total of more than 1100 MRMs. Individual lipid species were quantified by the ratio of the signal intensity of each target compound to that of its assigned internal standard, followed by the multiplication of the concentration of internal standard added to the sample. Lipid class concentrations were calculated from the sum of all molecular species within a class, and fatty acid compositions were determined by calculating the proportion of each class comprised by individual fatty acids. Total of 1331 lipids were measured in 324 samples.
NMR metabolomics—urine
1H-NMR spectra analysis of urine samples was conducted at Institute of Clinical Chemistry and Laboratory Medicine, University of Greifswald, Germany as previously described14,131. In brief, Bruker DRX-400 NMR spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany) operating at 400.13 MHz 1H frequency equipped with 4 mm selective inverse flow probe (FISEI, 120 µL active volume) was used to record the spectra. 500 µL of sample volume was delivered via automatic flow injection. The sample acquisition temperature was 300 K. A standard one-dimensional 1H NMR pulse sequence with suppression of water peak (NOESYPRESAT) was used (Budde et al. 131). The free induction decays (FIDs) were collected into data points using spectral width of 20.689 ppm. The FIDs were multiplied by an exponential function corresponding to 0.3 HZ line-boarding prior to Fourier-transformed (FT). For the assessment of the spectra quality, the line width and signal-to-noise ration of Trimethylsilylpropanoic acid (TSP) signal, used as a reference, was analyzed. Additionally, quality control was carried out by analyzing the standard error of creatinine concentration and the potential variability of selected signals. The obtained spectra were processed within TOPSPIN 1.3 (Bruker BioSpin GmbH) and the metabolites annotation and quantification was conducted in semi-automated manner by spectral pattern matching using Chnomx NMR suit 7.0 (Chenomx Inc.). A total of 60 lipid molecules were measured in 353 samples.
NMR metabolomics—plasma
The EDTA-plasma (300 µL) was used for metabolite quantification by a high-throughput NMR metabolomics platform (Nightingale Ltd, Helsinki, Finland)14,132. The sample preparation was conducted automatically using Gilson Liquid Handler 215. Each sample after brief centrifugation was transferred to SampleJet NMR tubes and mixed with 300 µL of sodium phosphate containing 0.08% of TSP. The measurements were conducted on Bruker AVANCE III 500 MHz and Bruker AVANCE III HD 600 MHz spectrometers. The lipoprotein (LIPO) and low-molecular-weight metabolites (LMWM) were measured in the samples using either 500 MHz or 600 MHz spectrometers. The same samples were further extracted with multiple extraction steps as previously detailed133. The extracted lipid (LIPID) data was evaluated in full automation with the 600 MHz instrument using a standard parameter set133 (Soininen et al. 133). The FT and automated phasing of NMR spectra was conducted followed by automated spectral processing and quality control steps132. The subclasses for the lipoproteins are categorized according to size following this classification: chylomicrons and extremely large VLDL particles (average particle diameter at least 75 nm); five VLDL subclasses—very large VLDL (average particle diameter of 64.0 nm), large VLDL (53.6 nm), medium VLDL (44.5 nm), small VLDL (36.8 nm) and very small VLDL (31.3 nm); intermediate-density lipoprotein (IDL; 28.6 nm); three LDL subclasses—large LDL (25.5 nm), medium LDL (23.0 nm) and small LDL (18.7 nm); and four HDL subclasses—very large HDL (14.3 nm), large HDL (12.1 nm), medium HDL (10.9 nm) and small HDL (8.7 nm). Total of 225 molecules were measured in 350 samples.
Statistical data analysis
All statistical analyses were conducted using R (version 4.1.0 and above) and Rstudio (version 1.4.1717 and above). If not otherwise stated, the omics data was converted “as-received” into R Summarized Experiment format, representing processed final data. The saliva metabolomics data has been further normalized by saliva osmolality, and the urine metabolomics data has been normalized by urine creatinine.
Cross-platform correlations (omicsMBHs)
The association between each two platforms was described using mutual best hit (MBH) aiming to identify pairs of features (e.g., genes, proteins) that exhibited a significant correlation with each other. Spearman correlation coefficients between unscaled raw omics data were computed. Next, mutual best hits were identified; only those pairs demonstrating a significant and reciprocal relationships were retained. Reciprocity implies a two-way relationship, where the correlation is bidirectional. Platform-pairwise Bonferroni significance cutoffs (p < 0.05/(nPLTA1 * nPLAT2/2)) were obtained after cross-platform correlation.
Within-trait partial correlations (GGMs)
Partial correlations within platforms were computed as follows: Saliva and urine metabolites were normalized by saliva osmolality and urine creatinine obtained from the respective platform, respectively. The omicsdata was then inverse-normal scaled. Metabolites and then samples with more than 50% missing values were removed. Association statistics and residuals were then computed using the linear model “lm(OMICS ~ AGE + SEX + BMI + DIAB + genoPC1 + genoPC2 + genoPC3 + somaPC1 + somaPC2 + somaPC3)”. Missing values were imputed using the K-nearest-neighbors method134. Partial correlation coefficients were computed using the pcor function from the R-package GeneNet (version 1.2.15). Platform-wise Bonferroni significant correlations (p < 0.05/(NPLAT*(NPLAT-1)/2)), where NPLAT represents the number of traits measured on the respective platform, were retained.
