Abstract
With climate change and anthropic influence on the coastal ecosystems, mangrove ecosystems are disappearing at an alarming rate. Accordingly, it becomes important to track, study, record and store the mangrove microbial community considering their ecological importance and potential for biotechnological applications. Here, we provide information on mangrove fungal community composition and diversity in mangrove ecosystems with different plant species and from various locations differing in relation to anthropic influences. We describe twelve newly assembled genomes, including four chromosomal-level genomes of fungal isolates from the mangrove ecosystems coupled with functional annotations. We envisage that these data will be of value for future studies including comparative genome analysis and large-scale temporal and/or spatial research to elucidate the potential mechanisms by which mangrove fungal communities assemble and evolve. We further anticipate that the genomes represent valuable resources for bioprospecting related to industrial or clinical uses.
Subject terms: Fungal genomics, Biodiversity
Background & Summary
Mangroves comprise a type of salt-tolerant plants predominantly occupying the intertidal regions along the tropical and subtropical coastal ecosystems, which are characterized by distinct physiochemical properties, such as high salinity, periodic immergence, fluctuating oxygen condition, strong wind, and tidal pressures1. Fungal communities have attracted considerable attention due to their significant contributions to the mangrove ecosystems, including but not limited to sustaining ecosystem stability2–4, interactions with mangrove plants5, involvement in biogeochemical cycles6, degradation of a wide range of persistent organic pollutants7,8, and for being used as a source for isolation of various bioactive compounds and enzymes with potential industrial or clinical applications2,9. However, mangrove areas worldwide are becoming smaller and more fragmented, and in 2007 it was predicted that the mangrove forests, along with their microbial diversity and potential benefits, might be lost within the next 100 years10. Many studies have examined the composition and diversity of mangrove fungal communities inhabiting different ecological inches with varying degrees of interaction with the associated plants11–13. These studies not only deepened our understanding of the enormous diversity of mangrove fungi, but also facilitated the study of community assembly mechanism, unveiling major forces driving the dynamics and evolution of mangrove fungal communities11,14. A previous study reported that stochastic processes, mainly dispersal limitation, were the major forces driving fungal community assembly in the sediment of mangrove ecosystems along the southeast coastal line of China14, while another study demonstrated that host selection was playing a more significant role in shaping the endophytic fungal communities compared to that of epiphytic communities11. However, our understanding of the mangrove fungal communities is far from comprehensive and integrated due to several challenges in exploring this unique ecosystem, such as high fluctuation of the coastal environments, relatively low biomass of fungal populations, potential biases in experimental processes and limitations of previous databases for annotation. Diverse basal fungal lineages have been detected only in recent years14. Accordingly, we still lack detailed information on the interactions between different mangrove plants and their corresponding fungal communities, and how and to what extent mangrove plants may shape fungal communities in their rhizosphere. Thus, a comprehensive study design with proper control of potential confounding factors would be necessary to elucidate the underlying mechanisms shaping mangrove fungal communities.
From the perspective of microbial genome evolution, microorganisms living in such harsh environments have evolved special physiological and metabolic strategies to cope with adverse conditions8,9. Many studies have reported on the discovery of valuable enzymes with extraordinary properties for industrial uses2,15 and bioactive compounds with antibiotic, anti-cancer, anticholesterolemic, or immunosuppressive functions with a potential for pharmaceutical applications2,9,16–18. For example, a highly modified fatty acid amide, showing cytotoxicity against the HCT-116 cell line and inhibitory effects against protein tyrosine kinases, was identified from a fungus Penicillium variabile, which was originally isolated from the mangrove ecosystem19. Biosynthetic gene clusters (BGCs) were also identified on the genome of this Penicillium variabile. However, the “uncultivability” of most fungal members has been an obstacle for the discovery of new bioactive metabolites. Reconstruction of fungal genomes directly from short sequencing reads may result in highly fragmented genomes20, and is not as straightforward and reliable as that for prokaryotes21, making fungal genomic data underutilized. A recently published study reported the isolation of more than 700 fungal strains of 149 species from the mangrove ecosystem by enrichment and FiChip in situ cultivation methods22, expanding the availability of culturable fungal strains from the mangrove ecosystem. However, genome information of these isolated strains is not available, impeding potential applications of these strains through genome mining.
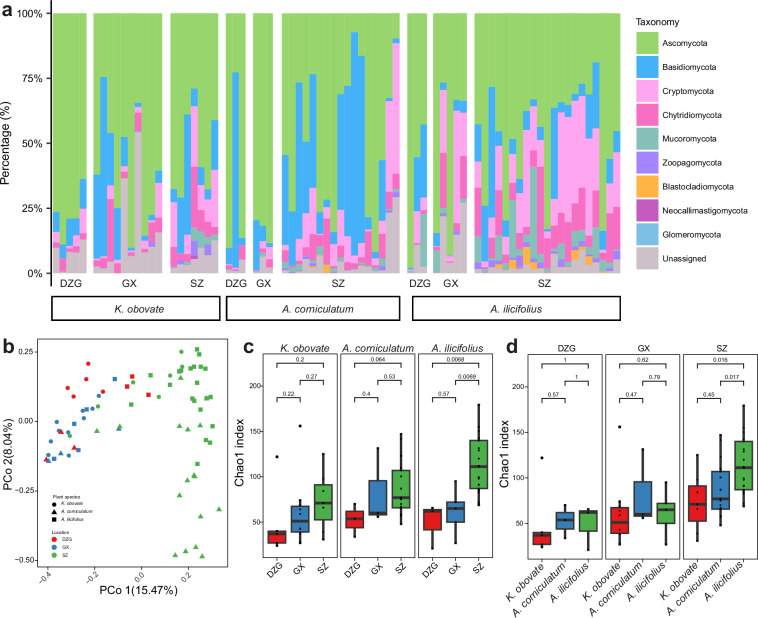
Here, in order to thoroughly interrogate how different locations and mangrove plant species may influence the structure and diversity of mangrove fungal communities, we collected rhizosphere sediment samples of three different mangrove plant species including Acanthus ilicifolius, Kandelia obovate, and Aegiceras corniculatum from three different mangrove ecosystems, i.e., Guangxi Mangrove Nature Reserve (GX), Shenzhen Futian Mangrove National Nature Reserve (SZ), and Dongzhaigang Mangrove National Nature Reserve (DZG). Relative abundance analysis based on 18S rDNA amplicon sequencing revealed that Ascomycota, Basidiomycota, Cryptomycota, Chytridiomycota and Mucoromycota were the top five abundant fungal populations (Fig. 1a), congruent with previously published studies13,14,23. Abundance profiles across different plant species and locations indicated that mangrove plants played a major role in selecting some of the dominant phyla, such as Mucoromycota, while locations seemed more determinant in selecting other phyla, such as Cryptomycota and Chytridiomycota, on top of the combined effects of both factors. Principle coordinate analysis (PCoA) based on Bray-Curtis dissimilarity matrix showed clear separation of fungal communities located in SZ from those of GX and DZG, regardless of the plant species, where relatively low anthropic activities were expected compared to SZ (Fig. 1b). Fungal community diversity based on Chao1 indices displayed an ascending trend of alpha diversity of samples from DZG, through GX to SZ (Fig. 1c). Interestingly, when we controlled the confounding factor of location, consistently higher averaged diversities were observed for communities from the rhizosphere of mangrove plant A. ilicifolius as compared to A. corniculatum and K. obovata plants (Fig. 1d), although the difference was not significant for DZG and GX samples.
