Abstract
Dietary restriction (DR), the process of decreasing overall food consumption over an extended period of time, has been shown to increase longevity across evolutionarily diverse species and delay the onset of age-associated diseases in humans. In Caenorhabditis elegans, the Myc-family transcription factors (TFs) MXL-2 (Mlx) and MML-1 (MondoA/ChREBP), which function as obligate heterodimers, and PHA-4 (orthologous to FOXA) are both necessary for the full physiological benefits of DR. However, the adaptive transcriptional response to DR and the role of MML-1::MXL-2 and PHA-4 remains elusive. We identified the transcriptional signature of C. elegans DR, using the eat-2 genetic model, and demonstrate broad changes in metabolic gene expression in eat-2 DR animals, which requires both mxl-2 and pha-4. While the requirement for these factors in DR gene expression overlaps, we found many of the DR genes exhibit an opposing change in relative gene expression in eat-2;mxl-2 animals compared to wild-type, which was not observed in eat-2 animals with pha-4 loss. Surprisingly, we discovered more than 2000 genes synthetically dysregulated in eat-2;mxl-2, out of which the promoters of down-regulated genes were substantially enriched for PQM-1 and ELT-1/3 GATA TF binding motifs. We further show functional deficiencies of the mxl-2 loss in DR outside of lifespan, as eat-2;mxl-2 animals exhibit substantially smaller brood sizes and lay a proportion of dead eggs, indicating that MML-1::MXL-2 has a role in maintaining the balance between resource allocation to the soma and to reproduction under conditions of chronic food scarcity. While eat-2 animals do not show a significantly different metabolic rate compared to wild-type, we also find that loss of mxl-2 in DR does not affect the rate of oxygen consumption in young animals. The gene expression signature of eat-2 mutant animals is consistent with optimization of energy utilization and resource allocation, rather than induction of canonical gene expression changes associated with acute metabolic stress, such as induction of autophagy after TORC1 inhibition. Consistently, eat-2 animals are not substantially resistant to stress, providing further support to the idea that chronic DR may benefit healthspan and lifespan through efficient use of limited resources rather than broad upregulation of stress responses, and also indicates that MML-1::MXL-2 and PHA-4 may have distinct roles in promotion of benefits in response to different pro-longevity stimuli.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-024-01197-x.
Keywords: Dietary restriction, Myc-family, Transcription factors, Gene expression
Introduction
Dietary restriction (DR) is one of few known interventions that results in delayed onset of aging-associated disease and improves longevity in a broad range of evolutionarily distinct animals ranging from invertebrates to monkeys, and has been found to improve health biomarkers in humans [54, 97, 98, 125, 67, 103, 104, 173, 188, 136, 83]. DR is a broad term encompassing a growing number of treatment paradigms, including chronic restriction of overall energy intake or reduction of specific nutrients (e.g., methionine, carbohydrates) and intermittent fasting: scheduled short-term food withdrawal as with alternate-day feeding or time-restricted feeding [47, 68, 172]. Recent clinical trials in humans have found significant improvement in biomarkers of cardiovascular health in both short term alternate-day fasting [175] and reduced average energy intake reduction over several years [83, 103]. Although the benefits of DR on organismal health are widely conserved, the full mechanistic picture of how various types of DR influences physiological decline during aging remains far from clear, due to complex interactions between genetics, diet, and environment that influence aging [122].
C. elegans is a preeminent model for aging studies, in part due to its normally short lifespan, fast generation time, and wealth of genetic tools. Working with an intact metazoan model enables identification of cell non-autonomous interactions across distinct tissues, including those coordinated by a nervous system [19]. Adult wild-type hermaphrodite animals have 959 somatic cells that arise from an invariant and well-characterized lineage, without contribution of stem or progenitor cells to the maintenance of adult somatic tissues [177]. This less-complex environment allows focus on elucidating genetic determinants of health and maintenance of differentiated somatic cells that act within an integrated biological system, without interference from regenerative cellular quality control mechanisms that preserve tissue homeostasis. Notably, the discovery of long-lived genetic mutants of C. elegans combined with the development of feeding-based RNAi has led to uncovering more than 1000 genes that influence lifespan when perturbed, which we refer to as “gerogenes” [11, 33, 71, 73, 89, 90, 99, 113, 135, 153, 183].
Many gerogenes are nodes in networks that connect nutrient-associated signals to organismal longevity [42, 59, 60, 73, 87, 138]. For example, loss-of-function mutations in daf-2, orthologous to the insulin and IGF1 receptors, extends longevity through activation of the DAF-16 transcription factor (FOXO homolog) [99, 118]. Increasing DAF-16 expression is sufficient to extend longevity and improve survival to a myriad of cellular stresses [134, 135]. Studies into genetic variants associated with healthy aging in human populations have identified polymorphisms over-represented in human centenarians, such as FOXO3, a homolog of daf-16 [191]. In addition to Insulin and IGF1 signaling (IIS), most gerogenes have evolutionarily conserved roles in the control of longevity including: the Target of Rapamycin (TOR) complexes, which regulate the balance between anabolism and catabolism based on amino acid and carbohydrate availability [48, 87, 110, 171], and AMP kinase (AMPK), which responds to insufficient ATP levels [5, 68, 69, 190]. Overall, nearly 75% of gerogenes in C. elegans show considerable homology with mammals and humans, to the extent of functional substitution in some cases [33, 76, 100, 179, 194]. However, the majority of gerogenes remain under-studied and have not been placed in an interaction network or pathway [33].
The eat-2 genetic mutant is a common DR model in C. elegans; eat-2 encodes a nicotinic acetylcholine receptor expressed in neuromuscular junctions of two pharyngeal muscle cells that contributes to control of pharyngeal pumping rate, thereby influencing food consumption [128]. Reduction of eat-2 function results in decreased feeding rates and increased longevity, which provides a genetic model for studying the molecular and cellular basis of DR [108, 128, 150]. A number of transcriptional effectors of DR have been identified using this model, most notably the FOXA homolog PHA-4 [143], which is specific to DR, whereas the FOXO homolog DAF-16 is not required for eat-2 longevity [108].
The MYC network of basic helix-loop-helix leucine zipper (bHLH Zip) transcription factors is perhaps best known for its namesake, MYC, which is a key regulator of metabolism, cell growth, and survival. MYC is also a well-characterized proto-oncogene that contributes to tumorigenic transformation in a range of cancers [29, 167]. MYC functionally regulates transcription as part of a heterodimeric complex with MAX. MAX binding regulates MYC activity, as MAX competitively binds to MXD and MNT factors, which in turn also compete for binding to MLX. These interactions further serve to regulate the activity of an additional set of metabolism-regulating transcription factors, MLXIP and MLXIPL (formerly known as MONDOA and MONDOB/CHREBP, respectively) [29]. All of these proteins form heterodimers at enhancer-box (E-box) sites [3, 41, 167]. The general properties of the MYC network are broadly conserved through metazoan evolution, although not all animals have a representative homolog for each of the core proteins found in vertebrates [126]. C. elegans in particular does not have a canonical MYC or MNT homolog, but does possess homologs for the other network members [126].
The C. elegans MYC network, and the MML-1::MXL-2 complex (mammalian MLXIP::MLX) in particular, increasingly appears to be a convergence point in multiple interaction networks involved in longevity and homeostatic maintenance. All of the Myc-family of transcription factors have been implicated in C. elegans longevity (mdl-1, mxl-1, mml-1, mxl-2, mxl-3) [88, 137, 140]. MDL-1 (MXD) heterodimerizes with MXL-1 (MAX); loss of either mdl-1 or mxl-1 increases longevity and is dependent upon the MML-1::MXL-2 complex [88, 145]. Interestingly, in humans, loss of a homolog of MML-1- MLXIPL- has been implicated in Williams-Beuren syndrome, a disease with progeric symptoms [22], and reduced MYC expression increases longevity in mice [79]. We previously demonstrated that the C. elegans MML-1::MXL-2 complex was necessary for the extended longevity conferred by chronic DR (eat-2) or reduced IIS; conversely, loss of mdl-1 or mxl-1 does not further increase the longevity of eat-2 or IIS mutants [88, 153]. mml-1 and mxl-2 are also required for the extended longevity conferred via cell non-autonomous signals from the germline, TORC1 inhibition, and increased activity of homeodomain-interacting protein kinase hpk-1, a neuronal transcriptional regulator of autophagy and longevity induced in response to TORC1 inhibition [39, 111, 137, 164]. Given our previous finding that MML-1::MXL-2 is required for the longevity of eat-2(ad465) animals [88], the importance of this complex in the integration of multiple pro-longevity signals, and the lack of known targets in the context of DR, we sought to define the adaptive transcriptional response to DR and the role of MML-1::MXL-2 therein [88, 137].
Materials and methods
C. elegans strains
N2 Bristol was used as the wild-type strain, and all other strains were back-crossed at least six times to N2 before proceeding. The mxl-2(tm1516) strain was created by and obtained from Dr. Shohei Mitani with the Japanese Bioresource Project. Strains were confirmed to be WT for fln-2(OT611), a background longevity mutation found in some commonly used wild-type strains [201].
The following C. elegans strains were utilized for this work:
| Genotype | Strain |
|---|---|
| wild-type | N2 Bristol (CGCM) |
| daf-2(e1370) | AVS1 |
| eat-2(ad465) | AVS518 |
| mxl-2(tm1516) | AVS385 |
| eat-2(ad465); mxl-2(tm1516) | AVS273 |
C. elegans culture
Nematodes were maintained using standard laboratory culture techniques [19]. Animals were normally maintained on plates with E. coli OP50-1 as a food source. Animals for assays were raised on the RNAi-compatible E. coli strain HT115(DE3), transformed with empty vector (EV) plasmid or plasmid with an insert sequence complementary to the given gene of interest for feeding-based RNAi [94, 183]. For assay plates, RNAi clones were grown overnight with shaking in LB and seeded onto agar plates containing 5 mM isopropylthiogalatoside (IPTG) and allowed to induce dsRNA expression overnight at room temperature; additional details can be found in [34].
Animal preparation and RNA isolation for RNA-sequencing
Plates prepared for the RNA-Seq experiment were 10 cm RNAi plates seeded with 1 mL of 20x concentrated bacteria from an overnight culture. Plates were allowed to dry for 1–2 days at room temperature, also providing time for the induction of dsRNA production before adding animals. Approximately 3000 L1 animals were then added, split across multiple plates to prevent starvation and overcrowding, and kept at 16 °C until the L4 stage. When animals reached L4, 600 µL of 4 mg/mL 5-fluorodeoxyuridine (FUdR) was added to each plate to prevent production of progeny, and the plates were transferred to 20 °C (“day 0”). Developmental stage was assessed separately for each strain to account for shifts in developmental timing.
At adult day 2, animals were collected in M9, washed twice in M9, finally washed with DEPC-treated RNase-free water. Approximately two times the volume of the animal pellet of TRIzol® reagent was added to each animal preparation, followed by brief mixing and freezing overnight at − 80 °C. Tubes were then allowed to partially thaw and were vortexed 5 min to assist with disrupting the cuticle. Each prep was transferred to a new tube, and 200 µL of chloroform per 1 mL of TRIzol was added, followed by vortexing for 20 s, and allowed to settle at room temperature for 10 min. Supernatants were transferred to new tubes, and an equal amount of 100% ethanol was added and mixed before proceeding to column purification with the Qiagen RNAeasy Mini kit (catalog 74104) according to the manufacturer’s instructions. Samples were eluted with 30–50 µL of DEPC-treated water and checked for initial concentration and quality with a Nanodrop ND-1000.
RNA-sequencing
Isolated RNA was provided to the University of Rochester Genomics Research Center for library preparation and sequencing. Prior to library preparation, an Agilent 2100 Bioanalyzer was used to confirm the RNA integrity of all samples. Libraries for sequencing were prepared with the Illumina TruSeq Stranded mRNA kit, according to the manufacturer’s instructions. Quality of the resulting libraries was checked again with a Bioanalyzer prior to sequencing to ensure integrity of the input material. Sequencing was performed on an Illumina HiSeq2500 v4, yielding an average of approximately 31 million single-ended 100 bp reads per sample. Quality of the output was summarized with FastQC [4], and reads were trimmed and filtered with Trimmomatic [16]. After filtering out low-quality reads, an average of 30.2 million reads per sample remained and were used for the rest of the analysis.
RNA-sequencing data pre-processing
Samples were aligned to the C. elegans genome assembly WBcel235 with STAR 2.4.2a [44] using gene annotation from Ensembl (version 82) [37]. An average of 94.7% of reads per sample were uniquely mapped to the genome. These alignments were used as input to RSEM (version 1.2.23) for transcript-level quantification [115] and featureCounts (version 1.4.6-p5) for gene-level counts [116]. Ambiguous or multi-mapping reads, comprising an average of approximately 5% of the reads per sample, were not included in the gene-level count results.
Downstream analyses were performed using custom scripts run in R 4.0.2 (R Core Team) [146]. Gene identifiers were updated from the Ensembl annotation used for mapping (version 82) to version 106 for further analysis. Genes with little evidence of expression were removed prior to analysis of the count-based data by filtering out genes with fewer than 10 reads in every sample. Removing the unexpressed genes left 17,907 genes across five gene types considered (16,883 protein coding, 794 pseudogenes, 136 ncRNA, 87 lincRNA, and 7 snRNA). For exploratory analysis of expression trends across samples or genes, expression values normalized for library size were utilized, either TPM from RSEM or VST-normalized counts from DESeq2, as indicated.