Association between DNA – RNA – METH (eQTLs, eQTMs, meQTLs)
Genetic variants (SNPs) were coded 0, 1, 2 for major allele homozygotes, heterozygotes, and minor allele homozygotes, respectively. Expression data was log-scaled, with all values off-set by the smallest occurring value in the dataset in order to avoid taking the log of zero, and z-scored. Methylation d(CpG) were b-values. The following linear models were used to compute the associations:
eQTL: lm(transcriptomics ~ SNP + AGE + SEX + BMI + DIAB + genoPC1 + genoPC2 + genoPC3)
meQTL: lm(CpG ~ SNP + AGE + SEX + BMI + DIAB + genoPC1 + genoPC2 + genoPC3)
eQTM: lm(transcriptomics ~ CpG + SNP + AGE + SEX + BMI + DIAB + genoPC1 + genoPC2 + genoPC3)
A significance cut-off of p < 5 × 10−8 was used.
Genetic variation—omicsdata associations (omicsQTLs)
Omicsdata was inverse-normal scaled and residual were computed using the linear model “lm(Omicsdata ~ SEX + AGE + BMI + DIAB + genoPC1 + genoPC2 + genoPC3 + somaPC1 + somaPC2 + somaPC3)”. After QC, excluding non-autosomal SNPs, MAF < 5%, HWE pvalue < 10-6, or genotyping rate <98%15, 1,221,345 SNPs for 353 samples were available. Additive linear models using Plink version 1.9135 were computed. Genomic inflation was lambda<1.04 for all traits. All associations with p < 5 × 10−8 were lumped, treating variants with R2 < 0.1 as independent15. Phenoscanner136, accessed 9 April 2019, was used to annotate the sentinel variants with GWAS hits, metabolomics and proteomics QTLs, and genes encodes at the locus, using R2 > 0.8 (LD from EUR), and limiting associations to p < 5 × 10−8. Genetic variants were annotated to human genome build 37.
Methylation—omicsdata association (omicsQTMs)
Residuals of methylation beta values (CpG) were computed using the linear model “lm (CpG ~ AGE + SEX + BMI + DIAB + Gran + NK + CD4T + CD8T + Mono + Bcell + genoPC1 + genoPC2 + genoPC3)” and then z-scored. Saliva and urine metabolites were first normalized by saliva and urine osmolality, respectively. All omics variables were then inverse normal-scaled and residuals computed using the linear model “lm (Omicsdata ~ AGE + SEX + BMI + DIAB + genoPC1 + genoPC2 + genoPC3)” and then z-scored. Association statistics were then computed using the linear model “lm(CpG_residual ~ Omicsdata_residual)”. Associations reaching an ad hoc significance level of 5×10-8 were retained. CpG sites were annotated for gene names and CpG position relative to the genes using the Illumina provided HumanMethylation 450k annotation file.
RNA expression—omicsdata association (omicsQTRs)
RNA expression data with less than 100 valid data points or median expression levels below 1 TPM were removed. Expression data was log-scaled, with all values off-set by the smallest occurring value in the dataset in order to avoid taking the log of zero, and z-scored. Saliva and urine metabolites were normalized by saliva osmolality and urine creatinine obtained from the respective platform, respectively. The omicsdata was then inverse-normal scaled. Metabolites and then samples with more than 50% missing values were removed. Association statistics were then computed using the linear model “lm(OMICS ~ transcriptomics + AGE + SEX + BMI + DIAB + genoPC1 + genoPC2 + genoPC3 + CD8 + CD4 + NK + Bcell + Mono + Gran + Eos)”. Associations reaching an ad hoc significance level of 5 × 10−8 were retained.
Potential to predict age, sex, BMI, and diabetes state by platform
For continuous variables we use the “pseudo-R 2” reported by the R package randomForest as an estimate of how well a given omics phenotype can predict age, sex, BMI, and diabetes state. For continuous variables this “pseudo-R 2” is defined as one minus the mean square error of the regression divided by the variance of the dependent variable. Note that the “pseudo-R 2” is not a strict measure of the explained variance and is used here to provide an intuition for the quality of the model fit that can be obtained using the different omics datasets.
For categorial variables we use the “Out-Of-Bag Error” (OOBErr) estimate from R, scaled to range between zero and one (1-OOBerr)/(1-OOBerrrnd), where OOBErrrnd was estimated by randomizing the sample identifiers.
Disease/trait associations from the GWAS catalog
We downloaded the GWAS catalog (gwas_catalog_v1.0.2-associations_e100_r2021-01-14.tsv) and identified 6,694 variants that are in LD (r2 > 0.8) with one of the 587 sentinel SNPs (incl. the SNPs themselves). We then identified 2294 records in the GWAS catalog that reported on one of the 6,694 SNPs. Where multiple associations with a same trait were reported for a same locus, we kept only the strongest association.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Acknowledgements
We are grateful to all participants of QMDiab for providing their time and blood, and to the late Prof. Mohammed M. El-Din Selim for enabling the sample collection at Hamad Medical Corporation, Doha, Qatar. K.S. is supported by the Biomedical Research Program at Weill Cornell Medicine in Qatar, a program funded by the Qatar Foundation. K.S. is also supported by the Qatar National Research Fund (QNRF) grant NPRP11C-0115-180010 and ARG01-0420-23000. A.H. is supported by the Qatar National Research Fund (QNRF) grant NPRP12S-0205-190042 and NPRP11S-0122-180359. The statements made herein are solely the responsibility of the authors.