Fig. 1.
Taxonomic composition and diversity distribution of the mangrove fungal community. (a) Relative abundance of the fungal community at the phylum level. (b) Principal Coordinate analysis (PCoA) of the fungal community based on the Bray-Curtis dissimilarity matrix (PERMANOVA test: 999 permutations, p = 0.001). (c,d) Comparison of Chao1 diversity indices between different locations of the same plant species (c) and between different plant species at the same location (d). The significance of differential alpha diversity was based on the Wilcoxon rank-sum test.
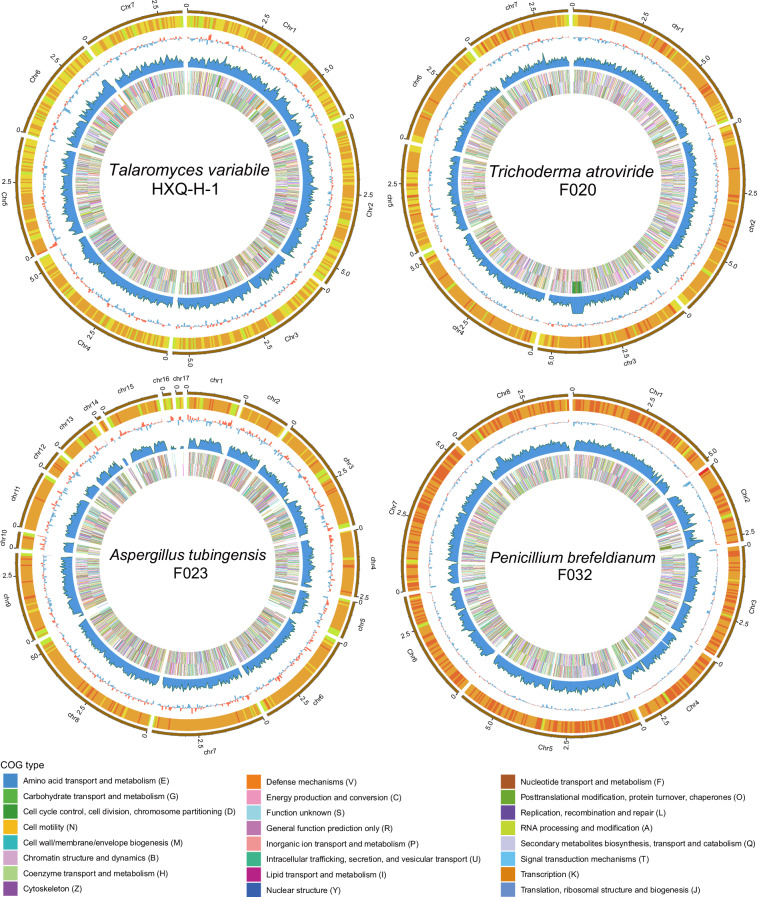
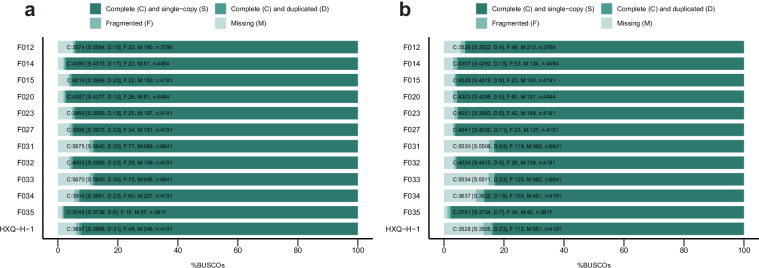
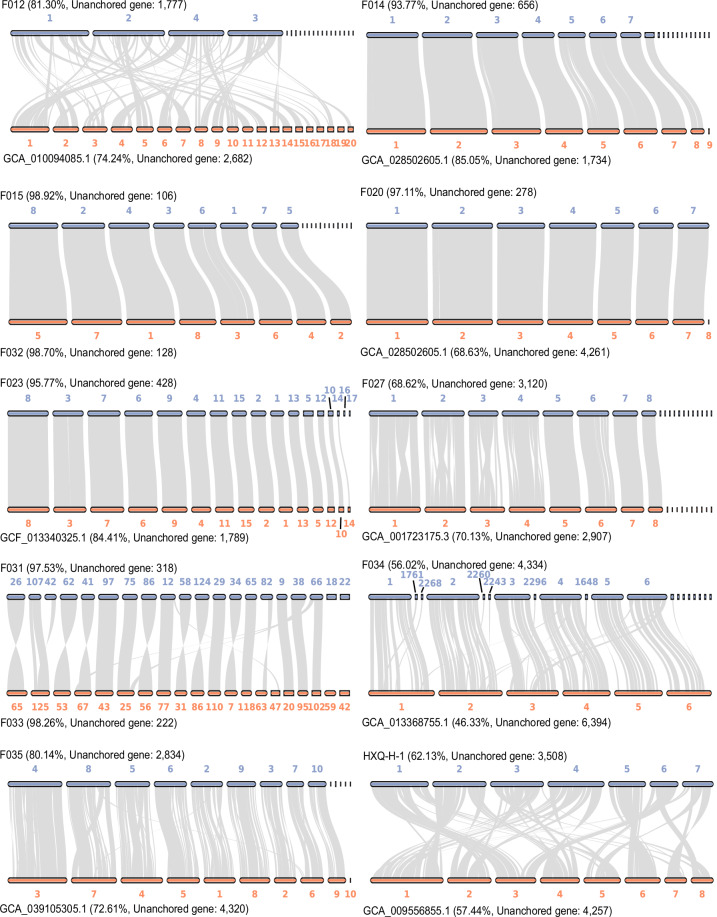
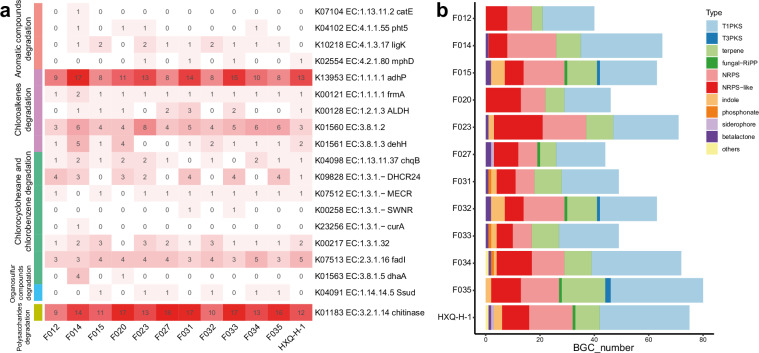
In previous studies, we have reported eight assembled genomes of fungal strains isolated from mangrove ecosystems24,25. However, assembly and scaffolding algorithms available at that time limited the genome quality that could be achieved, resulting in the low integrity and contiguity draft genomes previously. Here, in the current study, facilitated by the ever-developing sequencing technologies and bioinformatic algorithms, we report an updated set of eight mangrove fungal genomes with significantly higher quality and another four novel fungal genomes from mangrove environments, making a total of twelve high quality genomes available for future comparative genomics and genome mining studies (Table 1). In addition to using the most updated bioinformatic algorithms generating eight pseudo chromosomal-level genomes, we further refined four (Trichoderma atroviride F020, Aspergillus tubingensis F023, Penicillium brefeldianum F032, and Talaromyces variabile HXQ-H-1) of the twelve genomes using long-read sequencing technologies to improve the integrity of genomes to boost their biotechnological application20,26. Single-Tube Long Fragment Read (stLFR) sequencing strategy and Oxford Nanopore sequencing technology (ONT) were applied, and we obtained four chromosomal-level genomes of mangrove fungi with 7 to 17 chromosomes (Table 1 and Fig. 2). Both the updated genomes assembled from the whole genome sequencing (WGS) reads and the chromosomal-level genomes generated from the long-read fragment sequencing data showed high completeness based on Benchmarking Universal Single-Copy Orthologs (BUSCO) assessment with >88% of the conserved genes been covered in the genomes (Fig. 3a). The four chromosomal-level genomes contain fewer or no gaps with significantly longer scaffold N50 (>3 Mb) as compared to the other eight genomes26, although most of the updated genomes having a scaffold N50 size over 1 Mb (Table 1). For genomes with closely related reference genomes (same genus at least) available in public database, genome collinearity analysis between these newly assembled genomes and reference genomes of the same species (or genus where no reference of the same species is available) from public database revealed obvious difference between them, indicating these genomes may represent novel fungal species or sub-species (Fig. 4). Genomes (Penicillium brefeldianum strains F015 and F032, and the Roussoella solani strains F031 and F033) with no references from the same genus might represent novel fungal genomes at genus level from the mangrove ecosystem. Gene prediction of these genomes revealed a total of 9,200 to more than 14,200 genes encoded in each genome, representing an average coding density of 43.31% (Table 2). We performed BUSCO analysis to evaluate the completeness of the predicted genesets and the results indicated that more than 