Differential expression analysis
Differential expression analysis was performed with DESeq2 1.30.1 [120] in R, for all combinations of strain and treatment against WT animals with empty-vector (EV) RNAi. Foldchange shrinkage procedures were applied to moderate fold-changes of low-expression genes. We considered significantly differentially expressed genes to be those with an FDR-corrected p-value < 0.05 and a foldchange magnitude of at least twofold (|log2 fold-change|≥ 1). Fold-changes are for comparisons against an N2 reference population, unless otherwise specified. In a few instances, p-value results were smaller than R can output in the default configuration; these are reported here in supplemental tables as 2.225074e-308, the smallest non-zero floating point number supported.
Transcription factor target prediction
A workflow for prediction of transcription factor binding sites from positional weight matrix (PWM) motifs for C. elegans transcription factors was adapted from TargetOrtho 2 [64]. Motifs were curated from CIS-BP (313), JASPAR (10), UniProbe (12), and manually from publications (7) for coverage of over one-third of known C. elegans TFs [3, 23, 56, 82, 162]. Of motifs from CIS-BP, 202 are based on direct evidence from binding experiments (ChIP-Seq, PBM, etc.) and the remainder are inferred by close homology to a TF in another species with direct experimental evidence. Motifs from JASPAR and UniProbe are based on experimental binding profiles. FIMO [66] was used to scan for motif matches in the C. elegans genome and seven other nematodes with available genomes: C. brenneri, C. briggsae, C. remanei, C. japonica, P. pacificus, P. exspectatus, and A. lumbricoides. TargetOrtho scripts were used to execute FIMO and pre-process the output. Initial identification of predicted binding within promoter regions was based on a symmetrical threshold distance from transcriptional start sites (TSS) of − 700/ + 700, and later filtered to − 700/ + 100, within the range used for binding prediction to promoter regions in previous studies [151, 182]. For FIMO match specificity, predicted associations with p < 0.0001 were considered, except for two of the manually curated E-box motifs, which had minimum p-values of 0.000195 and 0.00016; p < 0.0002 was used for these cases, as FIMO p-values are biased against short motifs. Results were read into R, and after filtering for the aforementioned − 700/ + 100 TSS distance, two sets of motif matches were produced: (1) all C. elegans matches without further filtering, and (2) matches found in C. elegans and in the same region (upstream or downstream of the TSS) of a homologous gene in two or more of the other nematode species genomes; these are referred to as “all C. elegans motif matches” and “species homology-filtered motif matches,” respectively.
Pathway and gene set enrichment
Gene sets for functional enrichment analysis were curated from the Gene Ontology, KEGG, Reactome, WormBase, WormExp, DIANA microT-CDS (miRNA binding predictions), and our TF target prediction set [14, 63, 76, 95, 144, 192]. The R package GOSeq (1.42.0) was utilized for batch enrichment across many genes [193]. In some cases, over-representation analysis was performed for a smaller subset of gene sets of interest using Fisher’s exact test (hypergeometric test) either through the R package GeneOverlap (1.34.0) or the R function phyper.
Analysis of publicly available datasets
Gene count results from multiple public datasets were re-analyzed in the same manner as our own dataset for differential expression. The public datasets and samples considered include eat-2 and N2 animals on OP50 or HT115 bacteria at multiple timepoints in SRA SRP089617 [178] and eat-2 and N2 animals at the L4 stage in GEO GSE125718 [25]. All differential expression analyses were only performed for samples within each dataset.
Oxygen consumption
Animals were synchronized by bleach prep and added to RNAi plates with HT115 EV bacteria as a food source. FUdR was added at the L4 stage for each strain to prevent production of embryos, and animals were kept at 20 °C. O2 consumption was assayed for whole animals at day 2 of adulthood with a Clark-type O2 electrode (S1 electrode disc, DW2/2 electrode chamber, and Oxy-Lab control unit, Hansatech Instruments, Norfolk UK). Animals were collected from plates in M9 buffer, pelleted briefly by centrifugation, and added to the electrode chamber in 0.5 mL of M9. After measurement of baseline respiration rate, FCCP (160 μM final concentration) was added to obtain maximal respiration measurements, followed by sodium azide (40 mM final concentration) for consumption rate with inhibited respiration. Each measurement continued until readings were stable or up to 10 min. Post-measurement, animals were collected and frozen for protein quantification by Bradford assay to normalize oxygen consumption rates by total protein.
Lifespan
For lifespan experiments, animals were kept from the L1 to L4 stage at 16 °C, at which point FUdR was added to a final concentration of 400 μM; animals were then transferred to 20 °C. Replica Set lifespan experiments were performed as detailed in [32, 33, 153]. Briefly, in Replica Set experiments, individual animals are not followed over time, but rather a large population is split across 24-well “replica” plates, and a new sub-population of animals is scored at each time point and then discarded. Enough “replica” plates are set up at the outset of the experiment for each planned observation based on typical lifespan characteristics of the strains already established. Resulting data was fit to logistic function curve for determination of median lifespan, as previously described [32, 33].
Oxidative stress survival
Oxidative stress survival was performed similarly to [88] but with observation and scoring facilitated by our version of the C. elegans Lifespan Machine [176]. Briefly, synchronized L1 animals were raised on HT115 bacteria with empty vector (EV) RNAi at 15 °C from L1 to L4 on 50 mm plates (Corning Falcon 351,006). When animals reached the L4 stage, FUdR was added (400 μM final concentration) and then kept at 20 °C for the remainder of the experiment. When animals reached day 2 of adulthood, 7.7 mM tert-butylhydroperoxide (tBOOH) was added to the plates, and they were transferred to the lifespan machine scanners, with the plate lids in place, for imaging and analysis. The scan schedule was set such that every animal was analyzed approximately once per hour.
Brood size
For each strain, animals were synchronized, and parent hermaphrodites were picked to their own plate at L4. Starting from 24 h after L4, parents were picked to a new plate every 24 h until no further eggs were observed. Larval animals were counted after 48 h, allowing enough time for all viable eggs to hatch. Eggs remaining unhatched after this time were considered dead and recorded. Animals were kept at 20 °C on HT115 EV E. coli as a food source. Two independent trials were conducted. Pairwise t-tests were performed to assess the statistical significance of differences between conditions, and p-values were FDR-adjusted to account for multiple testing [10].
Results
DR induces an adaptive transcriptional response to a chronic reduction in food availability
We sought to identify changes in gene expression associated with dietary restriction, with and without perturbation of the MML-1::MXL-2 complex via the mxl-2(tm1516) null mutation. We conducted transcriptome-wide RNA-sequencing of chronologically age-matched day 2 adult animals, including wild-type (N2), eat-2(ad465), and mxl-2(tm1516) single mutant animals, as well as eat-2;mxl-2 double mutant animals. Additionally, we included loss of pha-4 (ortholog of FoxA, a forkhead transcription factor), a modulator of gene expression in response to nutrient stress [141, 202], which is required for the increased lifespan of eat-2 mutant animals [143]. Furthermore, we included loss of daf-16 (FoxO) as a putative negative control, as daf-16 has been shown to be dispensable for the increased lifespan of eat-2 mutant animals, but is a central regulator of longevity in other contexts [108, 182].
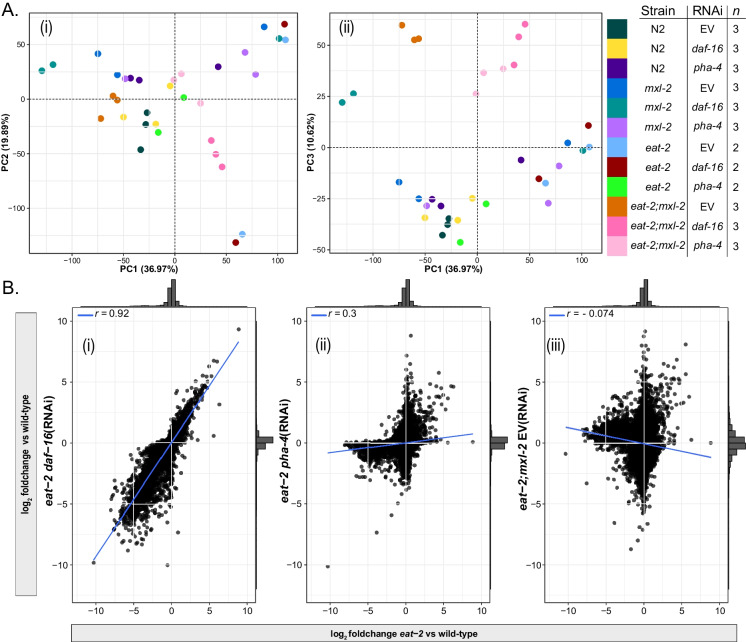
We first assessed the overall quality and fidelity of the gene expression for each condition. We were able to reliably assess gene expression for 17,907 genes, after removing consistently unexpressed genes and gene types without evidence of polyadenylation. We conducted principal component analysis (PCA) across all 17,907 genes and found at least one principal component that could account for substantial variability in the overall dataset in which biological replicate samples cluster together (Fig. 1A). The Pearson r for pairwise correlations among intra-group biological replicates is higher than 0.9 for all but two groups, and not lower than 0.85 in any case, implying reliably consistent patterns of gene expression between biological trials (Figure S1A). We found that the variability between biological replicates is associated with PC2 for eat-2(ad465) controls and eat-2(ad465);daf-16(RNAi); PC1 is associated with mxl-2(tm1516) control RNAi, daf-16(RNAi), and pha-4(RNAi). We also observed that this variability is not associated with known batch variables (Figure S1B). As expected, we found inactivation of daf-16 in eat-2 animals had very little effect on gene expression, as fold-changes between eat-2 controls and eat-2 daf-16(RNAi) are highly correlated (Pearson r = 0.92, Fig. 1B (i)), consistent with conclusions based on genetic interactions. In contrast, loss of either mxl-2 or pha-4 dramatically disrupted the gene expression profile of eat-2 mutant animals (Fig. 1B (ii) and (iii), respectively). The impact of mxl-2 and pha-4 loss on eat-2 gene expression aligns with previous genetic analysis: pha-4 and mxl-2 are required for eat-2-mediated increases in longevity [88, 143]. Collectively, we conclude our gene expression profiles have high fidelity and consistency, and genetic perturbation of mxl-2, pha-4, and daf-16 alters gene expression in eat-2 mutant animals as predicted based on previously characterized genetic interactions.
Fig. 1.
Changes in gene expression after dietary restriction are broadly disrupted by loss of pha-4 and mxl-2 but not daf-16. A Principal component analysis on variance-stabilized (VST) normalized count data revealed distinct clusters between most sample groups and, that biological replicate samples cluster together in at least one principal component explaining a substantial proportion of the variance of the overall dataset. The Pearson r for pairwise correlations of VST gene expression between replicate samples across all expressed genes was not lower than 0.85 in any case (Figure S1A). Variability between biological replicates was not associated with sample batches (Figure S1B). The key shows the correspondence between color and sample group (strain, RNAi treatment) and the number of biological replicate samples for that group. At least two biological replicates were analyzed across 17,907 genes expressed in any sample. B Loss of pha-4 or mxl-2, but not daf-16, dramatically shifts changes in gene expression in eat-2 mutant animals. Comparison of relative expression changes (log2 fold-change) across 17,907 genes expressed in eat-2 DR animals relative to WT (x-axis), and daf-16(RNAi) (i), pha-4(RNAi) (ii), or eat-2;mxl-2 double mutant animals (iii). Pearson correlation indicates a strong and significant positive linear association between eat-2 vs WT and eat-2;daf-16(RNAi) vs WT (r = 0.92, p < 2.2 × 10−16), but a much weaker positive trend after loss of pha-4 (r = 0.3, p < 2.2 × 10−16), and a weak negative association after mxl-2 loss (r = − 0.074, p < 2.2 × 10−16). The histograms indicate the density of genes
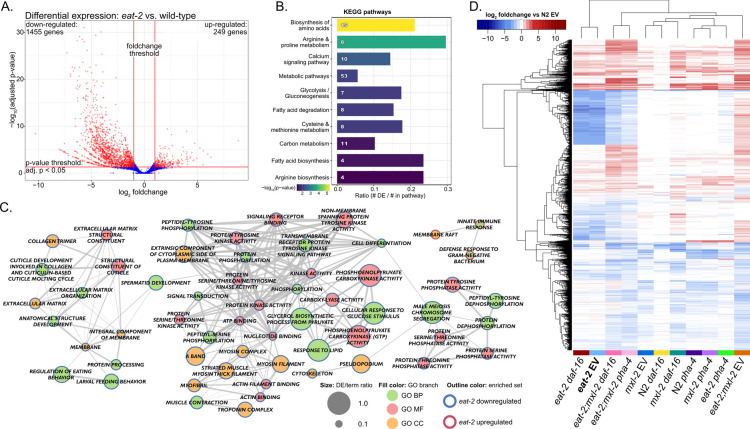
We sought to identify the transcriptional adaptive response to DR and compared the gene expression signature of eat-2 mutant animals to wild-type (N2) animals through differential expression (DE) analysis. We found 1704 genes with significantly altered expression and at least a moderate fold-change in eat-2 animals, of which 249 genes (14.6%) were up-regulated and the majority—1455 (85.4%)—were down-regulated (Fig. 2A) (DESeq2 FDR-corrected p-value < 0.05, absolute log2 fold-change ≥ 1). Among DE genes in eat-2 mutant animals, 75 had prior associations to lifespan phenotypes (based on Wormbase WS282 [40]): most in down-regulated genes, with more than half associated with extended longevity with loss of function, concordant with down-regulation in long-lived DR animals. As about 15% of C. elegans genes are co-transcribed in operons [15], we also looked for enrichment of annotated operons and found enrichment of five operons corresponding to 11 genes among all differentially expressed genes in eat-2, indicating that regulation of transcription at the level of operons has little influence on the DR transcriptional profile. The complete set of differential expression results can be found in Table S1.