Author contributions
Study design: A.H., K.S.; Sample collection: S.K., W.Al.M., M.M.-K., Conducted Experiments: H.S., Y.A.M., S.A., M.P.B., C.P., J.A., N.F., U.V., M.W., G.L., S.H.N.-S., J.A.M., S.A., J.G., D.M.-K., F.S., Data analysis: K.S., S.Z.; Data interpretation: A.H., K.S.; Provided Materials: A.H., N.S., G.T., S.K., W.Al.M., M.M.-K., J.K., J.M.S.; Manuscript writing: A.H, K.S.; Manuscript editing: A.H., K.S., J.M.S., F.S. All authors contributed to the critically reviewed manuscript.
Peer review
Peer review information
Nature Communications thanks Jessica Lasky-Su, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The source of data generated by each platform is provided on Figshare (10.6084/m9.figshare.25975627.v2). The data can be downloaded as an excel file. Additionally, we are also providing data in.rda format for which we prepared R script that downloads the rda. data. The genetic data and methylation data access is not deposited because the informed consent given by the study participants does not cover posting of participant genotype and methylation data in public databases. Researcher affiliated with a research institution may request access to genetic data on an individual basis from the corresponding author (Karsten Suhre and Anna Halama, Weill Cornell Medicine—Qatar, Doha, Qatar). Access is subject to approval by the institutional research board of Weill Cornell Medicine—Qatar. The data sets deployed in this study were previously utilized as follows: Genomics data depicted as DNA14–16,20,137,138; Methylation data depicted as MET14,16; Transcriptomics data depicted as RNA139; Proteomics data measured on SOMA platform depicted as SOMA14–16,20,53,137,140; Glycomics data reflecting on total plasma N-glycosylation depicted as PGP14,53; Glycomics data reflecting on plasma IgG levels depicted as IgG14,138; Lipoproteomics data depicted as BRAIN14; The broad lipidomics data depicted as LD130; The targeted lipidomics data depicted as BM14,130; The untargeted metabolomics measured on HD4 platform depicted as HDF19,75,141; PM12–14,16–18,141,142; SM12–14,16–18,142; UM12–14,16–18,142; and CM16. Transcriptomics data covering microRNA depicted as miRNA, proteomics data measured on OLINK platform depicted as OLINK, and Glycomics data reflecting on plasma IgA levels depicted as IgA were not published before.
Code availability
We are also providing access to the source code used to generate COmics Server. The source code and a docker image could be accessed via GitHub at https://github.com/karstensuhre/comics and referenced using 10.5281/zenodo.11487725.
Competing interests
The authors declare the following conflicts of interest: M.P.B. and G.L. are working for or have stakes in Genos Ltd., a private company specialized in glycomics analyses. All the other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anna Halama, Email: amh2025@qatar-med.cornell.edu.
Karsten Suhre, Email: kas2049@qatar-med.cornell.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51134-x.
References
- 1.Borges, L., Kubin, M. & Kuhlman, T. LIR9, an immunoglobulin-superfamily-activating receptor, is expressed as a transmembrane and as a secreted molecule. Blood101, 1484–1486 (2003). 10.1182/blood-2002-05-1432 [DOI] [PubMed] [Google Scholar]
- 2.Campeau, A. et al. Multi-omics of human plasma reveals molecular features of dysregulated inflammation and accelerated aging in schizophrenia. Mol. Psychiatry27, 1217–1225 (2022). 10.1038/s41380-021-01339-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marabita, F. et al. Multiomics and digital monitoring during lifestyle changes reveal independent dimensions of human biology and health. Cell Syst.13, 241–255.e247 (2022). 10.1016/j.cels.2021.11.001 [DOI] [PubMed] [Google Scholar]
- 4.Sailani, M. R. et al. Deep longitudinal multiomics profiling reveals two biological seasonal patterns in California. Nat. Commun.11, 4933 (2020). [DOI] [PMC free article] [PubMed]
- 5.Benson, M. D. et al. Protein-metabolite association studies identify novel proteomic determinants of metabolite levels in human plasma. Cell Metab.35, 1646–1660.e1643 (2023). 10.1016/j.cmet.2023.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikaeloff, F. et al. Network-based multi-omics integration reveals metabolic at-risk profile within treated HIV-infection. Elife12, e82785 (2023). [DOI] [PMC free article] [PubMed]
- 7.Shi, L. et al. Multiomics profiling of human plasma and cerebrospinal fluid reveals ATN-derived networks and highlights causal links in Alzheimer’s disease. Alzheimers Dement19, 3350–3364 (2023). 10.1002/alz.12961 [DOI] [PubMed] [Google Scholar]
- 8.Garrett-Bakelman, F. E. et al. The NASA Twins Study: a multidimensional analysis of a year-long human spaceflight. Science364, eaau8650 (2019). [DOI] [PMC free article] [PubMed]
- 9.Contrepois, K. et al. Molecular choreography of acute exercise. Cell181, 1112–1130 e1116 (2020). 10.1016/j.cell.2020.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell148, 1293–1307 (2012). 10.1016/j.cell.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tebani, A. et al. Integration of molecular profiles in a longitudinal wellness profiling cohort. Nat. Commun.11, 4487 (2020). 10.1038/s41467-020-18148-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mook-Kanamori, D. O. et al. 1,5-Anhydroglucitol in saliva is a noninvasive marker of short-term glycemic control. J. Clin. Endocrinol. Metab.99, E479–E483 (2014). 10.1210/jc.2013-3596 [DOI] [PubMed] [Google Scholar]