83% of the conserved genes were predicted (Fig. 3b). Only around half of these genes could be annotated against the KEGG database, indicating that there is still much of the mangrove fungi functional potential awaiting to be explored (Table 2). Considering fungal contribution to the carbon cycle in mangrove ecosystems, we also annotated the predicted genes against the KEGG and CAZyme databases and identified diverse and abundant genes involved in lignocellulosic biomass degradation (Table 2). Intriguingly, key genes in pathways of organic pollutants degradation, including genes involved in the degradation of aromatic compounds, chloroalkenes, chlorocyclohexane, and chlorobenzene were identified in these assembled fungal genomes (Table 2 and Fig. 5a). These findings are congruent with previous studies demonstrating the potential of mangrove fungi in bioremediation applications2. In addition, we identified a vast number of chitin degrading genes in all fungal genomes (average ~14 genes per genome) (Table 2 and Fig. 5a), which may be involved in the degradation of exogenous chitin and fungal cell wall27. Moreover, all the recovered fungal strains contain diverse BGCs in their genomes (Fig. 5b), indicating their potential to be used for genome mining of bioactive natural products for pharmaceutical usage. With their pure cultured strains being available with highly contiguous, or even chromosomal-level assembled genomes with detailed annotations, these fungi provide valuable resources for future research and biotechnological applications.
Table 1.
Detailed statistics information of 12 mangrove fungal genomes.
| Species | Plants | Sequencing Strategy | Scaffold Number | Scaffold Length (bp) | Scaffold N50(bp) | Contig Number | Contig Length (bp) | Contig N50 (bp) | GC (%) | ITS blast accesion | ITS Identity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Westerdykella dispersa F012 | K. obovata | WGS | 56 | 29,392,879 | 7,963,268 | 136 | 29,385,205 | 727,844 | 52.71 | NR_111187.1 | 98.88 |
| Trichoderma lixii F014 | K. obovata | WGS | 331 | 40,841,950 | 6,313,746 | 471 | 40,827,264 | 832,935 | 49.05 | NR_131264.1 | 99.68 |
| Penicillium brefeldianum F015 | A. ilicifolius | WGS | 46 | 32,622,330 | 3,839,637 | 302 | 32,596,404 | 329,369 | 51.53 | NR_138263.1 | 100 |
| Trichoderma atroviride F020 | A. corniculatum | WGS, ONT | 7 | 36,450,509 | 5,495,881 | 15 | 36,449,709 | 3,047,778 | 49.63 | NR_077207.1 | 98.98 |
| Aspergillus tubingensis F023 | A. ilicifolius | WGS, stLFR | 17 | 37,041,838 | 3,354,248 | 263 | 36,172,923 | 757,164 | 49.39 | NR_131293.1 | 100 |
| Penicillium raperi F027 | A. corniculatum | WGS | 194 | 35,597,292 | 4,229,135 | 298 | 35,587,169 | 754,312 | 50.47 | NR_121230.1 | 99.84 |
| Roussoella solani F031 | A. ilicifolius | WGS | 236 | 48,087,133 | 632,907 | 295 | 48,081,117 | 485,642 | 49.01 | NR_145198.1 | 99.49 |
| Penicillium brefeldianum F032 | A. ilicifolius | WGS, ONT | 8 | 33,894,595 | 3,974,671 | 8 | 33,894,595 | 3,974,671 | 51.4 | NR_138263.1 | 100 |
| Roussoella solani F033 | A. ilicifolius | WGS | 283 | 48,279,305 | 665,785 | 364 | 48,271,385 | 429,658 | 48.78 | NR_145198.1 | 99.49 |
| Talaromyces fuscoviridis F034 | K. obovata | WGS | 2,331 | 36,207,399 | 3,583,865 | 2,562 | 36,184,447 | 112,298 | 47.07 | NR_153227.1 | 99.46 |
| Arthrinium guizhouense F035 | A. ilicifolius | WGS | 14 | 45,875,886 | 5,326,493 | 144 | 45,863,246 | 612,443 | 54.33 | NR_157468.1 | 99.58 |
| Talaromyces variabile HXQ-H-1 | / | WGS, ONT | 7 | 33,780,117 | 5,544,665 | 25 | 33,772,867 | 2,127,226 | 46.93 | KT429657.1 | 100 |
Fig. 2.