Fig. 2.
Broad metabolic reprogramming through changes in gene expression under conditions of DR. A Volcano plot for eat-2 differential gene expression analysis. The x-axis shows log2 fold-changes for eat-2 EV relative to wild-type (N2 EV), and the y-axis shows -log10 FDR-adjusted p-values from DESeq2. Vertical red lines indicate the threshold for fold-change magnitude (|log2 FC| ≥ 1), and the horizontal red line indicates the p-value threshold (adjusted p-value < 0.05). 1455 genes were significantly down-regulated, and 249 were upregulated in eat-2 (red points). The same criteria were applied to all other comparisons. B KEGG pathways significantly enriched under DR conditions. Pathways associated with down-regulated genes include multiple aspects of metabolism, including amino acid biosynthesis, fatty acid metabolism, and energy-associated pathways. The bar length is the fraction of the pathway genes overlapping with the DE genes, the number in the bar is the number of overlapping genes, and color shows the −log10 adjusted p-values (GOSeq). All pathways shown were significantly enriched (adjusted p-value < 0.05). C Significantly enriched Gene Ontology terms for DR signature genes. Network representation allows visualization of related terms which share member genes. Up- and down-regulated genes were treated as separate gene sets (red and blue node outline, respectively). Node fill color indicates the GO sub-ontology a term belongs to: biological process (BP), molecular function (MF), or cellular component (CC). Node size indicates the proportion of genes associated with the term that were present in the gene set. Connections between nodes (edges) show terms with shared member genes, with thicker edges representing a greater degree of overlap. D Heatmap of log2 fold-changes across the 4762 genes significantly DE in at least two comparisons relative to wild-type. Rows and columns are ordered by hierarchical clustering
In order to identify functionally how expression changes may contribute toward the aging benefits of DR, we performed over-representation analysis to identify significant associations with pathways and gene sets. Among the genes downregulated in eat-2 mutant animals, metabolic reprogramming, including reduced expression of genes involved with amino acid and fatty acid biosynthesis, was significantly enriched. In contrast, analysis of eat-2 upregulated genes produced no significantly enriched KEGG or Reactome pathways, indicating a lack of broadly coordinated activation of gene expression across the organism in response to DR (Fig. 2B). Macromolecule metabolites have been previously observed to be less abundant in eat-2 through metabolomics experiments [58, 133], and overall protein content is lower in eat-2 animals [58, 195], which is consistent with our findings. We also found downregulated expression of genes regulating fatty acid metabolism,eat-2 mutant animals are thin with reduced fat stores [6, 7, 78]. A switch to fatty acids as an energy source is important in DR [78, 190], and eat-2 mutant animals have largely depleted reserves of lipids. Interestingly, elo-5, a fatty acid elongase involved in monomethyl branched-chain fatty acid synthesis, which is important for lifespan of wild-type and daf-2 animals, as well as glucose-induced stress resistance, is significantly upregulated in our eat-2 samples, indicating possible adaptive upregulation of specific fatty acids to maintain membrane integrity in the context of low overall lipid availability [101, 153, 187] (Table S1). We conclude eat-2 mutant animals globally downregulate significant components of metabolic gene expression to match the diminished availability of nutrients and optimize energy utilization, which suggests that expression levels of metabolic genes are causally linked with nutrient availability.
Metabolic remodeling during DR re-allocates energy away from costly processes toward somatic health via changes in gene expression
We next assessed enrichment of broader functional terms from the Gene Ontology in eat-2 animals. We found the only terms enriched for eat-2 upregulated genes are associated with innate immune and bacterial defense responses (Fig. 2C, Table S2); however, we also noted that the upregulation of a small set of innate immune response genes was not specific to eat-2 and also occurred with mxl-2 mutation or pha-4 RNAi under both basal conditions and in eat-2 (Figure S2). The lack of concerted regulation of functions associated with eat-2 upregulated genes suggests that chronic DR is not activating transcription as part of a specific adaptive response to nutrient stress. Instead, we observed reduced gene expression associated with several biological processes consistent with a shift from reproduction to survival. For example, there is significant enrichment for downregulation of sperm-associated gene sets, including spermatid development, sperm meiosis chromosome segregation, sperm motility via pseudopodia, and Major Sperm Proteins (MSPs, 31 of 47). MSPs are the most abundant proteins in C. elegans sperm and necessary for reproduction, both as structural components and signals for oocyte maturation [75, 163] (Fig. 2C). We posit this represents an adaptive shift away from reproductive fecundity in favor of survival. The total number of sperm within the hermaphrodite spermatheca limits the progeny number in unmated hermaphrodites, which typically form prior to the onset of adulthood. eat-2 animals have smaller total brood sizes and extended reproductive-span [2, 36, 102, 121]. Our results suggest down regulation of gene expression associated with sperm production provides a simple but effective mechanism for animals undergoing chronic DR to prioritize survival over investing in the energetically costly process of producing progeny; this is in contrast to the response to acute starvation in reproductive adult wild-type animals in which animals retain eggs, leading to internal hatching and matricide to promote survival of progeny [26].
We found a significant number of genes related to muscular function down-regulated in eat-2 mutant animals. For example, decreased expression of genes associated with the A band of the sarcomere and the myosin complex (Fig. 2C), which is consistent with thinning of body wall muscles observed in eat-2 mutant animals [128]. Additional genes include those broadly expressed in the body wall-muscle, including unc-54, myo-3 as well as mlc-3 (log2 foldchange −1.32 to −2.3), which encode myosin heavy chain and light chain proteins, respectively [9]. We posit downregulation of muscle gene expression results in the maintenance of a smaller volume of muscle, thereby limiting the drain of energy reserves from functions vital to maintaining viability (i.e., an overall reduction in metabolic cost). Concurrently, protein from skeletal muscle provides energy during severe nutrient restriction [159].
Increased expression of collagens has been associated with longevity, particularly under reduced insulin-like signaling in daf-2 animals [53, 147, 155]. However, we found collagen-associated genes significantly down-regulated in eat-2 mutant animals (e.g., collagen trimer, constituent of cuticle, ECM constituent). Despite this, eat-2 animals do not exhibit compromised cuticle integrity and retain properties of “young” cuticle into old age [52]. Decreased expression of collagen genes is also consistent across analysis of public eat-2 gene expression datasets in late-larval animals at L4 [25] and young adults at days 1 and 3 [178]. Downregulation of collagens has been observed in mice under DR, suggesting an evolutionarily conserved mechanism [27]. As collagen is the most abundant protein in the C. elegans cuticle and covers the entire outside of the animal [155], we posit synthesis of new collagen to be highly regulated in eat-2 due to restricted energy availability. Collagen usually consists of trimeric repeats of glycine and two variable amino acids, often proline and hydroxyproline [35, 91]. While these are not among the most bioenergetically costly to synthesize and are also not among the diet-derived essential amino acids in C. elegans, the total amount of collagen in the cuticle and ECM still makes collagen maintenance an energetic burden [1, 155, 199]. Interestingly, supplementing WT animals with low concentrations of proline or glycine is sufficient to extend lifespan but does not further increase lifespan of eat-2(ad1116) animals [48]. Overall, the consistent downregulation of collagens across multiple eat-2 datasets again suggests a reduction in metabolic cost, which allows directing the limited available resources to the most acute needs and provides an additional energy resource, though it remains unclear if collagen synthesis would be upregulated to heal wounds in DR animals subject to cuticle damage [131].
A large number of genes with kinase or phosphatase activity are down-regulated in eat-2 mutant animals, 111 and 76, respectively (Figure S3A). To determine whether gene products were associated to specific signaling cascades, or enriched in specific tissues, we looked for pathways and functions associated with this gene subset. Looking at all associations, we found kinases associated with arginine metabolism (F46H5.3, F32B5.1, W10C8.5), glycolysis and gluconeogenesis (pfk-1.2, pck-2, pck-1), and phosphatases linked to mRNA surveillance (C09H5.7, C34D4.2, F52H3.6, ZK938.1, gsp-4, gsp-3, T16G12.7) (Figure S3B–C). Arginine kinases play a role in energy homeostasis by helping to meet acute increases in energy demands [55], for example, overexpression of argk-1 extends C. elegans lifespan through activation of AAK-2 (AMPK) [38, 129]. C. elegans possesses more than 25 phosphatases with substantial homology to yeast GLC7 (WormBase WS282), which is important for mRNA export from the nucleus, an aspect of mRNA surveillance [40, 62, 84]. Of the seven GLC7 homologs with reduced expression in eat-2, GSP-3/4 are the most well-studied, with roles in spermatogenesis that mirror the function of homologs expressed in male mice [28]. More broadly, both the kinases and phosphatases down-regulated in eat-2 mutant animals are enriched in functions related to the development of sperm (Figure S3D); kinase and phosphatase expression is a putative mechanism to regulate spermatid function without translation of new proteins through post-translational modulation of protein activity, as spermatids lack ribosomes [50, 148].
MXL-2 regulates expression of a distinct set of genes during dietary restriction
We next sought to identify genes with altered levels of expression in our other sample groups. We proceeded to run differential expression analysis for single mutant genetic backgrounds: eat-2(ad465), mxl-2(tm1516), along with eat-2(ad465);mxl-2(tm1516) double mutants after treatment with either control RNAi (empty vector), pha-4(RNAi), or daf-16(RNAi). These genetic perturbations were compared to wild-type animals treated with empty vector RNAi in a pairwise manner, using the same criteria for significant expression changes (Table 1, S1). Across these 11 contrasts, a total of 4762 genes were differentially expressed relative to N2 EV in at least two comparisons (Fig. 2D). Loss of daf-16 has little effect on the transcriptional response to chronic DR, as expected. Also, in accordance with genetic analysis of requirements for eat-2 lifespan, loss of pha-4 effectively negated the gene expression signature of eat-2 animals: the eat-2;pha-4 profile more closely resembles pha-4 inactivation in wild-type and mxl-2 mutant animals (Fig. 2D).
Table 1.
Summary of significantly differentially expressed (DE) genes for all considered samples compared to wild-type EV
| Strain | RNAi | Compared to: | # significant DE genes (p-adj < 0.05, |LFC|> = 1) | ||
|---|---|---|---|---|---|
| Total # DE | Up-regulated | Down-regulated | |||
| N2 | daf-16 | Wild-type EV | 318 | 49 | 269 |
| N2 | pha-4 | 1066 | 400 | 666 | |
| mxl-2(tm1516) | EV | 824 | 219 | 605 | |
| mxl-2(tm1516) | daf-16 | 1609 | 714 | 895 | |
| mxl-2(tm1516) | pha-4 | 1043 | 584 | 459 | |
| eat-2(ad465) | EV | 1704 | 249 | 1455 | |
| eat-2(ad465) | daf-16 | 1627 | 238 | 1389 | |
| eat-2(ad465) | pha-4 | 879 | 478 | 401 | |
| eat-2(ad465);mxl-2(tm1516) | EV | 3728 | 1922 | 1806 | |
| eat-2(ad465);mxl-2(tm1516) | daf-16 | 4168 | 1631 | 2537 | |
| eat-2(ad465);mxl-2(tm1516) | pha-4 | 2759 | 998 | 1761 | |
Summary of differential expression analysis for genes with significant changes in expression compared to wild-type (N2 EV). Significant DE genes were based on the following criteria after analysis with DESeq2: adjusted p-value < 0.05 and log2 fold-change magnitude ≥ 1. Up-regulated genes exhibited higher expression (positive fold-change) in the subject condition compared to N2 EV, and vice-versa for down-regulated genes (negative fold change)
Comparison of the mxl-2(tm1516) null mutation alone to wild-type animals yielded significant expression changes to more than 800 genes, with 75% of those down-regulated (Table 1, S1). These genes were not strongly enriched for distinct pathways or biological functions (Table S2), though we did note the moderate repression of five heat shock proteins (hsp-16.2, hsp-16.41, hsp-12.3, hsp-12.6, and hsp-1.1), which encode small molecular chaperones that are upregulated in response to stress and prevent protein misfolding [112, 114, 189]. Genes with increased expression in mxl-2 mutant animals were enriched in diverse functions related to lipid metabolism and carbohydrate binding, similar to some of the regulatory functions of orthologous MML-1::MXL-2 complexes in other species [77, 149]. However, genes dysregulated in mxl-2 mutants under normal feeding conditions were largely divergent from eat-2 signature genes: only 6% of mxl-2 and 3% of eat-2 genes were found commonly differentially expressed relative to WT. mxl-2 loss was also distinct from that of either pha-4 or daf-16, the latter of which yielded the fewest total DE genes of any of these comparisons (318 total, 49 up and 269 down), which may reflect typically minimal DAF-16 activity under basal conditions [160]. Thus, MXL-2 impacts expression of distinct genes under normal and DR conditions (explored in more detail later).
We looked for tissue-associated biases in differential gene expression, based on sets of genes enriched in muscle, neurons, hypodermis, intestine [93], or in sperm [148] (Figure S4A). In agreement with our Gene Ontology analysis of eat-2 expression changes, we found a substantial and significant set of downregulated genes in eat-2 mutant animals to be enriched in sperm, as well as significant over-representation of muscle-associated genes. The intestine is commonly referred to as the most metabolically active tissue in C. elegans, and both the MML-1::MXL-2 complex and PHA-4 act in intestinal cells in adult animals [80, 88, 137, 143]; we found enrichment for intestinally expressed genes in both the up- and down-regulated transcriptional response to eat-2, mxl-2, or pha-4 single-perturbation conditions. We also found enrichment of neuron-associated genes among those down-regulated in mxl-2 null mutant animals, which is consistent with a recent report that demonstrated MML-1 and MXL-2 activity within the nervous system is sufficient to alter longevity [164]. Collectively, these results suggest Myc-family TFs may regulate adaptive longevity responses from distinct tissues in response to a particular longevity signal.