- 13.Yousri, N. A. et al. A systems view of type 2 diabetes-associated metabolic perturbations in saliva, blood and urine at different timescales of glycaemic control. Diabetologia58, 1855–1867 (2015). 10.1007/s00125-015-3636-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaghlool, S. B. et al. Deep molecular phenotypes link complex disorders and physiological insult to CpG methylation. Hum. Mol. Genet.27, 1066–1121 (2018). 10.1093/hmg/ddy006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suhre, K. et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun.8, 14357 (2017). [DOI] [PMC free article] [PubMed]
- 16.Zaghlool, S. B. et al. Epigenetics meets proteomics in an epigenome-wide association study with circulating blood plasma protein traits. Nat. Commun.11, 15 (2020). [DOI] [PMC free article] [PubMed]
- 17.Do, K. T. et al. Network-based approach for analyzing intra- and interfluid metabolite associations in human blood, urine, and saliva. J. Proteome Res14, 1183–1194 (2015). 10.1021/pr501130a [DOI] [PubMed] [Google Scholar]
- 18.Do, K. T., Rasp, D. J. N. P., Kastenmüller, G., Suhre, K. & Krumsiek, J. MoDentify: phenotype-driven module identification in metabolomics networks at different resolutions. Bioinformatics35, 532–534 (2019). 10.1093/bioinformatics/bty650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomari, D. P. et al. Variational autoencoders learn transferrable representations of metabolomics data. Commun. Biol.5, 645 (2022). 10.1038/s42003-022-03579-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudmundsdottir, V. et al. Circulating protein signatures and causal candidates for type 2 diabetes. Diabetes69, 1843–1853 (2020). 10.2337/db19-1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharapov, S. Z. et al. Defining the genetic control of human blood plasma N-glycome using genome-wide association study. Hum. Mol. Genet.28, 2062–2077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krumsiek, J., Suhre, K., Illig, T., Adamski, J. & Theis, F. J. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst. Biol.5, 21 (2011). 10.1186/1752-0509-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overbeek, R., Fonstein, M., D’Souza, M., Push, G. D. & Maltsev, N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA96, 2896–2901 (1999). 10.1073/pnas.96.6.2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tatusov, R. L. et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res29, 22–28 (2001). 10.1093/nar/29.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suhre, K. & Zaghlool, S. Connecting the epigenome, metabolome and proteome for a deeper understanding of disease. J. Intern Med290, 527–548 (2021). 10.1111/joim.13306 [DOI] [PubMed] [Google Scholar]
- 26.Tam, V. et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet20, 467–484 (2019). 10.1038/s41576-019-0127-1 [DOI] [PubMed] [Google Scholar]
- 27.Wainberg, M. et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet51, 592–599 (2019). 10.1038/s41588-019-0385-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner, G. P. The biological homology concept. Annu. Rev. Ecol. Syst.20, 51–69 (1989). 10.1146/annurev.es.20.110189.000411 [DOI] [Google Scholar]
- 29.Brown, T. A. The HumanGenome, (Oxford: Wiley-Liss, 2002). [PubMed]
- 30.Elemento, O., Gascuel, O. & Lefranc, M.-P. Reconstructing the duplication history of tandemly repeated genes. Mol. Biol. Evol.19, 278–288 (2002). 10.1093/oxfordjournals.molbev.a004081 [DOI] [PubMed] [Google Scholar]
- 31.Fitch, W. M. Homologya personal view on some of the problems. Trends Genet16, 227–231 (2000). 10.1016/S0168-9525(00)02005-9 [DOI] [PubMed] [Google Scholar]
- 32.Gabaldón, T. & Koonin, E. V. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet.14, 360–366 (2013). 10.1038/nrg3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krumsiek, J. et al. Gender-specific pathway differences in the human serum metabolome. Metabolomics11, 1815–1833 (2015). 10.1007/s11306-015-0829-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miike, K. et al. Proteome profiling reveals gender differences in the composition of human serum. Proteomics10, 2678–2691 (2010). 10.1002/pmic.200900496 [DOI] [PubMed] [Google Scholar]
- 35.Singmann, P. et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenetics Chromatin8, 43 (2015). 10.1186/s13072-015-0035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristic, J. et al. Glycans are a novel biomarker of chronological and biological ages. J. Gerontol. A Biol. Sci. Med. Sci.69, 779–789 (2014). 10.1093/gerona/glt190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bocklandt, S. et al. Epigenetic predictor of age. PLoS ONE6, e14821 (2011). 10.1371/journal.pone.0014821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hertel, J. et al. Measuring biological age via metabonomics: the metabolic age score. J. Proteome Res.15, 400–410 (2016). 10.1021/acs.jproteome.5b00561 [DOI] [PubMed] [Google Scholar]
- 39.Lehallier, B. et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med.25, 1843–1850 (2019). 10.1038/s41591-019-0673-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters, M. J. et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun.6, 8570 (2015). 10.1038/ncomms9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson, O. et al. Determinants of accelerated metabolomic and epigenetic aging in a UK cohort. Aging Cell19, e13149 (2020). [DOI] [PMC free article] [PubMed]