Circos plots displaying the genomic features of four chromosomal-level fungal genomes. Circles from the outer to the inner are: GC-content heatmap (light-green: GC < 40%, yellow: 40% ≤ GC < 45%, orange: 45% ≤ GC < 50%, tangerine: 50% ≤ GC < 55%, red: GC ≥ 55%), GC skew histogram (red: G > C, blue: G < C), gene density in chromosomes, COG functional genes distribution (outside: plus strand, inside: minus strand), respectively. The legend is shown in the bottom.
Fig. 3.
BUSCO assessment scores of assembled genomes (a) and predicted genesets (b) for the twelve mangrove fungal isolates.
Fig. 4.
Genome collinearity analysis between the newly assembled genomes and their reference genomes. The gene collinearity percentage and the unanchored gene number for each genome are showed in the figure.
Table 2.
Overall description of protein coding gene prediction and functional annotation.
| Species | Gene annotation | KEGG annotation | CAZyme annotation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene number | Average gene length (bp) | Average coding length (bp) | Coding density (%) | Annotated genes | Annotated KOs | Chitinase | Organic pollutants degradation | All | AA | CBM | CE | GH | GT | PL | |
| Westerdykella dispersa F012 | 9,505 | 1,722.57 | 1,557.36 | 50.36 | 4,581 | 4,308 | 9 | 24 | 609 | 112 | 49 | 74 | 249 | 122 | 3 |
| Trichoderma lixii F014 | 10,521 | 1,767.56 | 1,538.66 | 39.64 | 5,155 | 4,436 | 14 | 49 | 623 | 81 | 43 | 61 | 305 | 126 | 7 |
| Penicillium brefeldianum F015 | 9,797 | 1,800.14 | 1,561.21 | 46.89 | 5,141 | 4,338 | 11 | 27 | 707 | 82 | 49 | 80 | 363 | 124 | 9 |
| Trichoderma atroviride F020 | 9,599 | 1,789.22 | 1,549.99 | 40.82 | 4,870 | 4,389 | 17 | 32 | 587 | 76 | 43 | 51 | 286 | 123 | 8 |
| Aspergillus tubingensis F023 | 10,116 | 1,780.99 | 1,528.97 | 41.76 | 5,438 | 4,368 | 13 | 39 | 700 | 112 | 46 | 83 | 301 | 147 | 11 |
| Penicillium raperi F027 | 9,942 | 1,742.65 | 1,530.33 | 42.74 | 5,190 | 4,345 | 18 | 25 | 748 | 65 | 54 | 93 | 402 | 123 | 11 |
| Roussoella solani F031 | 12,876 | 1,635.84 | 1,471.19 | 39.39 | 5,730 | 4,493 | 17 | 36 | 935 | 222 | 70 | 111 | 394 | 119 | 19 |
| Penicillium brefeldianum F032 | 9,819 | 1,839.11 | 1,557.17 | 45.11 | 5,138 | 4,345 | 10 | 27 | 707 | 82 | 48 | 81 | 364 | 123 | 9 |
| Roussoella solani F033 | 12,780 | 1,637.75 | 1,473.03 | 39.00 | 5,723 | 4,472 | 17 | 36 | 918 | 214 | 69 | 110 | 388 | 118 | 19 |
| Talaromyces fuscoviridis F034 | 9,847 | 1,825.83 | 1,579.03 | 42.94 | 5,111 | 4,179 | 13 | 30 | 656 | 81 | 35 | 79 | 337 | 120 | 4 |
| Arthrinium guizhouense F035 | 14,243 | 1,653.63 | 1,491.46 | 46.31 | 5,590 | 4,689 | 16 | 28 | 884 | 205 | 63 | 116 | 353 | 137 | 10 |
| Talaromyces variabile HXQ-H-1 | 9,240 | 1,988.05 | 1,573.12 | 43.03 | 4,939 | 4,144 | 12 | 31 | 653 | 88 | 33 | 74 | 331 | 120 | 7 |
Fig. 5.
Functional characterization of the twelve mangrove fungal genomes. (a) Key genes involved in the degradation of organic pollutants and chitin identified in the twelve genomes. (b) The biosynthesis gene clusters identified in the twelve genomes.
Methods
Rhizosphere soil sample collection
The rhizosphere soil samples of mangrove plants Acanthus ilicifolius, Kandelia obovate and Aegiceras corniculatum were collected in three Chinese southern provinces with different environmental conditions including Hainan, Guangxi, and Guangdong in the summer of 2017 or 2018, using the same method described in the previous studies28,29. A total of 11 samples were collected from Dongzhaigang Mangrove National Nature Reserve (DZG) (Hainan, China), which is the most diverse and sustainable mangrove ecosystem in China. In addition, a total of 18 samples were collected from Guangxi Mangrove Nature Reserve areas (including Beihai, Fangchenggang and Maoweihai Mangrove Nature Reserves) (GX) which are considered as pristine forests in mainland China. Additionally, 45 samples were collected from Shenzhen Futian Mangrove National Nature Reserve (SZ) (Guangdong, China) located in an inland urban area affected by industries and anthropogenic activities. The rhizosphere soil within 1–2 mm tightly around the roots was collected in sterile bags. All samples were stored in a refrigerator at a low temperature (0 °C) immediately after collection and then transferred to the laboratory and stored at −40 °C until used.