MXL-2 and PHA-4 are required for the majority of gene expression reprogramming associated with chronic dietary restriction
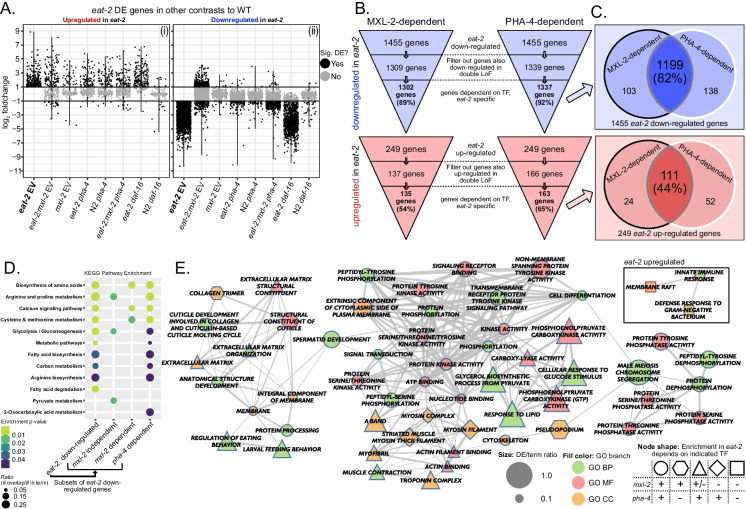
We next identified the portion of the eat-2 expression signature that required mxl-2 and/or pha-4. We defined mxl-2-dependent genes as (1) differentially expressed in eat-2, (2) not similarly differentially expressed in eat-2;mxl-2 (i.e., not changing in the same direction), and (3) specific to the DR background (i.e., unchanged in mxl-2), all compared to wild-type. This strategy was applied to both eat-2 up-regulated and down-regulated genes (Fig. 3A (i, ii)), for dependence on mxl-2, pha-4, and daf-16 (Fig. 3B, Table S3). A total of 89% of genes down-regulated in eat-2 mutant animals required mxl-2 and 92% required pha-4, while only 19% required daf-16. The pattern is similar for genes up-regulated in eat-2, but less pronounced, with 54% of eat-2 upregulated genes requiring mxl-2, 65% requiring pha-4, and only 31% requiring daf-16. Overall, the vast majority of genes down-regulated in eat-2 require both pha-4 and mxl-2, but not daf-16, to maintain transcriptional suppression (Fig. 3C).
Fig. 3.
mxl-2 and pha-4 are required for the majority of gene expression changes after DR. A Scatter plots of relative fold change in gene expression under the indicated condition. Only genes that were positively (i) or negatively (ii) DE in eat-2 mutant animals are shown. For each sub-panel, the specific genes represented in each column are the same, but the fold changes are for the indicated comparison. Horizontal lines at 1 and −1 indicate the foldchange threshold. mxl-2-dependent genes were DE in eat-2 but not significantly regulated in a similar manner in eat-2;mxl-2 animals. Genes with significant and opposite fold-change in mxl-2 single mutant animals were filtered out for specificity to the DR context. An analogous strategy was applied for investigating pha-4-dependent genes. B A schematic representation of the filter steps and the numbers of DE genes at each level (also see Table S3). C A slightly larger proportion of the eat-2 signature depends on pha-4, but the vast majority of the downregulated portion of the signature requires both mxl-2 and pha-4 (C). D, E Pathway (D) and Gene Ontology (E) enrichment for TF-dependent eat-2 genes. Over-representation analysis was run on each list independently. E shows the same network representation for eat-2 GO term enrichment as Fig. 2C, with node shapes indicating TF dependency (see legend on bottom-right). Triangle-shaped nodes are dependent on pha-4 but lacked significant enrichment in both the mxl-2-dependent and -independent sets. The only GO terms with significant enrichment in the DR upregulated genes are independent of both factors (see also Figure S2)
We conducted functional enrichment analysis with the eat-2 gene sets that were either dependent or independent of mxl-2 or pha-4 (Fig. 3D, E). The only GO terms with significant enrichment in the DR-upregulated genes are independent of both mxl-2 and pha-4. However, for the transcriptional signature of repressed expression in eat-2 mutants, many pathways and functions are dependent on both factors, with an overall greater proportion dependent on pha-4. We also conducted the converse analysis to see whether significant pathway enrichment remained in the subsets of DE genes that were independent of either pha-4 or mxl-2. No KEGG pathways were significantly enriched in pha-4-independent genes, implying that all transcriptional reprogramming conferred by chronic DR requires pha-4. Notably, some muscle-associated terms (myosin filament/complex) are mxl-2-independent but pha-4-dependent (Fig. 3E); while myosin filament/complex are mxl-2-independent, eat-2 DEGs associated with other muscle components were mxl-2-dependent (Fig. 3E). This hints that MXL-2 is still required for the proper reallocation of resources away from some aspects of muscle function or maintenance during extended periods of DR. Consistently, MXL-2 was required for down-regulation of genes in amino acid biosynthesis pathways under DR, which would provide the foundational material for protein synthesis associated with maintaining muscle mass in ad libitum conditions. In contrast, glycolysis, gluconeogenesis, as well as arginine and proline metabolism pathways were largely mxl-2 independent (Fig. 3D), suggesting that changes in overall energy metabolism are not directly regulated by MXL-2. We noted that the four mxl-2-independent eat-2 downregulated genes in arginine/proline metabolism function in catabolism of these amino acids; while proline catabolism has been linked to longevity in other contexts [142, 198], down-regulation in eat-2 may be concomitant with downregulation of collagen synthesis genes, as collagen is rich in proline/hydroxyproline [35, 91]. A few GO terms were independent of pha-4 but dependent on mxl-2, including pseudopodium (associated with sperm development and motility) and collagen trimers. Sperm-enriched genes down-regulated in eat-2 mutant animals are broadly dependent on mxl-2 (Figure S4B). Taken together, we posit the de-repression of some of these energy intensive processes with loss of the mxl-2 or pha-4 transcription factors prevents the adaptive allocation of resources toward somatic maintenance, resulting in compromised health and lifespan.
C. elegans Myc-family members possess unexpected transactivation domains
In C. elegans, the consensus to date is that the MML-1::MXL-2 complex activates and the MDL-1::MXL-1 complex represses transcription [145, 194]. Consistent with their different activities, we previously found loss of mdl-1 or mxl-1 increased longevity, while loss of mml-1 or mxl-2 decreased longevity, and that these genetic interactions are epistatic [88]. These results suggested that under otherwise basal conditions, the two complexes converge on shared target sites and compete to regulate longevity-associated gene expression. In agreement with the Y2H studies of MML-1::MXL-2 and MDL-1::MXL-1 complex activity [145], mammalian Mondo proteins contain a transactivation domain (TAD) [24, 29, 127] and mammalian Mad complexes oppose the transcriptional activity of Myc [107, 126, 157]. Thus, we were surprised to find mxl-2 dependence for the downregulation of gene expression in eat-2 animals; the clear prediction based on molecular annotation, known function, and genetic analysis suggested that the MML-1::MXL-2 complex would be necessary for activating transcription under conditions of DR. While the mxl-2 dependence for the vast downregulation of gene expression in eat-2 could be indirect, we considered the possibility that Myc-family members may not unidirectionally regulate transcription in every context. Using recently developed neural network models for TAD prediction, we applied these models to the C. elegans Myc-family members and their human orthologs [51, 154]. As expected, we found a high-confidence predicted TAD in MML-1 (Figure S5A), within amino acid residues 245–276, which parallels the presence of the TAD in Mondo-conserved regions IV and V in vertebrates, and is also predicted by ADPred (Figure S5A) [24, 127]. To our surprise, a high-confidence TAD was also predicted on the C-terminus of MDL-1 (Figure S5C). In contrast, human MXD/MAD family proteins show much weaker confidence of a predicted TAD in MXD1 and MXD3, which suggests in some instances MDL-1 may activate transcription, in contrast to the mammalian homologs. We also found potential TAD regions of lower confidence on the C-terminal side of MXL-2 and MXL-3, but not MXL-1 (Figure S5B, D). Intriguingly, the wider presence of TADs within C. elegans Myc-family members may explain why C. elegans lack a true Myc-ortholog, and loss of any particular Myc-family member still results in viable animals, yet loss of Myc is lethal in higher metazoans.
Dysregulated gene expression in eat-2;mxl-2 compromises reproductive fitness
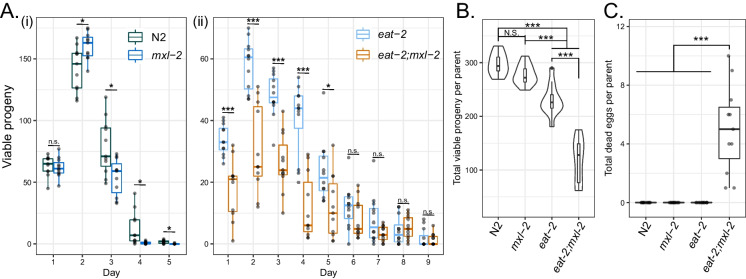
Given the alterations in gene expression linked to reproduction we observed in eat-2 animals, we assessed the impact of mxl-2 loss on eat-2 fecundity. We quantified progeny production through reproductive lifespan (i.e., the period of active egg laying in reproductive hermaphrodites) and whether animals produced unhatched eggs. eat-2 animals are known to have smaller brood sizes and exhibit extended reproductive lifespan [36]. In the absence of mxl-2, eat-2 mutants produce a significantly reduced total brood size—almost half that of eat-2 alone—without affecting the duration of the egg-laying period (Fig. 4A, B, Table S4). In contrast, mxl-2 loss in ad libitum animals did not significantly affect brood size (Fig. 4B). Furthermore, dead eggs were only observed from eat-2;mxl-2 hermaphrodites (Fig. 4C, Table S4). Interestingly, pha-4 inactivation is known to reduce eat-2 reproductive lifespan [121], in contrast, in the absence of mxl-2, we find the duration of the reproductive lifespan of eat-2 mutant animals is intact, but negatively impacts total fecundity. Given the limited nutrient resources of animals experiencing chronic DR, and our observation of de-repression of many metabolic and reproduction-associated genes in eat-2;mxl-2 mutant animals, we posit that aberrant reallocation of resources in eat-2;mxl-2 yields uneven provisioning to progeny and thus critically compromises the fitness of a portion of embryos (see Figure S6 and Table S4 for additional trial data).
Fig. 4.
Loss of MXL-2 in DR but not ad libitum animals compromises fecundity and embryo viability. Brood size assays were performed to determine how reproduction-associated gene expression changes with mxl-2 loss in eat-2 affected reproductive fitness. A Viable progeny from each day over the course of the reproductive lifespan were counted for singled parent hermaphrodites for (i) N2 and mxl-2 and (ii) eat-2 and eat-2;mxl-2 animals. B Total brood size aggregated from across the observations in A for each strain. C Unhatched eggs were counted for each strain, but only found for eat-2;mxl-2 animals. Data is shown from a representative trial. See Table S4 for complete results and statistical analysis, and Figure S6 for the plotted results from an additional trial. Stars indicate FDR-corrected p-values: * < 0.05, ** < 0.01, *** < 0.005
Lower food intake in DR does not significantly alter respiratory rate and is not affected by MXL-2 loss in young adult animals
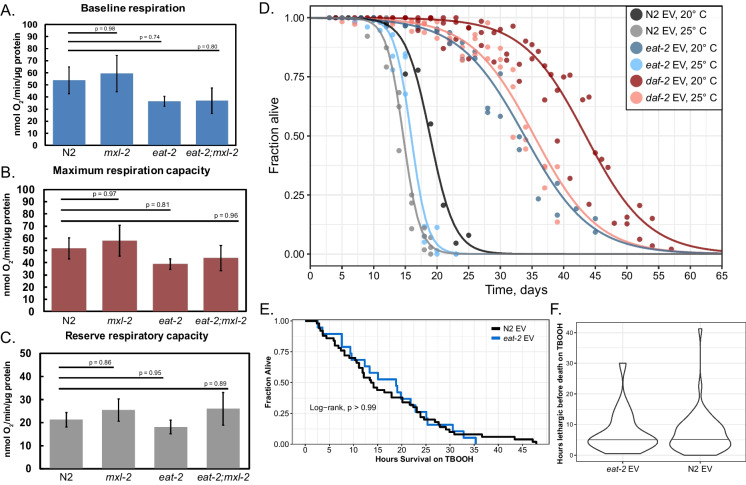
We ascertained whether other physiological aspects of a DR-optimized metabolism were disrupted in the absence of mxl-2. Multiple groups have shown, perhaps counter-intuitively, that respiratory rates are as high or even higher in eat-2 mutant animals than in wild-type [49, 81, 117]. Surprisingly, we found no significant difference in baseline respiratory rate, maximal, or reserve respiratory capacity between N2, mxl-2, eat-2, or eat-2;mxl-2 mutant animals (Fig. 5A–C). Given that only a subset of the lipid metabolism genes that are part of our DR expression signature are dependent on mxl-2, a normal respiratory rate may be maintained by a switch to fatty acid oxidation in DR that does not require MXL-2. It remains to be determined if pha-4 loss alters DR metabolic rate. Age may also play a role; recent work reported that eat-2 animals maintain “youth-like” mitochondrial membrane polarity at more advanced age than wild-type [12]. As such, it is possible that older eat-2;mxl-2 animals might not be able to maintain a normal respiratory rate.