- 42.Tanaka, T. et al. Plasma proteomic signature of age in healthy humans. Aging Cell17, e12799 (2018). [DOI] [PMC free article] [PubMed]
- 43.Pena, M. J., Mischak, H. & Heerspink, H. J. Proteomics for prediction of disease progression and response to therapy in diabetic kidney disease. Diabetologia59, 1819–1831 (2016). 10.1007/s00125-016-4001-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schrader, S. et al. Novel subgroups of type 2 diabetes display different epigenetic patterns that associate with future diabetic complications. Diab. Care45, 1621–1630 (2022). 10.2337/dc21-2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang-Sattler, R. et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol.8, 615 (2012). 10.1038/msb.2012.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lelo, A., Kjellen, G., Birkett, D. J. & Miners, J. O. Paraxanthine metabolism in humans: determination of metabolic partial clearances and effects of allopurinol and cimetidine. J. Pharm. Exp. Ther.248, 315–319 (1989). [PubMed] [Google Scholar]
- 47.Rybak, M. E., Sternberg, M. R., Pao, C. I., Ahluwalia, N. & Pfeiffer, C. M. Urine excretion of caffeine and select caffeine metabolites is common in the U.S. population and associated with caffeine intake. J. Nutr.145, 766–774 (2015). 10.3945/jn.114.205476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeffcoate, S. L. Diabetes control and complications: the role of glycated haemoglobin, 25 years on. Diabet. Med. 21, 657–665 (2004). [DOI] [PubMed]
- 49.Rahbar, S., Blumenfeld, O. & Ranney, H. M. Studies of an unusual hemoglobin in patients with diabetes mellitus. Biochem. Biophys. Res. Commun.36, 838–843 (1969). 10.1016/0006-291X(69)90685-8 [DOI] [PubMed] [Google Scholar]
- 50.Lever, M. et al. Variability of plasma and urine betaine in diabetes mellitus and its relationship to methionine load test responses: An observational study. Cardiovasc. Diabetol.11, 34 (2012). [DOI] [PMC free article] [PubMed]
- 51.Mardinoglu, A. et al. Plasma mannose levels are associated with incident type 2 diabetes and cardiovascular disease. Cell Metab.26, 281–283 (2017). [DOI] [PubMed]
- 52.Contreras, P., Generini, G., Michelsen, H., Pumarino, H. & Campino, C. Hyperprolactinemia and galactorrhea: Spontaneous versus iatrogenic hypothyroidism. J. Clin. Endocrinol. Metab.53, 1036–1039 (1981). 10.1210/jcem-53-5-1036 [DOI] [PubMed] [Google Scholar]
- 53.Suhre, K. et al. Fine-mapping of the human blood plasma N-glycome onto its proteome. Metabolites9, 122 (2019). [DOI] [PMC free article] [PubMed]
- 54.Gilly, A. et al. Whole-genome sequencing analysis of the cardiometabolic proteome. Nat. Commun.11, 6336 (2020). [DOI] [PMC free article] [PubMed]
- 55.Huan, T. et al. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat. Commun.10, 4267 (2019). [DOI] [PMC free article] [PubMed]
- 56.Kettunen, J. et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun.7, 11122 (2016). [DOI] [PMC free article] [PubMed]
- 57.Suhre, K., McCarthy, M.I. & Schwenk, J. M. Genetics meets proteomics: perspectives for large population-based studies. Nat. Rev. Genet. 22, 19–37 (2021). [DOI] [PubMed]
- 58.Suhre, K. et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature477, 54–62 (2011). 10.1038/nature10354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet47, 1091–1098 (2015). 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet48, 245–252 (2016). 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin, W. D. et al. Sialylation of CD55 by ST3GAL1 facilitates immune evasion in cancer. Cancer Immunol. Res.9, 113–122 (2021). 10.1158/2326-6066.CIR-20-0203 [DOI] [PubMed] [Google Scholar]
- 62.Wu, X. et al. Sialyltransferase ST3GAL1 promotes cell migration, invasion, and TGF-β1-induced EMT and confers paclitaxel resistance in ovarian cancer. Cell Death Dis.9, 1102 (2018). [DOI] [PMC free article] [PubMed]
- 63.Steffen, U. et al. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat. Commun.11, 120 (2020). 10.1038/s41467-019-13992-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huan, T. et al. Genome-wide identification of microRNA expression quantitative trait loci. Nat. Commun.6, 6601 (2015). [DOI] [PMC free article] [PubMed]
- 65.Wagschal, A. et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med.21, 1290–1297 (2015). 10.1038/nm.3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonder, M. J. et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet.49, 131–138 (2017). 10.1038/ng.3721 [DOI] [PubMed] [Google Scholar]
- 67.Granjon, A. et al. The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes58, 2555–2564 (2009). 10.2337/db09-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Volkmar, M. et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J.31, 1405–1426 (2012). 10.1038/emboj.2011.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersen, I. K. N. et al. Upregulated PDK4 expression is a sensitive marker of increased fatty acid oxidation. Mitochondrion49, 97–110 (2019). 10.1016/j.mito.2019.07.009 [DOI] [PubMed] [Google Scholar]
- 70.Karlsson, M. et al. A single-cell type transcriptomics map of human tissues. Sci. Adv.7, eabh2169 (2021). [DOI] [PMC free article] [PubMed]