18S rDNA sequencing and bioinformatics process
Total DNA was extracted using the PowerSoil DNA isolation kit (Mobio Labs, USA) from ~0.5 g of each rhizosphere soil sample28. The 18S rRNA gene V4 region universal primers 528F-706R (528 F: GCGGTAATTCCAGCTCCAA, 706 R: AATCCRAGAATTTCACCTCT) were used for PCR amplification. The first-step PCR procedure was performed as follows: 95 °C for 3 min, followed by 14 cycles at 98 °C for 20 s, 58 °C for 30 s, and 72 °C for 30 s with a final extension at 72 °C for 5 min. The PCR products were diluted 5 times and then used as templates for the second-step PCR using the primers with sample barcode and the DNBSEQ sequencing adapter: 95 °C for 3 min, followed by 15 cycles at 98 °C for 20 s, 58 °C for 30 s, and 72 °C for 30 s with a final extension at 72 °C for 5 min. Finally, the two-step PCR products were verified and purified to construct the amplicon sequencing library and sequenced on a DNBSEQ-G400 platform (BGI-Qingdao, China) using 200 bp paired-end sequencing model30. Sequencing reads with adapter contaminations and low-quality were filtered out by SOAPnuke (v1.5.6) and then clean reads were merged into tags by FLASH (v1.2.11) as described previously30. The clean tags were clustered into operational taxonomic units (OTUs) using the USEARCH (v10.0.240) with sequence identity ≥ 0.97 and OTU abundance profile was generated31. After removing chimeric OTUs, the OTU representative sequences were taxonomically classified using the QIIME2 “feature-classifier” algorithm (v2023.7.0)32 against the Silva 18S rRNA database (v138). Only OTUs annotated to the “Fungi” domain were retained for downstream analysis. The EasyAmplicon pipeline (v1.2.0) was used to visualize the fungal taxonomy composition and calculate alpha diversity (Chao1 index) and beta diversity (Bray-Curtis dissimilarity)33.
Fungi isolation
A total of 11 fungal strains were isolated from the Dongzhaigang Mangrove National Nature Reserve (Hainan, China), including six strains (F015, F023, F031, F032, F033, and F035) isolated from one rhizosphere soil sample of A. ilicifolius, and another three strains (F012, F014 and F034) isolated from one rhizosphere soil sample of K. obovata, and the remaining two strains (F020 and F027) isolated from one rhizosphere soil sample of A. corniculatum. The strain HXQ-H-1 (currently named Talaromyces variabilis) was isolated from the mangrove rhizosphere soil in the Fujian province (Fuzhou, China)25. All strains were cultivated on Potato Dextrose Agar (PDA) media as described previously24,25 and deposited at Qingdao Key Laboratory of Marine Genomics (Qingdao), BGI-Qingdao, China.
Whole genome shotgun sequencing and assembly
Genomic DNA of each isolated fungal strain was extracted using the CTAB (cetyl trimethylammonium bromide) method and sheared into fragments between 100 to 800 bp in size using a E220 ultrasonicator (Covaris, Brighton, UK). The DNA fragments was used to construct the whole genome shotgun sequencing (WGS) library by the MGIEasy Micro DNA Library Preparation Kit (MGI, China) following the manufacturer’s instructions. The single-strand circular DNA libraries were sequenced on the BGISEQ-500 platform (BGI-Qingdao, China) which is based on DNA nanoball (DNB) and probe-anchor synthesis (cPAS) technology using 100 bp paired-end sequencing model. The WGS sequencing data was filtered and assembled to obtain the draft fungal genomes by SOAPnuke (v1.5.6) with parameters “-l 20 -q 0.2 -n 0.05 -d” and SPAdes (v3.10.1) with the k-mer parameter ranging from 33 to 83 and a step size of 10.
Long-Read sequencing and genome assembly
For the fungal strains F020, F032 and HXQ-H-1, the extracted DNA was used to construct the Oxford Nanopore Technologies (ONT) long fragment sequencing libraries using the Oxford Nanopore Ligation Sequencing Kit SQK-LSK109 and then sequenced on a GridION platform. The ONT sequencing reads with quality score > 7.0 and lengths longer than 8 Kb were used to assemble the genomic contigs using SMARTdenovo (v1.0) with default parameters34,35. Pilon (v1.23) was used to polish the assembled contigs three times to fix the INDEL and SNP errors with the BGISEQ-500 high-quality NGS data36.
For the strain F023, we used the single-tube long-fragment reads (stLFR) sequencing strategy to obtain the chromosome-level genome37. Briefly, MGIEasy stLFR Library Prep kit v1.1 (PN: 1000005622) was used to construct the stLFR library and then the library was sequenced on a BGISEQ-500 platform38. Based on de novo assembled result by SPAdes (v3.10.1), we further use the stLFR corresponding barcode information to improve the genomic contiguity and integrity38,39. After filtering the low-quality, PCR duplication and adapter contamination reads, the high-quality stLFR data was applied for chromosome-scale scaffolding based on the draft genome using SLR-superscaffolder (v0.9.0)40.
Genome scaffolding and chromosome construction
To improve the integrity and continuity of the draft genomes, we downloaded the chromosomal reference genomes from NCBI which were the same species or same genus as the five WGS assembled genomes (F012, F014, F027, F034, and F035) (Fig. 4). Then the draft genomes were mapped to their reference genomes by using RagTag (v2.1.0) with default parameters, respectively. For the Penicillium brefeldianum strains F015 and F032, and the Roussoella solani strains F031 and F033, since no reference genome of the same genus has been published, these two genomes were compared to each other and then we constructed the super-scaffolds by using RagTag (v2.1.0)42 with default parameters, respectively.
To obtain chromosome-level genomes based on the long-read sequencing assembled results, the HXQ-H-1 assembled contigs were anchored to chromosomes through HiC-Pro (v2.8.0_devel) and 3D-DNA pipelines (v170123) using previously generated Hi-C sequencing reads25, whereas the chromosomes of F020 were anchored to the public chromosomal-level genomes Trichoderma atroviride P1 (GCA_020647795.1) using JCVI (v1.0.9)41. The super-scaffolds of F023 were aligned to the reference genome Aspergillus tubingensis WU-2223L (GCA_013340325.1) using JCVI (v10.9) and short scaffolds without alignments and gene annotation were removed.
The genome collinearity analysis between our newly assembled genomes and reference genomes was conducted by using JCVI (v1.0.9). The links of all the syntenic gene blocks between the two genomes were plotted. The genes with C-score < 0.7 were marked as unanchored genes, and then the unanchored gene ratio was calculated to evaluate the difference between the two genomes.