Fig. 5.
Chronic DR does not alter respiratory rate, nor confer stress resistance. A–C Young adult eat-2(ad465) animals do not exhibit reduced respiratory rate, which remains unaltered by loss of mxl-2. Oxygen consumption of whole animals was measured and normalized by total protein. Results show mean ± SEM from three biological replicates, with p-values from ANOVA followed by the Tukey post-hoc test. A Baseline respiratory rate before treatment with FCCP or sodium azide. B The maximum respiratory capacity is the difference between the uncoupled rate after FCCP treatment and the inhibited rate after sodium azide treatment. C The reserve respiratory capacity is the difference between the uncoupled rate after FCCP treatment and the baseline rate. D eat-2(ad465) animals are not long-lived under chronic mild heat stress. Replica-set lifespan experiments for N2, eat-2(ad465), and daf-2(e1370) mutant animals kept at 20 and 25 °C post-development. Points represent observations of independent sub-populations of animals, fit with a logistic function to obtain a survival curve [32, 33]. daf-2 mutant animals (decreased insulin/IGF-1 signaling) are included as a positive control. Data represent at least two trials for each condition. E, F eat-2(ad465) animals are not more resistant to oxidative stress than wild-type. Young adult (day 2) animals were treated with tert-butyl hydroperoxide (TBOOH), and survival was analyzed from time-series images taken with an automated longitudinal imaging platform. Data represents two biological replicate plates per condition (N2 n = 50, eat-2 n = 19). E No significant difference was found in median survival in DR animals, indicating eat-2 does not provide increased resistance to oxidative stress in young adults. Data were analyzed non-parametrically with Kaplan–Meier, followed by the log-rank test in R. F The duration between when animals cease broad locomotion and time of death, based on automated longitudinal imaging, was also consistent between N2 and eat-2. Wilcoxon rank-sum test p-value = 0.8193
DR in eat-2 increases lifespan without a concomitant increase in stress resistance
We next sought to determine whether mxl-2 was required for enhanced stress resistance of eat-2 mutant animals. Stress resistance broadly declines during normal aging [45, 106, 169, 189]; eat-2 mutant animals have previously been shown to have increased resistance to proteotoxic stress in aggregation-prone models [124, 174]. We chose to assess how a chronic state of DR would impact mild but persistent proteotoxic stress; wild-type animals maintained at 25 °C are slightly short-lived, have accelerated proteostatic decline, but lack the severe fecundity deficits of animals maintained at higher temperatures [21, 65]. We previously showed that maintaining wild-type C. elegans at 25 °C is sufficient to induce a mild hormetic heat shock [39]. We reasoned if chronic DR extended longevity through improved stress resistance, then eat-2 mutant animals should have a similar increase in lifespan at 25 °C. To our surprise, we found that keeping eat-2 animals at 25 °C instead of 20 °C is sufficient to completely suppress DR lifespan (Fig. 5D, Table S5), suggesting chronic DR does not extend longevity via improved resilience to heat stress.
Dietary restriction reduces oxidative stress in rodent models and even biomarkers of oxidative stress in humans [168, 83]. To test whether eat-2 mutant animals had increased resistance to acute oxidative stress, we treated animals with tert-Buytl-hydroperoxide (TBOOH) and assayed survival. To our surprise, we found no significant difference in median survival between wild-type and eat-2 animals (Fig. 5E). To determine whether a more subtle improvement was occurring, we ascertained the time at which animals cease large translational body movements and enter a lethargic state for a period in advance of death. The duration between lethargy and death for eat-2 animals under oxidative stress was similar to wild-type (Fig. 5F). Thus, it appears that eat-2 animals do not exhibit improved oxidative stress resistance (OSR) by day 2 adulthood.
Autophagy is critical for maintenance of organismal health, both under stress and normal conditions, and has also been implicated in DR in C. elegans, specifically in the intestine [60, 72, 110]. We were thus surprised to find a lack of transcriptional evidence for autophagy induction in our eat-2 animals (Figure S7). Even though eat-2 expressed genes associated with autophagy machinery at wild-type levels, when combined with mxl-2 mutation, we found significant down-regulation of two genes associated with autophagy: lgg-1 (orthologous to LC3/Atg8), an important component of autophagosomes involved in cargo recruitment, and unc-51 (orthologous to ULK1/2) which initiates autophagosome formation, both are necessary for long lifespan in a different eat-2 mutant [60, 119, 130, 184]. Thus, eat-2 mutant animals do not increase expression of autophagy genes by day two of adulthood, but mxl-2 is still required for maintenance of critical autophagy gene expression specifically in DR. While others have shown that autophagy genes are required for eat-2 lifespan and that LGG-1 puncta and autophagosome turnover are increased in a different allele of eat-2, ad1116 [60, 72], we posit that while basal autophagy activity is maintained in eat-2, by early adulthood, most available metabolic resources have been mobilized and energy metabolism has been optimized such that further induction of autophagy is no longer beneficial to organismal homeostasis.
Overall, our data aligns with previous work indicating that eat-2(ad465) animals are not as broadly stress-resistant as other long-lived mutants [46, 57, 81, 92]. Intriguingly, eat-2(ad1116) and eat-2(ad453) animals exhibit an even slower pumping and feeding rate than eat-2(ad465) [18, 108] and enhanced resistance to thermal and oxidative stress [74, 165, 170], which may parallel experiments that have shown a non-linear relationship between the degree of DR and the resulting aging benefits [123]. We considered whether the use of FUdR might suppress increased stress resistance in eat-2 mutant animals, as chemically preventing progeny production could potentially impact adaptive responses; we surveyed publications that assessed stress resistance in eat-2 mutant animals and found no correlation between stress resistance and the use of FUdR [57, 74, 92, 139, 165, 170]. In our hands, stress resistance of eat-2(ad465) animals is similar to wild-type animals, indicating that lifespan and healthspan benefits from chronic and constitutive restricted food availability in this model do not arise from raising general barriers against stress.
Loss of MXL-2 yields synthetic dysregulation of gene expression and inverts the expression of hundreds of genes normally repressed in DR
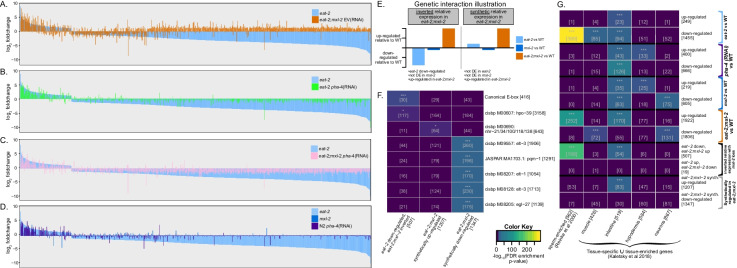
Strikingly, the eat-2;mxl-2 double mutant animals yielded two additional distinct gene expression patterns. First, we observed a synthetic gene expression phenotype: a number of genes at wild-type levels in either eat-2 or mxl-2 single mutant animals were differentially expressed in eat-2;mxl-2 (Fig. 2D, Table S6): 1207 and 1347 genes were synthetically up- or down-regulated in [139] eat-2;mxl-2, respectively. Second, we observed a pattern of inversion in expression within eat-2;mxl-2 animals: 507 genes normally repressed in eat-2 became significantly upregulated in eat-2;mxl-2 (Fig. 6A, Table S6). In contrast, inactivation of pha-4 in DR animals results in the majority of genes that were previously differentially down-regulated in eat-2 reverting to basal wild-type-like levels, without synthetic changes in expression (Fig. 6B). A schematic illustration of the synthetic and inverted genetic interactions is provided in Fig. 6E.
Fig. 6.
The Myc-family member MXL-2 is essential to coordinate changes in gene expression induced by dietary restriction. A–D Loss of mxl-2 inverts expression of genes normally downregulated by DR. Differentially expressed eat-2 genes, ordered by foldchange compared to wild-type (blue bars). Foldchanges for the same genes in the indicated comparisons versus wild-type (N2 EV) are overlaid: A eat-2;mxl-2 (orange), B eat-2 pha-4(RNAi) (green), C eat-2;mxl-2 pha-4(RNAi) (pink), and D mxl-2 or pha-4 loss in otherwise wild-type animals (dark blue and purple, respectively). A Loss of mxl-2 in eat-2 inverts the relative expression of 507 significantly downregulated DR genes. B PHA-4 is required for repression of gene expression by DR. C PHA-4 is epistatic to MXL-2 in the regulation of gene expression after DR: loss of pha-4 suppresses the inverted pattern of relative expression that occurs in eat-2;mxl-2 mutant animals. E Schematic illustrating the inverted and synthetic patterns of eat-2 DE genes. Left: cases where a large number of DE genes in eat-2 animals were significantly differentially expressed in the opposite direction in eat-2;mxl-2 animals (inverted), but did not change expression after only a single genetic perturbation. Right: a large class of genes produced significant expression changes only in the presence of both the eat-2 and mxl-2 mutation but remain at WT-like levels in eat-2 and mxl-2 single mutant animals (synthetic). F The inverted gene set was significantly enriched for canonical Myc-family TF binding sites (E-boxes). Enrichment for TF motif-based predicted binding within the promoter regions of genes in the inverted and the synthetic gene sets. Promoters of the inverted gene set were enriched for CACGTG canonical E-box motifs. Synthetically upregulated gene promoters are enriched for the TF motif of nuclear hormone receptors (nhr-21, 34, 100, 118, and 138, paralogs). Synthetically down-regulated gene promoters are enriched for binding of GATA factors elt-1, elt-3, egl-27, and pqm-1. G Tissue enrichment for significantly differentially expressed genes (top four sets of rows), and genes with inverted expression and synthetic gene expression (bottom two sets of rows). Somatic tissue gene sets are based on the union of tissue-specific and tissue-enriched gene lists from cell type-specific bulk RNA-Seq in [93]. Sperm-specific genes are from [148]. For both F and G numbers in brackets indicate the size of the gene set or the number of genes in the intersection. Adjusted enrichment p-values from the hypergeometric test are indicated by cell color and stars: “***” p < 0.001 “**” 0.01 “*” 0.05 “.” 0.1
Opposing Myc-family members, nuclear hormone receptors, and GATA TFs putatively regulate changes in gene expression when MXL-2 is unable to respond to chronic DR
We reasoned the aforementioned inverted and synthetic changes in gene expression may be revealing novel transcriptional responses that are germane to understanding how biological systems maintain homeostasis under conditions of metabolic stress when one regulatory component is impaired. To identify putative upstream regulators and tissue-associated biases in expression for the inverted and synthetic gene sets, we analyzed each set for over-representation of TF binding sites in promoters and for tissue specificity. As only a fraction of the known C. elegans transcription factors have been directly profiled to map context-specific binding sites, we were able to perform in silico binding predictions based on motif matching for more than one-third of C. elegans TFs [56]. Concurrently, we analyzed the presence of E-box (CACGTG or CANNTG) and E-box-like sequences within the C. elegans genome, as the Myc-family of TFs bind to these sequences, including the C. elegans MML-1::MXL-2 and MDL-1::MXL-1 complexes [3, 61, 70, 145, 162]. Overall, we found 695 genes with E-box-like motifs in promoter regions when restricted to matches also found in homologous genes of at least two other nematode species, and 3108 gene matches in C. elegans when homology was not considered; about half of these matches were for the canonical CACGTG E-box (Table S7).
As proof-of-principle, we first analyzed single genetic perturbations and found specificity in predicted motif sites among the promoters of down-regulated genes. Three different E-box motifs were enriched among all down-regulated genes in eat-2 animals: canonical CACGTG E-boxes, MDL-1 and CRH-2 (a bHLH transcription factor known to bind at degenerate E-box sites) (Figure S8A). As expected, the daf-16 DNA binding motif was the most significantly enriched with daf-16(RNAi) treatment. Neither PHA-4 nor E-box sites were enriched in the down-regulated DE genes of pha-4(RNAi) or mxl-2(Ø) animals, which may be due to limited activity of these factors under well-fed conditions [88, 161]. Interestingly, we did not observe enrichment of PHA-4 sites even in the subset of pha-4-dependent DR genes, indicating a few possible modalities for PHA-4 function in DR: 1) PHA-4 acts indirectly in eat-2, 2) PHA-4 binds at motifs that are distinct from those that have already been characterized in other contexts, or 3) PHA-4 acts primarily at earlier developmental stages. The latter is consistent with the role of PHA-4 (FOXA) as a pioneer TF, which keep enhancer nucleosomes accessible in chromatin, thereby allowing other TFs to bind and regulate transcription particularly in development [196, 197].
We found distinct significant TF motif enrichment between the inverted, synthetically upregulated, and synthetically downregulated gene sets. The canonical CACGTG E-box was significantly enriched in the inverted gene set; tissue-associated enrichment revealed significant expression in sperm and intestine (Fig. 6F, G). Transcriptionally activating and repressing heterodimers of bHLH Myc-family TFs are known to compete for binding at E-box elements [29, 145], and previous results suggested that the MML-1::MXL-2 and MDL-1::MXL-1 heterodimeric complexes may function as a rheostat in the transcriptional regulation of longevity [88]. We posit that loss of mxl-2, and therefore loss of MML-1::MXL-2 complex binding, facilitates MDL-1::MXL-1 binding in eat-2 for overlapping loci, yielding the inverted gene expression pattern. Collectively, it is tempting to speculate that the convergence of opposing Myc-family heterodimers at these loci may be key for coupling metabolic changes to reproductive fecundity.