- 71.Cho, J. H. & Gregersen, P. K. Genomics and the multifactorial nature of human autoimmune disease. N. Engl. J. Med.365, 1612–1623 (2011). 10.1056/NEJMra1100030 [DOI] [PubMed] [Google Scholar]
- 72.Pearson, E. R. et al. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet362, 1275–1281 (2003). 10.1016/S0140-6736(03)14571-0 [DOI] [PubMed] [Google Scholar]
- 73.Wu, S., Zhu, W., Thompson, P. & Hannun, Y. A. Evaluating intrinsic and non-intrinsic cancer risk factors. Nat. Commun.9, 3490 (2018). 10.1038/s41467-018-05467-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahlqvist, E. et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diab. Endocrinol.6, 361–369 (2018). 10.1016/S2213-8587(18)30051-2 [DOI] [PubMed] [Google Scholar]
- 75.Zaghlool, S. B. et al. Metabolic and proteomic signatures of type 2 diabetes subtypes in an Arab population. Nat. Commun.13, 7121 (2022). 10.1038/s41467-022-34754-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pohjolainen, V. et al. Noncollagenous bone matrix proteins as a part of calcific aortic valve disease regulation. Hum. Pathol.39, 1695–1701 (2008). 10.1016/j.humpath.2008.04.015 [DOI] [PubMed] [Google Scholar]
- 77.Ress, C. et al. Circulating Wnt inhibitory factor 1 levels are associated with development of cardiovascular disease. Atherosclerosis273, 1–7 (2018). 10.1016/j.atherosclerosis.2018.03.045 [DOI] [PubMed] [Google Scholar]
- 78.Sun, Y. et al. Inhibition of fap promotes cardiac repair by stabilizing BNP. Circ. Res.132, 586–600 (2023). 10.1161/CIRCRESAHA.122.320781 [DOI] [PubMed] [Google Scholar]
- 79.Adeva-Andany, M. et al. Insulin resistance and glycine metabolism in humans. Amino Acids50, 11–27 (2018). 10.1007/s00726-017-2508-0 [DOI] [PubMed] [Google Scholar]
- 80.Rekhter, M. D. Collagen synthesis in atherosclerosis: too much and not enough. Cardiovasc. Res.41, 376–384 (1999). 10.1016/S0008-6363(98)00321-6 [DOI] [PubMed] [Google Scholar]
- 81.Rhee, E. P. et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab.18, 130–143 (2013). 10.1016/j.cmet.2013.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liso, A., Capitanio, N., Gerli, R. & Conese, M. From fever to immunity: a new role for IGFBP-6? J. Cell Mol. Med.22, 4588–4596 (2018). 10.1111/jcmm.13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Velenosi, T. J. et al. Untargeted metabolomics reveals N, N, N-trimethyl-L-alanyl-L-proline betaine (TMAP) as a novel biomarker of kidney function. Sci. Rep.9, 6831 (2019). 10.1038/s41598-019-42992-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Powell, D. R. et al. Insulin-like growth factor-binding protein-6 levels are elevated in serum of children with chronic renal failure: a report of the Southwest Pediatric Nephrology Study Group. J. Clin. Endocrinol. Metab.82, 2978–2984 (1997). [DOI] [PubMed] [Google Scholar]
- 86.Alicic, R. Z., Rooney, M. T. & Tuttle, K. R. Diabetic kidney disease: challenges, progress, and possibilities. Clin. J. Am. Soc. Nephrol.12, 2032–2045 (2017). 10.2215/CJN.11491116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rieckmann, J. C. et al. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol.18, 583–593 (2017). 10.1038/ni.3693 [DOI] [PubMed] [Google Scholar]
- 88.Mitchell, A. et al. LILRA5 is expressed by synovial tissue macrophages in rheumatoid arthritis, selectively induces pro-inflammatory cytokines and IL-10 and is regulated by TNF-alpha, IL-10 and IFN-gamma. Eur. J. Immunol.38, 3459–3473 (2008). 10.1002/eji.200838415 [DOI] [PubMed] [Google Scholar]
- 89.Oh, J. et al. Macrophage secretion of miR-106b-5p causes renin-dependent hypertension. Nat. Commun.11, 4798 (2020). 10.1038/s41467-020-18538-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hirayasu, K. & Arase, H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J. Hum. Genet60, 703–708 (2015). 10.1038/jhg.2015.64 [DOI] [PubMed] [Google Scholar]
- 91.Verschueren, E. et al. The immunoglobulin superfamily receptome defines cancer-relevant networks associated with clinical outcome. Cell182, 329–344 e319 (2020). 10.1016/j.cell.2020.06.007 [DOI] [PubMed] [Google Scholar]
- 92.Kissel, T., Toes, R. E. M., Huizinga, T. W. J. & Wuhrer, M. Glycobiology of rheumatic diseases. Nat. Rev. Rheumatol.19, 28–43 (2023). 10.1038/s41584-022-00867-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li, X. et al. Lactate metabolism in human health and disease. Signal Transduct. Target Ther.7, 305 (2022). 10.1038/s41392-022-01151-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mariappan, M. M., Feliers, D., Mummidi, S., Choudhury, G. G. & Kasinath, B. S. High glucose, high insulin, and their combination rapidly induce laminin-beta1 synthesis by regulation of mRNA translation in renal epithelial cells. Diabetes56, 476–485 (2007). 10.2337/db05-1334 [DOI] [PubMed] [Google Scholar]