Genome components prediction and functional annotation
Prediction of repetitive elements and gene sequences was performed as previously described24,25. Briefly, RepeatModeler (v1.0.8), Tandem Repeat Finder (v4.07) (v4.07), LTR_FINDER (v1.0.6) and RepeatMasker (v4.06) were used to predict repetitive elements in the twelve genomes. After marking the genomic repetitive sequences by bedtools (v2.29.2), GeneMark (v4.72) with default parameters, Augustus (v3.1) with parameters of “-ES -fungus -cores 10” was used for ab initio gene prediction. In addition, homologous protein sequences were downloaded from NCBI24,25 for homology-based gene prediction by Genewise (v2.4.1). To generate the final gene sequences of each genome, the ab initio predictions and the homolog gene prediction were meticulously merged, and redundant sequences were removed by EVidenceModeler43. Both the pseudo chromosomal-level genomes and chromosomal-level genomes were assessed using BUSCO (v5.2.2) with the closest evolutionary database (–auto_lineage model) to evaluate the genomic and gene set completeness44. Subsequently, protein sequences were subjected to functional annotation against the COG45, CAZyme (v8.0), and KEGG (v108.0) databases by NCBI BLAST + (v2.2.26), dbCAN2 (v2.0.11)46 and kofamscan (v1.3.0)47, respectively. Genes involved in the degradation of recalcitrant organic compounds were searched in the functional annotation results of the fungal genomes. To evaluate the production potential of natural bioactive compounds, the secondary metabolite BGCs were identified using antiSMASH (v6.1) with default parameters48.
Data Records
The 18S rDNA amplicon sequencing reads generated in this study are deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject number PRJNA111038249. The fungal OTU representative sequences, taxonomic annotation, abundance profile and diversity analysis results of 18S rDNA amplicon sequencing can be accessed through the Figshare repository50. All the 12 fungal genomes and their corresponding genomic sequencing data are deposited in the NCBI Genbank database under the BioProject number PRJNA111679451 with accession numbers JBEBND000000000-JBEBNO00000000052–63 and China National GeneBank Sequence Archive (CNSA) database with accession numbers CNP000048764 and CNP000091065. The genome and gene sequences are also available in the Figshare repository66.
Technical Validation
Raw sequencing data quality for both amplicon sequencing and WGS sequencing was checked using SOAPnuke (v1.5.6), and the results showed that the quality score of each dataset was above the required Q30 accuracy, suitable for further analyses. We performed pair-ends merging using FLASH (v1.2.11), and singleton and chimeric sequences were removed using USEARCH (v10.0.240). Fungal OTUs were identified using “feature-classifier” algorithm (v2023.7.0), resulting in 801 highly accurate OTUs. To confirm the quality of the 12 assembled fungal genomes and their predicted genesets, we selected assembled contigs with a length greater than 1000 bp for downstream analysis for each fungal genome. The NGS reads were mapped back to the final genomes using Bowtie2 (v2.2.5), and a fraction ranging from 92.19% to 97.93% of reads could be recalled, demonstrating the integrity of these genome assemblies. In addition, the quality of the fungal genomes was evaluated by BUSCO, and more than 85% conserved fungi genes were identified in all genomes, indicating high completeness and good quality of these genomes.
Acknowledgements
This work was supported by the grants of Joint Funds of the National Natural Science Foundation of China (Grant No. U2106228), National Natural Science Foundation of China (Grand No. 32100047), Major Scientific and Technological Innovation Projects of Qingdao West Coast New Area (Grand No. ZDKC-2022-03) and Thousands Marine Species Genome Sequencing Project of Qingdao Free Trade Zone Management Committee.
Author contributions
J.C. and Y.J. planned the study and wrote the manuscript. J.C., C.Z., L.P., L.L. Q.G. and C.S. performed the bioinformatics analysis and visualization. W.G. and T.C. collected the samples and conducted the amplicon sequencing and fungal genome sequencing. L.J., Z.Z., G.F., W.Z. and K.K. reviewed and revised the paper.
Code availability
No custom script was used to generate datasets in this study. The software with parameters and versions of all the bioinformatics tools used in this study are listed in the “Methods” section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jianwei Chen, Ling Peng, Changhao Zhou.
Contributor Information
Karsten Kristiansen, Email: kk@bio.ku.dk.
Yangyang Jia, Email: jiayangyang@genomics.cn.
References
- 1.He, Z. et al. Evolution of coastal forests based on a full set of mangrove genomes. Nat Ecol Evol6, 738–749, 10.1038/s41559-022-01744-9 (2022). 10.1038/s41559-022-01744-9 [DOI] [PubMed] [Google Scholar]
- 2.Jia, S. L., Chi, Z., Liu, G. L., Hu, Z. & Chi, Z. M. Fungi in mangrove ecosystems and their potential applications. Crit Rev Biotechnol40, 852–864, 10.1080/07388551.2020.1789063 (2020). 10.1080/07388551.2020.1789063 [DOI] [PubMed] [Google Scholar]
- 3.Alias, S. A., Zainuddin, N. & Jones, E. B. G. Biodiversity of marine fungi in Malaysian mangroves. Botanica Marina53, 10.1515/bot.2010.066 (2010).
- 4.Sridhar, K. R. Mangrove fungi in India. Current science86, 1586–1587 (2006). [Google Scholar]
- 5.Thatoi, H., Behera, B. C. & Mishra, R. R. Ecological role and biotechnological potential of mangrove fungi: a review. Mycology4, 54–71, 10.1080/21501203.2013.785448 (2013). 10.1080/21501203.2013.785448 [DOI] [Google Scholar]
- 6.Bohu, T. et al. The role of fungi in the biogeochemical cycling of supergene gold and satellite transition metals: A potential new exploration tool. Ore Geology Reviews140, 10.1016/j.oregeorev.2021.104595 (2022).