We next examined the synthetically differentially expressed genes (i.e., DE in eat-2;mxl-2, but not either single mutant). In contrast to the inverted gene set, synthetically up-regulated genes did not exhibit significant enrichment for E-box sequences, suggesting regulation by TFs other than Myc-family members. Significant motif enrichment for synthetically up-regulated genes was limited to a subset of nuclear hormone receptor paralogs—nhr-21, 34, 100, 118, 138—and enriched for predicted intestinal expression (Fig. 6F, G). Lastly, significant enrichment for GATA binding factors—elt-1, elt-3, pqm-1, and egl-27—were found in genes synthetically down-regulated (Fig. 6F). Interestingly, both pqm-1 and elt-3 are required for eat-2 lifespan, and PQM-1 acts antagonistically with DAF-16 for nuclear entry [20, 165, 182]. Further, an earlier transcriptomic study found GATA motifs enriched in genes with expression changes during aging and in gerogene mutant animals [20]. Whether or not these TFs functionally impact longevity under these conditions is unclear, yet a picture begins to emerge. Loss of pha-4 completely suppresses eat-2 lifespan [143]. In contrast, we find that while loss of mxl-2 significantly shortens eat-2 lifespan, suppression is only partial [88]. Yet loss of either pha-4 or mxl-2 is sufficient to suppress the vast majority of the DE signature we observe in eat-2 mutant animals, but only loss of mxl-2 produces synthetic changes in gene expression in eat-2 animals. It remains to be determined if NHR and GATA transcription factors may be activated to compensate for the breakdown in appropriate regulation of gene expression in chronic DR without mxl-2, or if this represents dysfunction of a transcriptional network in which MML-1 and MXL-2 normally play crucial roles.
Discussion
It has been known for centuries that caloric restriction extends longevity. Luigi Cornaro the Venetian wrote four Discorsi between 1550 and 1562 describing how his adoption of a temperate lifestyle and consuming only 350 g of food daily were the basis for his attaining a long and healthful life [30, 31]. Since the modern era of genetic discovery, indeed a common theme has emerged: evolutionarily conserved genes often act as “watchtowers” of nutrient and energy availability, which regulate pathways that dictate the progression of aging [112]. Why would there be deep evolutionary conservation causally linking mechanisms of nutrient sensing to organismal longevity? We posit that organisms able to couple physiology to energy resources had a survival advantage when food was scarce by conserving and recycling resources, while delaying energetically costly physiological processes, such as development and reproduction. We find the transcriptional signature of eat-2 mutant animals, a form of constitutive and chronic DR, broadly involves the down-regulation of gene expression associated with amino acid and lipid metabolism and other energetically expensive processes such as collagen production and maintenance of muscle mass.
Delaying the production of offspring has the benefit of limiting competition for limited resources. In C. elegans, many long-lived mutant animals, including those with eat-2 mutations, have slower development, reduced numbers of overall progeny, and an extended period of progeny production [156]. We find downregulation of genes that encode functions related to sperm production in eat-2 mutant animals were dependent upon mxl-2; in the absence of mxl-2, down-regulation of gene expression fails to occur, brood size from self-progeny is reduced, and non-viable embryos are produced. Thus, our work reveals that MXL-2 plays a key role in mediating a transcriptional adaptive response that links energy availability to reproductive fitness. C. elegans hermaphrodites produce a limited number of sperm: approximately 300, but mated animals can produce nearly 1400 progeny [166]. In response to limited food availability, decreased expression of genes required to generate sperm could provide a simple but elegant method to maximize reproductive fitness by limiting the total number of sperm. Importantly, refined genetic analysis have revealed that alterations in reproduction/development are separable from longevity. For example, early discoveries in C. elegans aging research using temperature-sensitive alleles in the insulin/IGF1 pathway (IIS) revealed that dauer formation and extended longevity were genetically separable [99], which implies strategies to improve healthy aging may not require a cost in developmental or reproductive fitness.
It has been suggested eat-2 animals are a model of dietary restriction when raised on E. coli due to intestinal bacteria colonization resulting from insufficient pharyngeal grinder function, in turn leading to upregulation of innate immune responses and food-avoidance behavior [105]. While we see a handful of innate-immune-related genes upregulated, we find that set is not specific to eat-2. Thus, gene expression changes in innate immunity are unlikely to be the result of intestinal colonization, but rather a low-level compensatory response to perturbations that affect the intestine. We have previously established that mxl-2 animals do not have pharyngeal pumping defects [88] and are indistinguishable from wild-type with respect to adult size and development rate. It seems unlikely that infection response is regulating gene expression in DR differently from other contexts. However, Myc-family TFs respond non-canonically to the microsporidian intestinal pathogen N. parisii: loss of mdl-1, mxl-1, or mxl-2 alleviated infection severity, while loss of mml-1 increased severity [17]. Whether these responses connect to DR signals by relaying intestinal stress remains unknown.
Different methods of generating DR in C. elegans, either through alteration in food availability, type or amount of bacteria, specific nutrient restriction, timing of restriction, or genetic perturbation, have vastly different genetic requirements for DR benefits [42, 68]. In comparison to non-genetic DR models in C. elegans, eat-2 differs substantially in that DR in eat-2 is constitutive and chronic, even when multiple generations are raised on adequate food. Genetic perturbation of the links between nutrient sensing and adaptive responses that extend longevity have a high degree of specificity to the nutrient, tissue of action, and cell-type [132, 181]. For instance, TOR responds to levels of amino acids and carbohydrates [72, 87, 96, 110, 158]; AMPK is a conserved energy sensor of increased levels of AMP and ADP [5, 68, 69, 152]; Sirtuins, a family of (NAD +)-dependent deacetylases, sense levels of NAD + [85, 86, 186]; and decreased IIS delays aging from C. elegans to humans [8, 180]. The aforementioned metabolic-longevity signals converge on a limited number of transcription factors, including skn-1 (ortholog of nuclear factor erythroid 2-related factor 2, NRF2), daf-16 (FOXO), pha-4 (FOXA), mml-1, mxl-2, hsf-1 (heat shock transcription factor), hif-1 (HIF1), and hlh-30 (TFEB) [88, 109, 112, 118, 137, 161, 185, 200] and other transcriptional regulators, such as hpk-1 [39, 111]. Collectively, a growing number of C. elegans studies have begun to unravel the complex integrated networks that maintain organismal homeostasis from an extensive array of diverse extrinsic and intrinsic signals, which converge on distinct but overlapping adaptive transcriptional responses [68, 42]. All the aforementioned signal transduction pathways act acutely via monitoring metabolic state; we find that eat-2 mutant animals regulate a distinct transcriptional program through MXL-2 to maintain homeostasis under conditions of chronic metabolic stress.
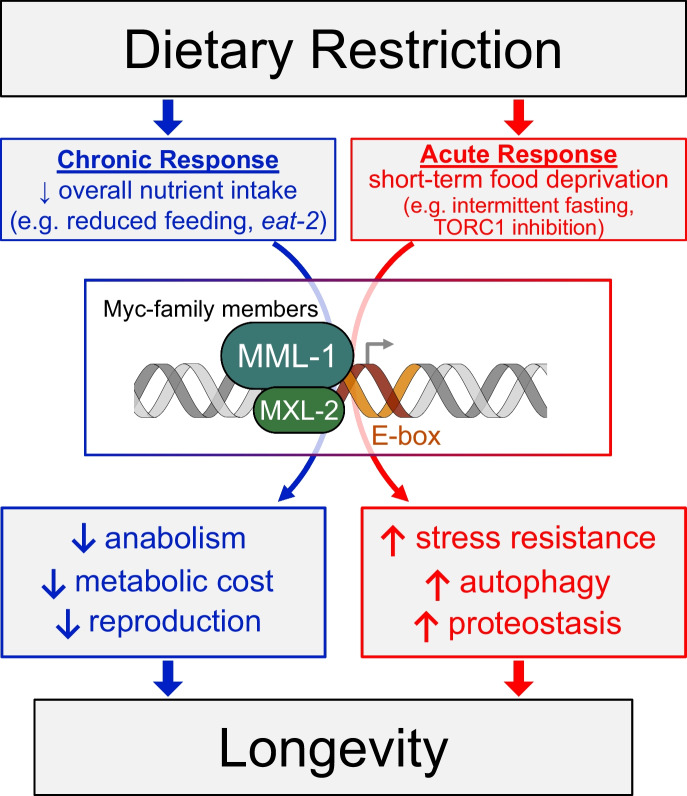
The C. elegans Myc-network is a key integration point for multiple longevity signals, including decreased IIS, decreased pharyngeal pumping and feeding of eat-2 mutants, TORC1 inhibition, glp-1 mutant germline-less animals, and HLH-30 (TFEB). All activate the MML-1::MXL-2 complex by promoting nuclear accumulation of MML-1 [88, 137, 164], consistent with known modes of regulation of mammalian MLXIP/MLXIPL and MLX [13, 43, 167]. We previously discovered that MML-1::MXL-2 complex function is necessary for the increased lifespan of not only daf-2 and eat-2 mutant animals, but also oxidative stress resistance and heat shock survival [88]. The MML-1::MXL-2 complex is also essential for the increased longevity conferred by TORC1 inhibition [111, 137]. MML-1::MXL-2 is further necessary for hpk-1, an essential component of TORC1-mediated longevity, to increase lifespan, induce autophagy, and maintain proteostasis during aging [39, 111]. Previous work characterizing stress resistance in eat-2 found that DR buffers against proteotoxic stress, specifically the accumulation of toxic aggregates [124, 165, 174], however, the eat-2 mutation does not protect against chronic mild thermal stress ([92] and this study). We also find a lack of broad stress resistance in eat-2 mutant animals, nor transcriptional changes associated with stress resistance, despite a significantly longer lifespan and healthspan. This supports the notion that the Myc-family of transcription factors not only integrate longevity signals [88, 137] by responding to acute forms of metabolic stress, but also coordinate unique signal-specific adaptive responses, which act in a key role to remodel transcription during extended periods of dietary deprivation (Fig. 7).
Fig. 7.
A model for the critical roles of Myc-family transcription factors in response to chronic and acute dietary restriction. Acute food deprivation and chronically reduced dietary intake are distinct stimuli that lead to longevity through different metabolic and physiological responses, but both require the Myc-family transcription factor complex MML-1::MXL-2 to regulate gene expression at E-box sites ([88, 137] and this work)
Supplementary Information
Below are the links to the electronic supplementary material.