- 95.Bomba, L. et al. Whole-exome sequencing identifies rare genetic variants associated with human plasma metabolites. Am. J. Hum. Genet109, 1038–1054 (2022). 10.1016/j.ajhg.2022.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kelly, R. S. et al. Metabolomic differences in lung function metrics: evidence from two cohorts. Thorax77, 919–928 (2022). 10.1136/thoraxjnl-2020-216639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu, Q. et al. Plasma metabolomics provides new insights into the relationship between metabolites and outcomes and left ventricular remodeling of coronary artery disease. Cell Biosci.12, 173 (2022). 10.1186/s13578-022-00863-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Fatima Cobre, A. et al. Diagnosis and prognosis of COVID-19 employing analysis of patients’ plasma and serum via LC-MS and machine learning. Comput Biol. Med.146, 105659 (2022). 10.1016/j.compbiomed.2022.105659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res.45, D362–D368 (2017). 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rusinova, I. et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res.41, D1040–D1046 (2013). 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang, C. R. et al. Prophylactic adenovirus-mediated human kallistatin gene therapy suppresses rat arthritis by inhibiting angiogenesis and inflammation. Arthritis Rheum.52, 1319–1324 (2005). 10.1002/art.20991 [DOI] [PubMed] [Google Scholar]
- 102.Raffield, L. M. et al. Comparison of proteomic assessment methods in multiple cohort studies. Proteomics20, e1900278 (2020). 10.1002/pmic.201900278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Richardson, C. R. et al. Management of Type 2 Diabetes Mellitus (Michigan Medicine University of Michigan, 2021). [PubMed]
- 104.Dai, C. et al. A proteomics sample metadata representation for multiomics integration and big data analysis. Nat. Commun.12, 5854 (2021). 10.1038/s41467-021-26111-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol.18, 83 (2017). 10.1186/s13059-017-1215-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Karczewski, K. J. & Snyder, M. P. Integrative Omics for Health and Disease Vol. 19, 299–310 (Nature Publishing Group, 2018). [DOI] [PMC free article] [PubMed]
- 107.Suhre, K. et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS ONE5, e13953–e13953 (2010). 10.1371/journal.pone.0013953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee, D. et al. CXCL5 secreted from macrophages during cold exposure mediates white adipose tissue browning. J. Lipid Res.62, 100117 (2021). 10.1016/j.jlr.2021.100117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sarmiento, U. et al. Morphologic and molecular changes induced by recombinant human leptin in the white and brown adipose tissues of C57BL/6 mice. Lab Invest77, 243–256 (1997). [PubMed] [Google Scholar]
- 110.Dodd, G. T. et al. Leptin and insulin act on POMC neurons to promote the browning of white fat. Cell160, 88–104 (2015). 10.1016/j.cell.2014.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bluher, M. Metabolically healthy obesity. Endocr. Rev.41, bnaa004 (2020). [DOI] [PMC free article] [PubMed]
- 112.Cooles, F. A. H. et al. Interferon-alpha-mediated therapeutic resistance in early rheumatoid arthritis implicates epigenetic reprogramming. Ann. Rheum. Dis.81, 1214–1223 (2022). 10.1136/annrheumdis-2022-222370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao, M. et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis.75, 1998–2006 (2016). 10.1136/annrheumdis-2015-208410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pan, H. et al. Integrative multi-omics database (iMOMdb) of Asian pregnant women. Hum. Mol. Genet31, 3051–3067 (2022). 10.1093/hmg/ddac079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saw, W. Y. et al. Establishing multiple omics baselines for three Southeast Asian populations in the Singapore Integrative Omics Study. Nat. Commun.8, 653 (2017). 10.1038/s41467-017-00413-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yousri, N. A., Albagha, O. M. E. & Hunt, S. C. Integrated epigenome, whole genome sequence and metabolome analyses identify novel multi-omics pathways in type 2 diabetes: a Middle Eastern study. BMC Med21, 347 (2023). 10.1186/s12916-023-03027-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE5, e15004 (2010). [DOI] [PMC free article] [PubMed]
- 118.Assarsson, E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE9, e95192 (2014). [DOI] [PMC free article] [PubMed]
- 119.Trbojević Akmačić, I. et al. High-throughput glycomics: optimization of sample preparation. Biochemistry80, 934–942 (2015). [DOI] [PubMed] [Google Scholar]
- 120.Wahl, A. et al. IgG glycosylation and DNA methylation are interconnected with smoking. Biochim. Biophys. Acta Gen. Subj.1862, 637–648 (2018). 10.1016/j.bbagen.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 121.Pučić, M. et al. High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol. Cell. Proteom.10, M111.010090 (2011). [DOI] [PMC free article] [PubMed]
- 122.Menni, C. et al. Glycosylation of immunoglobulin G: role of genetic and epigenetic influences. PLoS ONE8, e82558 (2013). [DOI] [PMC free article] [PubMed]
- 123.Dotz, V. et al. O- and N-glycosylation of serum immunoglobulin A is associated with IgA nephropathy and glomerular function. J. Am. Soc. Nephrol.32, 2455–2465 (2021). 10.1681/ASN.2020081208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Momcilovic, A. et al. Simultaneous immunoglobulin A and G glycopeptide profiling for high-throughput applications. Anal. Chem.92, 4518–4526 (2020). 10.1021/acs.analchem.9b05722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Evans, A. M., DeHaven, C. D., Barrett, T., Mitchell, M. & Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem.81, 6656–6667 (2009). 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- 126.Evans, A.M. High-resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics4, 1 (2014).