- 7.Behera, A. D., Chatterjee, S. & Das, S. Enzymatic degradation and metabolic pathway of phenanthrene by manglicolous filamentous fungus Trichoderma sp. CNSC-2. Microbiological research276, 127483, 10.1016/j.micres.2023.127483 (2023). 10.1016/j.micres.2023.127483 [DOI] [PubMed] [Google Scholar]
- 8.Deshmukh, R., Khardenavis, A. A. & Purohit, H. J. Diverse metabolic capacities of fungi for bioremediation. Indian J Microbiol56, 247–264, 10.1007/s12088-016-0584-6 (2016). 10.1007/s12088-016-0584-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ancheeva, E., Daletos, G. & Proksch, P. Lead compounds from mangrove-associated microorganisms. Mar Drugs16, 10.3390/md16090319 (2018). [DOI] [PMC free article] [PubMed]
- 10.Duke, N. C. et al. A world without mangroves? Science317, 41–42, 10.1126/science.317.5834.41b (2007). 10.1126/science.317.5834.41b [DOI] [PubMed] [Google Scholar]
- 11.Yao, H. et al. Phyllosphere epiphytic and endophytic fungal community and network structures differ in a tropical mangrove ecosystem. Microbiome7, 57, 10.1186/s40168-019-0671-0 (2019). 10.1186/s40168-019-0671-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang, W. et al. Diversity, function and assembly of mangrove root-associated microbial communities at a continuous fine-scale. NPJ Biofilms Microbiomes6, 52, 10.1038/s41522-020-00164-6 (2020). 10.1038/s41522-020-00164-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devadatha, B. et al. Occurrence and geographical distribution of mangrove fungi. Fungal Diversity106, 137–227, 10.1007/s13225-020-00468-0 (2021). 10.1007/s13225-020-00468-0 [DOI] [Google Scholar]
- 14.Zhang, Z. F., Pan, Y. P., Liu, Y. & Li, M. High-level diversity of basal fungal lineages and the control of fungal community assembly by stochastic processes in mangrove sediments. Applied and environmental microbiology87, e0092821, 10.1128/AEM.00928-21 (2021). 10.1128/AEM.00928-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, S., Cai, R., Liu, Z., Cui, H. & She, Z. Secondary metabolites from mangrove-associated fungi: source, chemistry and bioactivities. Nat Prod Rep39, 560–595, 10.1039/d1np00041a (2022). 10.1039/d1np00041a [DOI] [PubMed] [Google Scholar]
- 16.Cadamuro, R. D. et al. Bioactive compounds from mangrove endophytic fungus and their uses for microorganism control. J Fungi (Basel)7, 10.3390/jof7060455 (2021). [DOI] [PMC free article] [PubMed]
- 17.Xu, Z.-Y. et al. Secondary metabolites produced by mangrove endophytic fungus Aspergillus fumigatus HQD24 with immunosuppressive activity. Biochemical Systematics and Ecology93, 10.1016/j.bse.2020.104166 (2020).
- 18.Gozari, M., Alborz, M., El-Seedi, H. R. & Jassbi, A. R. Chemistry, biosynthesis and biological activity of terpenoids and meroterpenoids in bacteria and fungi isolated from different marine habitats. Eur J Med Chem210, 112957, 10.1016/j.ejmech.2020.112957 (2021). 10.1016/j.ejmech.2020.112957 [DOI] [PubMed] [Google Scholar]
- 19.He, X. et al. Varitatin A, a Highly modified fatty acid amide from Penicillium variabile cultured with a DNA methyltransferase Inhibitor. J Nat Prod78, 2841–2845, 10.1021/acs.jnatprod.5b00742 (2015). 10.1021/acs.jnatprod.5b00742 [DOI] [PubMed] [Google Scholar]
- 20.Voorhies, M. et al. Chromosome-level genome assembly of a human fungal pathogen reveals synteny among geographically distinct species. mBio13, e0257421, 10.1128/mbio.02574-21 (2022). 10.1128/mbio.02574-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Cobas, A. E., Gomez-Valero, L. & Buchrieser, C. Metagenomic approaches in microbial ecology: an update on whole-genome and marker gene sequencing analyses. Microb Genom6, 10.1099/mgen.0.000409 (2020). [DOI] [PMC free article] [PubMed]
- 22.Li, M. et al. Application of culturomics in fungal isolation from mangrove sediments. Microbiome11, 272, 10.1186/s40168-023-01708-6 (2023). 10.1186/s40168-023-01708-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pham, T. T., Dinh, K. V. & Nguyen, V. D. Biodiversity and enzyme activity of marine fungi with 28 new records from the tropical coastal ecosystems in Vietnam. Mycobiology49, 559–581, 10.1080/12298093.2021.2008103 (2021). 10.1080/12298093.2021.2008103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi, C. et al. Draft genomes and comparative analysis of seven mangrove rhizosphere-associated fungi isolated from kandelia obovata and acanthus ilicifolius. Frontiers in Fungal Biology2, 10.3389/ffunb.2021.626904 (2021). [DOI] [PMC free article] [PubMed]
- 25.Peng, L. et al. Chromosome-level comprehensive genome of mangrove sediment-derived fungus Penicillium variabile HXQ-H-1. J Fungi (Basel)6, 10.3390/jof6010007 (2019). [DOI] [PMC free article] [PubMed]
- 26.Naranjo-Ortiz, M. A. & Gabaldon, T. Fungal evolution: cellular, genomic and metabolic complexity. Biol Rev Camb Philos Soc95, 1198–1232, 10.1111/brv.12605 (2020). 10.1111/brv.12605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartl, L., Zach, S. & Seidl-Seiboth, V. Fungal chitinases: diversity, mechanistic properties and biotechnological potential. Applied microbiology and biotechnology93, 533–543, 10.1007/s00253-011-3723-3 (2012). 10.1007/s00253-011-3723-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao, S. et al. Deciphering the microbial taxonomy and functionality of two diverse mangrove ecosystems and their potential abilities to produce bioactive compounds. mSystems5, 10.1128/mSystems.00851-19 (2020). [DOI] [PMC free article] [PubMed]
- 29.Wang, Y. et al. Dynamics of rhizosphere microbial structure and function associated with the biennial bearing of moso bamboo. J Environ Manage351, 119977, 10.1016/j.jenvman.2023.119977 (2024). 10.1016/j.jenvman.2023.119977 [DOI] [PubMed] [Google Scholar]
- 30.Jia, Y. et al. Sequencing introduced false positive rare taxa lead to biased microbial community diversity, assembly, and interaction interpretation in amplicon studies. Environ Microbiome17, 43, 10.1186/s40793-022-00436-y (2022). 10.1186/s40793-022-00436-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, G., Li, T., Zhu, X., Zhang, X. & Wang, J. An independent evaluation in a CRC patient cohort of microbiome 16S rRNA sequence analysis methods: OTU clustering, DADA2, and Deblur. Front Microbiol14, 1178744, 10.3389/fmicb.2023.1178744 (2023). 10.3389/fmicb.2023.1178744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome6, 90, 10.1186/s40168-018-0470-z (2018). 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Y. X. et al. EasyAmplicon: An easy‐to‐use, open‐source, reproducible, and community‐based pipeline for amplicon data analysis in microbiome research. iMeta2, 10.1002/imt2.83 (2023). [DOI] [PMC free article] [PubMed]