Figure S1. RNA-Seq biological replicate correlations are strong, and variability is not associated with sample batches. (A) Pearson correlations for biological replicate sample are strong, and not lower than 0.847 in any case. Sample groups are organized by strain as indicated on the left side and RNAi condition as indicated on the top. All sample groups have at least two biological replicates. Histograms indicate the density of the points in the correlation dot plots. All 17,907 genes considered as expressed in any sample in the dataset are included. All correlations were significant. (B) Due to the size of the experiment, animals were prepared in six batches, indicated by the shape of the points in the PCA scatterplots. Variability between samples is not associated with these batches. All RNA-Seq libraries were sequenced at the same time to eliminate a common source of technical variation. Supplementary figure 1 (PDF 6403 KB)
Figure S2. A subset of innate immune response-associated genes are upregulated in eat-2, but not specific to DR. We observed enrichment of innate immune response Gene Ontology (GO) terms in eat-2 animals (Figure 2C), and thus further investigated expression of innate immune response-associated genes in the other samples. (A) Venn diagram indicating the number of C. elegans genes associated to GO terms for innate immune response, from annotation in WormBase (WS282). 391 genes are represented in the union of the three innate immune response terms. (B) The 391 immune response genes from (A) were intersected with the genes significantly up-regulated in any of eat-2, mxl-2, or N2 pha-4 RNAi, yielding 85 genes. The heatmap represents foldchanges for the indicated sample group compared to wild-type (N2) for these 85 genes. Red blocks on the left bar indicate GO terms associated with each gene. A subset of these genes is similarly upregulated in eat-2, mxl-2, and N2 pha-4 RNAi, and is thus not specific to DR. Supplementary file 2 (PDF 51 KB)
Figure S3. Genes with demonstrated or predicted functions in phosphorylation or dephosphorylation were significantly enriched among transcripts downregulated in eat-2. (A) We found 76 de-phosphorylation genes and 111 phosphorylation genes significantly downregulated in eat-2 based on Gene Ontology annotation. (B, C) We looked at other functions and pathways associated with these genes and represented the relative frequency of the functional terms as a treemap each for the de-phosphorylation genes (B) and phosphorylation genes (C) downregulated in eat-2. The box size represents the number of genes associated with the term relative to all the associations for the group. Only terms for which more than two genes were associated were considered, and phosphorylation cycle terms used to select the genes in (A) were removed. (D) Tissue enrichment for the phosphorylation and de-phosphorylation genes down-regulated in eat-2 from (A). Somatic tissue gene sets are based on the union of tissuespecific and tissue-enriched gene lists from cell type specific bulk RNA-Seq in (Kaletsky et al. 2018). Sperm-specific genes are from (Reinke et al. 2000). Both phosphorylation and dephosphorylation genes are significantly enriched in sperm, rather than being distinctly regulated in different somatic tissues. Numbers in brackets indicate the size of the gene set or the number of genes in the intersection. Hypergeometric test adjusted p-value significance level: ‘***’ p < 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1. Supplementary file 3 (PDF 53 KB)
Figure S4. DR down-regulation of sperm and muscle genes is dependent on mxl-2. (A.) Tissue enrichment for significantly differentially expressed in genes in eat-2, mxl-2, and pha-4 RNAi all compared to wild-type (N2 EV). We found strong enrichment of muscle- and spermassociated genes in eat-2 but not loss of mxl-2 or pha-4. (B.) Tissue enrichment for eat-2 differentially expressed genes that maintained eat-2-like expression with mxl-2 loss (“mxl-2-independent”, bottom set of rows), and mxl-2-dependent genes (middle set of rows). 394 of 395 sperm-associated genes downregulated in eat-2 were dependent on mxl-2 to maintain reduced expression in DR. Rather than returning to WT-like-levels, a substantial fraction of sperm-related genes are significantly upregulated in eat-2;mxl-2 animals (top set of rows). Somatic tissue gene sets are based on the union of tissue-specific and tissue-enriched gene lists from cell-typespecific bulk RNA-Seq from (Kaletsky et al 2018). Sperm-specific genes are from (Reinke et al 2000). Numbers in brackets indicate the size of the gene set, or the number of genes in the intersection. Adjusted p-value significance level: ‘***’ p < 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1. Supplementary file 4 (PDF 66 KB)
Figure S5. Putative high-confidence trans-activation domains (TAD) are predicted on the amino-terminal side of MML-1 and the carboxy-terminal side of MDL-1. Trans-activation domains (TADs) in C. elegans Myc-family TFs (blue headers) and their human homologs (grey headers) were predicted with ADpred; the red horizontal line in each plot indicates an ADpred score of 0.8, which was suggested as a threshold of high-confidence TAD prediction in the ADpred manuscript (Erijman et al 2020). Scores are predicted along the protein sequence with black lines: raw scores per amino acid, and blue lines: smoothed to highlight broader trends. (A.) ADpred analysis identified a high-confidence TAD from 245-276 on the amino terminal side of MML-1 between the “Mondo-conserved” domains IV and V (Ceballos, Esse, Grishok 2021) that parallels similar predicted TADs in the human homologs MLXIP and MLXIPL. (B.) There are TAD regions of lower confidence on the C-terminal side of MXL-2, which also correspond to a region of similar confidence on the C-terminal side of human homolog MLX. (C.) C. elegans MDL-1, thought to function primarily as a transcriptional repressor, harbors a high confidence predicted TAD on the C-terminal end, indicating that MDL-1 may have roles in transcriptional activation in some contexts, unlike its human counterparts. (D.) Similar to mammalian MAX, there were no predicted TADs in MXL-1 but we found a predicted TAD of lower confidence on the C-terminal side of MXL-3. Protein sequences were from WormBase for C. elegans and UniProt for human: MLXIP (Q9HAP2), MXLIPL (Q9NP71), MLX (Q9UH92), MXI1 (P50539), MXD1/MAD (Q05195), MXD3/MAD3 (Q9BW11), MXD4/MAD4 (Q14582), MAX (P61244). Supplementary file5 (PDF 88 KB)
Figure S6. Plot for additional trial for brood size assay. Loss of MXL-2 in DR but not ad libitum animals compromises fecundity and embryo viability. Additional trial data for brood size assay, analogous to the representative trial in Figure 4. Brood size assays were performed to determine how reproduction-associated gene expression changes with mxl-2 loss in eat-2 affected reproductive fitness. (A) Viable progeny from each day over the course of the reproductive lifespan were counted for singled parent hermaphrodites for (i) N2 and mxl-2, and (ii) eat-2 and eat-2;mxl-2 animals. (B) Total brood size aggregated from across the observations in (A) for each strain. (C) Unhatched eggs were counted for each strain, but only found for eat-2;mxl-2 animals. Data is shown from a representative trial. See Table S4 for complete results and statistical analysis. Stars indicate FDR-corrected p-values: *< 0.05, **< 0.01, ***< 0.005. Supplementary file6 (PDF 68 KB)
Figure S7. Key autophagy-associated genes were not differentially expressed in eat-2 but are dysregulated in eat-2;mxl-2. Heatmap of log2 foldchanges for the indicated sample group compared to wild-type (N2) for key genes involved in autophagy, with their canonical role indicated by the labeled sidebar on the left. Autophagy-related gene expression was not up-regulated in eat-2 animals but was dysregulated by loss of mxl-2 in the eat-2 background. Columns are ordered by hierarchical clustering. Significantly differentially expressed genes are indicated by red stars, adjusted pvalue significance level: ‘***’ p < 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1. Supplementary file7 (PDF 41 KB)
Figure S8. Enrichment for predicted transcription factor binding in promoters of genes differentially expressed in eat-2, mxl-2, daf-16 (RNAi) or pha-4 (RNAi). Heatmaps representing enrichment of transcription factor motifs in promoters of genes (A) downregulated or (B) up-regulated compared to wild-type. (A) Consistent with the expected binding motif of MML-1::MXL-2, the most enriched motifs for genes down-regulated under DR correspond to canonical CACGTG E-box sites and the motif for MDL-1. DAF-16 motifs are most significantly enriched for genes down-regulated with RNAi for daf-16, but we do not observe enrichment for PHA-4 predicted sites in samples with RNAi for pha-4, or in eat-2 DR conditions. E-box sites or motifs are not enriched in the genes downregulated in mxl-2 mutants, however MML-1::MXL-2 activity is low under such conditions (Johnson et al 2014). (B). An invariant set of transcription factor motifs represent those most significantly enriched across all single-perturbation conditions for up-regulated genes, corresponding to GATA-binding factors. Numbers in brackets are the size of the gene set, or the size of the intersection. Stars indicate significant enrichment results. Adjusted p-value significance level: ‘***’ p < 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1. Supplementary file8 (PDF 55 KB)
Acknowledgements
Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440), and by the National BioResource Project, Japan. We would like to acknowledge the UR Center for Integrated Research Computing (CIRC), the University of Rochester Genomics Research Center (GRC) for their resources and support. We would also like to thank members of the Biomedical Genetics department and the Western New York Worm Group for helpful discussions and feedback, particularly Dr. Doug Portman and Dr. Keith Nehrke. The oxygen consumption experiments were performed in conjunction with Brandon Berry of Dr. Andrew Wojtovich’s laboratory.
Author contribution
A.B.C. designed and conducted genomic analyses, performed survival experiments, and analyzed all experimental data. M.T. prepared animal populations and isolated RNA for RNA-sequencing. Y.Z. designed and conducted brood size experiments. J.T. advised genomic analyses and assisted with the development of the manuscript. A.V.S. oversaw the project, contributed to analysis and interpretation of results, and obtained funding. A.B.C. and A.V.S. wrote the manuscript.
Funding
Research reported in this publication was supported by the National Institute on Aging of the National Institutes of Health under Award Numbers RF1AG062593 and R01AG043421. A.B.C was also supported by a University of Rochester HSCCI fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data availability
The RNA-Seq dataset is available from the NCBI Gene Expression Omnibus (GEO) under accession GSE240821.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99. 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed]
- 2.Albert Hubbard EJ. Caenorhabditis elegans germ line: a model for stem cell biology. Dev Dyn. 2007;236:3343–57. 10.1002/dvdy.21335. 10.1002/dvdy.21335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allevato M, Bolotin E, Grossman M, et al. Sequence-specific DNA binding by MYC/MAX to low-affinity non-E-box motifs. PLoS ONE. 2017;12:1–20. 10.1371/journal.pone.0180147. 10.1371/journal.pone.0180147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 5.Apfeld J, O’Connor G, McDonagh T, et al. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes and Development. 2005;19:411. 10.1101/gad.1255404.3004. 10.1101/gad.1255404.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. 10.1093/genetics/133.4.897. 10.1093/genetics/133.4.897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar DZ, Charar C, Dorfman J, et al. Cell size and fat content of dietary-restricted Caenorhabditis elegans are regulated by ATX-2, an mTOR repressor. Proc Natl Acad Sci. 2016;113:E4620–9. 10.1073/pnas.1512156113. 10.1073/pnas.1512156113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbieri M, Bonafè M, Franceschi C, Paolisso G. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol-Endocrinol Metab. 2003;285:E1064–71. 10.1152/ajpendo.00296.2003. 10.1152/ajpendo.00296.2003 [DOI] [PubMed] [Google Scholar]
- 9.Benian GM, Epstein HF. Caenorhabditis elegans muscle: a genetic and molecular model for protein interactions in the heart. Circ Res. 2011;109:1082–95. 10.1161/CIRCRESAHA.110.237685. 10.1161/CIRCRESAHA.110.237685 [DOI] [PubMed] [Google Scholar]
- 10.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 11.Bennett CF, Vander Wende H, Simko M, et al. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat Commun. 2014;5:3483. 10.1038/ncomms4483. 10.1038/ncomms4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry BJ, Mjelde E, Carreno F, Gilham K, Hanson EJ, Na E, Kaeberlein M. Preservation of mitochondrial membrane potential is necessary for lifespan extension from dietary restriction. GeroScience. 2023;45(3):1573–81. 10.1007/s11357-023-00766-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billin AN, Ayer DE. The Mlx network: evidence for a parallel max-like transcriptional network that regulates energy metabolism. Curr Top Microbiol Immunol. 2006;302:255–78. 10.1007/3-540-32952-8_10. 10.1007/3-540-32952-8_10 [DOI] [PubMed] [Google Scholar]
- 14.Blake JA, Dolan M, Drabkin H, et al. Gene ontology annotations and resources. Nucleic Acids Res. 2013;41:530–5. 10.1093/nar/gks1050. 10.1093/nar/gks1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blumenthal T, Evans D, Link CD, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417. 10.1038/nature00831. [DOI] [PubMed]
- 16.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. 10.1093/bioinformatics/btu170. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botts MR, Cohen LB, Probert CS, et al. (2016) Microsporidia intracellular development relies on Myc interaction network transcription factors in the host. G3 Genes|Genomes|Genetics 6:2707–2716. 10.1534/g3.116.029983 [DOI] [PMC free article] [PubMed]
- 18.Boyd WA, McBride SJ, Freedman JH. Effects of genetic mutations and chemical exposures on Caenorhabditis elegans feeding: evaluation of a novel, high-throughput screening assay. PLoS ONE. 2007;2:e1259. 10.1371/journal.pone.0001259. [DOI] [PMC free article] [PubMed]
- 19.Brenner S, Brenner S. The genetics of Caenorήabditis elegans. Mol Biol. 1974;336:71–94. 10.1002/cbic.200300625. 10.1002/cbic.200300625 [DOI] [Google Scholar]
- 20.Budovskaya YV, Wu K, Southworth LK, et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. 10.1016/j.cell.2008.05.044. 10.1016/j.cell.2008.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byerly L, Cassada RC, Russell RL. The life cycle of the nematode Caenorhabditis elegans. Dev Biol. 1976;51:23–33. 10.1016/0012-1606(76)90119-6. 10.1016/0012-1606(76)90119-6 [DOI] [PubMed] [Google Scholar]
- 22.Cairo S, Merla G, Urbinati F, et al. WBSCR14, a gene mapping to the Williams – Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum Mol Genet. 2001;10:617–28. 10.1093/hmg/10.6.617 [DOI] [PubMed] [Google Scholar]
- 23.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50:D165–73. 10.1093/nar/gkab1113. 10.1093/nar/gkab1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceballos, A., Esse, R., & Grishok, A. (2021). The proline-rich domain of MML-1 is biologically important but not required for localization to target promoters. microPubl biol. 2021. [DOI] [PMC free article] [PubMed]
- 25.Chen D, Zou L (2019) GSE125718- Expression analysis of C. elegans wild-type N2, eat-2, acs-20 and eat-2; acs-20 mutants
- 26.Chen J, Caswell-Chen EP. Facultative vivipary is a life-history trait in Caenorhabditis elegans. J Nematol. 2004;36:107–13. [PMC free article] [PubMed] [Google Scholar]
- 27.Choi YJ (2020) Shedding light on the effects of calorie restriction and its mimetics on skin biology. Nutrients 12. 10.3390/nu12051529 [DOI] [PMC free article] [PubMed]
- 28.Chu DS, Liu H, Nix P, et al. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–5. 10.1038/nature05050. 10.1038/nature05050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4:1–24. 10.1101/cshperspect.a014357. 10.1101/cshperspect.a014357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornaro L. Sure and certain methods of attaining a long and healthful life. n.d.
- 31.Cornaro L. The art of living long. n.d.