- 127.Römisch-Margl, W. et al. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics8, 133–142 (2012). 10.1007/s11306-011-0293-4 [DOI] [Google Scholar]
- 128.Illig, T. et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet.42, 137–141 (2010). 10.1038/ng.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Löfgren, L. et al. The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res.53, 1690–1700 (2012). 10.1194/jlr.D023036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Quell, J. D. et al. Characterization of bulk phosphatidylcholine compositions in human plasma using side-chain resolving lipidomics. Metabolites9, 109–109 (2019). 10.3390/metabo9060109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Budde, K. et al. Quality assurance in the pre-analytical phase of human urine samples by 1H NMR spectroscopy. Arch. Biochem. Biophys.589, 10–17 (2016). 10.1016/j.abb.2015.07.016 [DOI] [PubMed] [Google Scholar]
- 132.Soininen, P. et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst134, 1781–1785 (2009). 10.1039/b910205a [DOI] [PubMed] [Google Scholar]
- 133.Soininen, P., Kangas, A. J., Würtz, P., Suna, T. & Ala-Korpela, M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ. Cardiovasc. Genet.8, 192–206 (2015). 10.1161/CIRCGENETICS.114.000216 [DOI] [PubMed] [Google Scholar]
- 134.Do, K. T. et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics14, 128 (2018). 10.1007/s11306-018-1420-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kamat, M. A. et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics35, 4851–4853 (2019). 10.1093/bioinformatics/btz469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zaghlool, S. B. et al. Revealing the role of the human blood plasma proteome in obesity using genetic drivers. Nat. Commun.12, 1279 (2021). 10.1038/s41467-021-21542-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sharapov, S. Z. et al. Defining the genetic control of human blood plasma N-glycome using genome-wide association study. Hum. Mol. Genet28, 2062–2077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Belkadi, A. et al. Identification of genetic variants controlling RNA editing and their effect on RNA structure stabilization. Eur. J. Hum. Genet28, 1753–1762 (2020). 10.1038/s41431-020-0688-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Matias-Garcia, P. R. et al. Plasma proteomics of renal function: a transethnic meta-analysis and mendelian randomization study. J. Am. Soc. Nephrol.32, 1747–1763 (2021). 10.1681/ASN.2020071070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Buyukozkan, M., Benedetti, E. & Krumsiek, J. rox: a statistical model for regression with missing values. Metabolites13, 127 (2023). [DOI] [PMC free article] [PubMed]
- 142.Sekula, P. et al. From discovery to translation: characterization of c-mannosyltryptophan and pseudouridine as markers of kidney function. Sci. Rep.7, 17400 (2017). 10.1038/s41598-017-17107-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
The source of data generated by each platform is provided on Figshare (10.6084/m9.figshare.25975627.v2). The data can be downloaded as an excel file. Additionally, we are also providing data in.rda format for which we prepared R script that downloads the rda. data. The genetic data and methylation data access is not deposited because the informed consent given by the study participants does not cover posting of participant genotype and methylation data in public databases. Researcher affiliated with a research institution may request access to genetic data on an individual basis from the corresponding author (Karsten Suhre and Anna Halama, Weill Cornell Medicine—Qatar, Doha, Qatar). Access is subject to approval by the institutional research board of Weill Cornell Medicine—Qatar. The data sets deployed in this study were previously utilized as follows: Genomics data depicted as DNA14–16,20,137,138; Methylation data depicted as MET14,16; Transcriptomics data depicted as RNA139; Proteomics data measured on SOMA platform depicted as SOMA14–16,20,53,137,140; Glycomics data reflecting on total plasma N-glycosylation depicted as PGP14,53; Glycomics data reflecting on plasma IgG levels depicted as IgG14,138; Lipoproteomics data depicted as BRAIN14; The broad lipidomics data depicted as LD130; The targeted lipidomics data depicted as BM14,130; The untargeted metabolomics measured on HD4 platform depicted as HDF19,75,141; PM12–14,16–18,141,142; SM12–14,16–18,142; UM12–14,16–18,142; and CM16. Transcriptomics data covering microRNA depicted as miRNA, proteomics data measured on OLINK platform depicted as OLINK, and Glycomics data reflecting on plasma IgA levels depicted as IgA were not published before.
We are also providing access to the source code used to generate COmics Server. The source code and a docker image could be accessed via GitHub at https://github.com/karstensuhre/comics and referenced using 10.5281/zenodo.11487725.