- 34.Liu, H., Wu, S., Li, A. & Ruan, J. SMARTdenovo: a de novo assembler using long noisy reads. GigaByte2021, gigabyte15, 10.46471/gigabyte.15 (2021). 10.46471/gigabyte.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res27, 722–736, 10.1101/gr.215087.116 (2017). 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, J. et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS one9, e112963, 10.1371/journal.pone.0112963 (2014). 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, O. et al. Efficient and unique cobarcoding of second-generation sequencing reads from long DNA molecules enabling cost-effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res29, 798–808, 10.1101/gr.245126.118 (2019). 10.1101/gr.245126.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, Z. et al. Comparison of different sequencing strategies for assembling chromosome-level genomes of extremophiles with variable GC content. iScience24, 102219, 10.1016/j.isci.2021.102219 (2021). 10.1016/j.isci.2021.102219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi, Y. et al. MetaTrass: A high‐quality metagenome assembler of the human gut microbiome by cobarcoding sequencing reads. iMeta, 10.1002/imt2.46 (2022). [DOI] [PMC free article] [PubMed]
- 40.Guo, L. et al. SLR-superscaffolder: a de novo scaffolding tool for synthetic long reads using a top-to-bottom scheme. BMC Bioinformatics22, 158, 10.1186/s12859-021-04081-z (2021). 10.1186/s12859-021-04081-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang, H. et al. JCVI: A versatile toolkit for comparative genomics analysis. iMeta10.1002/imt2.211 (2024). 10.1002/imt2.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alonge, M. et al. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol23, 258, 10.1186/s13059-022-02823-7 (2022). 10.1186/s13059-022-02823-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biology9, R7, 10.1186/gb-2008-9-1-r7 (2008). 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manni, M., Berkeley, M. R., Seppey, M., Simao, F. A. & Zdobnov, E. M. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Molecular biology and evolution38, 4647–4654, 10.1093/molbev/msab199 (2021). 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatusov, R. L. et al. The COG database: an updated version includes eukaryotes. BMC bioinformatics4, 41–41, 10.1186/1471-2105-4-41 (2003). 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, H. et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Research46, W95–W101, 10.1093/nar/gky418 (2018). 10.1093/nar/gky418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aramaki, T. et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics36, 2251–2252, 10.1093/bioinformatics/btz859 (2020). 10.1093/bioinformatics/btz859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medema, M. H. et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Research39, W339–W346, 10.1093/nar/gkr466 (2011). 10.1093/nar/gkr466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.NCBI Sequence Read Archive.https://identifiers.org/ncbi/insdc.sra:SRP507817 (2024).
- 50.Chen, J. Mangrove fungal community. figshare10.6084/m9.figshare.25060154 (2024). 10.6084/m9.figshare.25060154 [DOI]
- 51.NCBI Bioproject.https://identifiers.org/ncbi/bioproject:PRJNA1116794 (2024).
- 52.Chen, J. et al. Talaromyces variabilis strain HXQ-H-1, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBND000000000 (2024).
- 53.Chen, J. et al. Apiospora guizhouensis strain F035, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNE000000000 (2024).
- 54.Chen, J. et al. Talaromyces fuscoviridis strain F034, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNF000000000 (2024).
- 55.Chen, J. et al. Neoroussoella solani strain F033, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNG000000000 (2024).
- 56.Chen, J. et al. Penicillium brefeldianum strain F032, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNH000000000 (2024).
- 57.Chen, J. et al. Neoroussoella solani strain F031, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNI000000000 (2024).
- 58.Chen, J. et al. Penicillium raperi strain F027, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNJ000000000 (2024).
- 59.Chen, J. et al. Aspergillus tubingensis strain F023, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNK000000000 (2024).
- 60.Chen, J. et al. Trichoderma atroviride strain F020, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNL000000000 (2024).
- 61.Chen, J. et al. Penicillium brefeldianum strain F015, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNM000000000 (2024).
- 62.Chen, J. et al. Trichoderma lixii strain F014, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNN000000000 (2024).
- 63.Chen, J. et al. Westerdykella dispersa strain F012, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNO000000000 (2024).
- 64.China National GeneBank Sequence Archive.https://db.cngb.org/search/project/CNP0000487/ (2023).
- 65.China National GeneBank Sequence Archive.https://db.cngb.org/search/project/CNP0000910/ (2024).
- 66.Chen, J. Mangrove fungal genomes. figshare10.6084/m9.figshare.25053398 (2024). 10.6084/m9.figshare.25053398 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI Sequence Read Archive.https://identifiers.org/ncbi/insdc.sra:SRP507817 (2024).
- Chen, J. Mangrove fungal community. figshare10.6084/m9.figshare.25060154 (2024). 10.6084/m9.figshare.25060154 [DOI]
- NCBI Bioproject.https://identifiers.org/ncbi/bioproject:PRJNA1116794 (2024).
- Chen, J. et al. Talaromyces variabilis strain HXQ-H-1, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBND000000000 (2024).
- Chen, J. et al. Apiospora guizhouensis strain F035, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNE000000000 (2024).
- Chen, J. et al. Talaromyces fuscoviridis strain F034, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNF000000000 (2024).
- Chen, J. et al. Neoroussoella solani strain F033, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNG000000000 (2024).
- Chen, J. et al. Penicillium brefeldianum strain F032, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNH000000000 (2024).
- Chen, J. et al. Neoroussoella solani strain F031, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNI000000000 (2024).
- Chen, J. et al. Penicillium raperi strain F027, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNJ000000000 (2024).
- Chen, J. et al. Aspergillus tubingensis strain F023, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNK000000000 (2024).
- Chen, J. et al. Trichoderma atroviride strain F020, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNL000000000 (2024).
- Chen, J. et al. Penicillium brefeldianum strain F015, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNM000000000 (2024).
- Chen, J. et al. Trichoderma lixii strain F014, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNN000000000 (2024).
- Chen, J. et al. Westerdykella dispersa strain F012, whole genome shotgun sequencing project. GenBankhttps://identifiers.org/ncbi/insdc:JBEBNO000000000 (2024).
- China National GeneBank Sequence Archive.https://db.cngb.org/search/project/CNP0000487/ (2023).
- China National GeneBank Sequence Archive.https://db.cngb.org/search/project/CNP0000910/ (2024).
- Chen, J. Mangrove fungal genomes. figshare10.6084/m9.figshare.25053398 (2024). 10.6084/m9.figshare.25053398 [DOI]
Data Availability Statement
No custom script was used to generate datasets in this study. The software with parameters and versions of all the bioinformatics tools used in this study are listed in the “Methods” section.