- 32.Cornwell A, Llop JR, Salzman P, et al. The replica set method is a robust, accurate, and high-throughput approach for assessing and comparing lifespan in C. elegans experiments. Front Aging. 2022;3:1–21. 10.3389/fragi.2022.861701. 10.3389/fragi.2022.861701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornwell AB, Llop JR, Salzman P, Thakar J, Samuelson AV. The replica set method: a high-throughput approach to quantitatively measure Caenorhabditis elegans lifespan. JoVE (Journal of Visualized Experiments). 2018;(136):e57819. [DOI] [PMC free article] [PubMed]
- 34.Cornwell AB, Samuelson AV. Analysis of Lifespan in C. elegans: Low-and High-Throughput Approaches. Aging: Methods and protocols. 2020;7–27. [DOI] [PMC free article] [PubMed]
- 35.Cox GN, Kusch M, Edgar RS. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J Cell Biol. 1981;90:7–17. 10.1083/jcb.90.1.7. 10.1083/jcb.90.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crawford D, Libina N, Kenyon C. Caenorhabditis elegans integrates food and reproductive signals in lifespan determination. Aging Cell. 2007;6:715–21. 10.1111/j.1474-9726.2007.00327.x. 10.1111/j.1474-9726.2007.00327.x [DOI] [PubMed] [Google Scholar]
- 37.Cunningham F, Allen JE, Allen J, et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–95. 10.1093/nar/gkab1049. 10.1093/nar/gkab1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curtis R, O’Connor G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–26. 10.1111/j.1474-9726.2006.00205.x. 10.1111/j.1474-9726.2006.00205.x [DOI] [PubMed] [Google Scholar]
- 39.Das R, Melo JA, Thondamal M, et al. The homeodomain-interacting protein kinase HPK-1 preserves protein homeostasis and longevity through master regulatory control of the HSF-1 chaperone network and TORC1- restricted autophagy in Caenorhabditis elegans. PLoS Genet. 2017;1:1–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis P, Zarowiecki M, Arnaboldi V, et al. WormBase in 2022—data, processes, and tools for analyzing Caenorhabditis elegans. Genetics. 2022;220:iyac003. 10.1093/genetics/iyac003. 10.1093/genetics/iyac003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Martin X, Sodaei R, Santpere G. Mechanisms of binding specificity among bHLH transcription factors. IJMS. 2021;22:9150. 10.3390/ijms22179150. 10.3390/ijms22179150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denzel MS, Lapierre LR, Mack HID (2018) Emerging topics in C. elegans aging research: transcriptional regulation, stress response and epigenetics. Mech Ageing Dev 1–18. 10.1016/j.mad.2018.08.001 [DOI] [PMC free article] [PubMed]
- 43.Diolaiti D, McFerrin L, Carroll PA, Eisenman RN. Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim Biophys Acta - Gene Regul Mech. 2015;1849:484–500. 10.1016/j.bbagrm.2014.05.016. 10.1016/j.bbagrm.2014.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. 10.1093/bioinformatics/bts635. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dues DJ, Andrews EK, Schaar CE, et al (2016) Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways. Aging 8:777–795. 10.18632/aging.100939 [DOI] [PMC free article] [PubMed]
- 46.Dues DJ, Andrews EK, Senchuk MM, Van Raamsdonk JM. Resistance to stress can be experimentally dissociated from longevity. J Gerontol: Series A. 2019;74:1206–14. 10.1093/gerona/gly213. 10.1093/gerona/gly213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duregon E, Pomatto-Watson LCDD, Bernier M, et al. Intermittent fasting: from calories to time restriction. GeroScience. 2021;43:1083–92. 10.1007/s11357-021-00335-z. 10.1007/s11357-021-00335-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edwards C, Canfield J, Copes N, et al. Mechanisms of amino acid-mediated lifespan extension in Caenorhabditis elegans. BMC Genet. 2015;16:8. 10.1186/s12863-015-0167-2. 10.1186/s12863-015-0167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards CB, Copes N, Brito AG, et al. Malate and fumarate extend lifespan in Caenorhabditis elegans. PLoS ONE. 2013;8. 10.1371/journal.pone.0058345. [DOI] [PMC free article] [PubMed]
- 50.Ellis RE, Stanfield GM (2014) The regulation of spermatogenesis and sperm function in nematodes. Semin Cell Dev Biol 29. 10.1016/j.semcdb.2014.04.005 [DOI] [PMC free article] [PubMed]
- 51.Erijman A, Kozlowski L, Sohrabi-Jahromi S, et al. A high-throughput screen for transcription activation domains reveals their sequence features and permits prediction by deep learning. Mol Cell. 2020;78:890-902.e6. 10.1016/j.molcel.2020.04.020. 10.1016/j.molcel.2020.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Essmann CL, Martinez-Martinez D, Pryor R, et al (2020) Mechanical properties measured by atomic force microscopy define health biomarkers in ageing C. elegans. Nat Commun 11. 10.1038/s41467-020-14785-0 [DOI] [PMC free article] [PubMed]
- 53.Ewald CY, Landis JN, Abate JP, et al. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature. 2015;519:97–101. 10.1038/nature14021. 10.1038/nature14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fontana L, Partridge L, Longo VD. Extending healthy life span - from yeast to humans. Science. 2010;328:321–6. 10.1126/science.1172539. 10.1126/science.1172539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraga D, Aryal M, Hall JE, et al. Characterization of the arginine kinase isoforms in Caenorhabditis elegans. Comp Biochem Physiol B: Biochem Mol Biol. 2015;187:85–101. 10.1016/j.cbpb.2015.05.002. 10.1016/j.cbpb.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 56.Fuxman Bass JI, Pons C, Kozlowski L, et al. (2016) A gene-centered C. elegans protein–DNA interaction network provides a framework for functional predictions. Molec Syst Biol 12:1–19. 10.15252/msb.20167131 [DOI] [PMC free article] [PubMed]
- 57.Galbadage T, Hartman PS. Repeated temperature fluctuation extends the life span of Caenorhabditis elegans in a daf-16-dependent fashion. Mech Ageing Dev. 2008;129:507–14. 10.1016/j.mad.2008.04.012. 10.1016/j.mad.2008.04.012 [DOI] [PubMed] [Google Scholar]
- 58.Gao AW, Smith RL, Weeghel MV, et al. Identification of key pathways and metabolic fingerprints of longevity in C. elegans. Exp Gerontol. 2018;113:128–40. 10.1101/222554. 10.1101/222554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao AW, uit de Bos J, Sterken MG, et al. Forward and reverse genetics approaches to uncover metabolic aging pathways in Caenorhabditis elegans. Biochim Biophys Acta - Molec Basis Dis. 2017;0–1. 10.1016/j.bbadis.2017.09.006. [DOI] [PubMed]
- 60.Gelino S, Chang JT, Kumsta C, et al. Intestinal autophagy improves healthspan and longevity in C. elegans during dietary restriction. PLoS Genet. 2016;12:1–24. 10.1371/journal.pgen.1006135. 10.1371/journal.pgen.1006135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerstein MB, Lu ZJ, Van Nostrand EL, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Sci. 2010;330:1775–87. 10.1126/science.1196914. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–12. 10.1016/S1097-2765(04)00030-9. 10.1016/S1097-2765(04)00030-9 [DOI] [PubMed] [Google Scholar]
- 63.Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, D’Eustachio P. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50(D1):D687–92. 10.1093/nar/gkab1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glenwinkel L, Taylor SR, Langebeck-Jensen K, et al. In silico analysis of the transcriptional regulatory logic of neuronal identity specification throughout the C. elegans nervous system. Life. 2021;10:1–29. 10.7554/eLife.64906. 10.7554/eLife.64906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gouvea DY, Aprison EZ, Ruvinsky I (2015) Experience modulates the reproductive response to heat stress in C. elegans via multiple physiological processes. 1–27. 10.1371/journal.pone.0145925 [DOI] [PMC free article] [PubMed]
- 66.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. 10.1093/bioinformatics/btr064. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Green CL, Lamming DW, Fontana L. Molecular mechanisms of dietary restriction promoting health and longevity. Nat Rev Mol Cell Biol. 2022;23:56–73. 10.1038/s41580-021-00411-4. 10.1038/s41580-021-00411-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–27. 10.1111/j.1474-9726.2009.00459.x. 10.1111/j.1474-9726.2009.00459.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greer EL, Dowlatshahi D, Banko MR, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–56. 10.1016/j.cub.2007.08.047. 10.1016/j.cub.2007.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grove CA, De Masi F, Barrasa MI, et al. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–27. 10.1016/j.cell.2009.04.058. 10.1016/j.cell.2009.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hamilton B, Dong Y, Shindo M, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–55. 10.1101/gad.1308205. 10.1101/gad.1308205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen M, Chandra A, Mitic LL, et al. (2008) A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4. 10.1371/journal.pgen.0040024 [DOI] [PMC free article] [PubMed]
- 73.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:0119–28. 10.1371/journal.pgen.0010017. 10.1371/journal.pgen.0010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hansen M, Taubert S, Crawford D, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. 10.1111/j.1474-9726.2006.00267.x. 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- 75.Harris JE, Govindan JA, Yamamoto I, et al. Major sperm protein signaling promotes oocyte microtubule reorganization prior to fertilization in Caenorhabditis elegans. Dev Biol. 2006;299:105–21. 10.1016/j.ydbio.2006.07.013. 10.1016/j.ydbio.2006.07.013 [DOI] [PubMed] [Google Scholar]
- 76.Harris TW, Antoshechkin I, Bieri T, et al. Wormbase: a comprehensive resource for nematode research. Nucleic Acids Res. 2009;38:463–7. 10.1093/nar/gkp952. 10.1093/nar/gkp952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Havula E, Hietakangas V. Sugar sensing by ChREBP/Mondo-Mlx — new insight into downstream regulatory networks and integration of nutrient-derived signals. Curr Opin Cell Biol. 2018;51:89–96. 10.1016/j.ceb.2017.12.007. 10.1016/j.ceb.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 78.Heestand BN, Shen Y, Liu W, et al. Dietary restriction induced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003651. 10.1371/journal.pgen.1003651. [DOI] [PMC free article] [PubMed]
- 79.Hofmann JW, Zhao X, De Cecco M, et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell. 2015;160:477–88. 10.1016/j.cell.2014.12.016. 10.1016/j.cell.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horowitz BB, Nanda S, Walhout AJM (2023) A transcriptional cofactor regulatory network for the C. elegans intestine. G3 (Bethesda) 13:jkad096. 10.1093/g3journal/jkad096 [DOI] [PMC free article] [PubMed]
- 81.Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. No reduction of metabolic rate in food restricted Caenorhabditis elegans. Exp Gerontol. 2002;37(12):1359–69. 10.1016/S0531-5565(02)00172-9 [DOI] [PubMed] [Google Scholar]
- 82.Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: New tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015;43:D117–22. 10.1093/nar/gku1045. 10.1093/nar/gku1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Il’yasova D, Fontana L, Bhapkar M, et al. Effects of 2 years of caloric restriction on oxidative status assessed by urinary F2-isoprostanes: the CALERIE 2 randomized clinical trial. Aging Cell. 2018;17:1–9. 10.1111/acel.12719. 10.1111/acel.12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Izaurralde E. Directing mRNA export. Nat Struct Mol Biol. 2004;11:210–2. 10.1038/nsmb0304-210. 10.1038/nsmb0304-210 [DOI] [PubMed] [Google Scholar]
- 85.Jedrusik-Bode M. C. elegans sirtuin SIR-24 and its mammalian homolog SIRT6 in stress response. Worm. 2014;3:e29102. 10.4161/worm.29102. 10.4161/worm.29102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jedrusik-Bode M, Studencka M, Smolka C, et al (2013) The sirtuin SIRT6 regulates stress granules formation in C. elegans and in mammals. J Cell Sci jcs.130708. 10.1242/jcs.130708 [DOI] [PubMed]
- 87.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–906. 10.1242/dev.01255. 10.1242/dev.01255 [DOI] [PubMed] [Google Scholar]
- 88.Johnson DW, Llop JR, Farrell SF, et al. The Caenorhabditis elegans Myc-Mondo/Mad complexes integrate diverse longevity signals. PLoS Genet. 2014;10: e1004278. 10.1371/journal.pgen.1004278. 10.1371/journal.pgen.1004278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johnson T. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Sci. 1990;249:908–12. 10.1126/science.2392681. 10.1126/science.2392681 [DOI] [PubMed] [Google Scholar]
- 90.Johnson TE, Wood WB. Genetic analysis of life-span in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1982;79:6603–7. 10.1073/pnas.79.21.6603. 10.1073/pnas.79.21.6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnstone IL. Cuticle collagen genes. Expression in Caenorhabditis elegans Trends Genet. 2000;16:21–7. 10.1016/S0168-9525(99)01857-0 [DOI] [PubMed] [Google Scholar]
- 92.Kaeberlein TL, Smith ED, Tsuchiya M, et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–94. 10.1111/j.1474-9726.2006.00238.x. 10.1111/j.1474-9726.2006.00238.x [DOI] [PubMed] [Google Scholar]
- 93.Kaletsky R, Yao V, Williams A, et al. Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 2018;14. 10.1371/journal.pgen.1007559. [DOI] [PMC free article] [PubMed]
- 94.Kamath RS, Martinez-Campos M, Zipperlen P, et al. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2000;2(research0002):1–10. 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed]
- 95.Kanehisa M, Goto S, Furumichi M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–60. 10.1093/nar/gkp896. 10.1093/nar/gkp896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kapahi P, Chen D, Rogers AN, et al. With TOR, Less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–65. 10.1016/j.cmet.2010.05.001. 10.1016/j.cmet.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kapahi P, Kaeberlein M, Hansen M. Dietary restriction and lifespan: lessons from invertebrate models. Ageing Res Rev. 2017;39:3–14. 10.1016/j.arr.2016.12.005. 10.1016/j.arr.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cell Mol Life Sci. 2007;64:1323–8. 10.1007/s00018-007-6470-y. 10.1007/s00018-007-6470-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kenyon C, Chang J, Gensch E, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. 10.1038/366461a0. 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- 100.Kim W, Underwood RS, Greenwald I, Shaye DD. Ortholist 2: a new comparative genomic analysis of human and Caenorhabditis elegans genes. Genet. 2018;210:445–61. 10.1534/genetics.118.301307. 10.1534/genetics.118.301307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kniazeva M, Crawford QT, Seiber M, et al. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2004;2. 10.1371/journal.pbio.0020257. [DOI] [PMC free article] [PubMed]
- 102.Korta DZ, Tuck S, Hubbard EJA. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development. 2012;139:859–70. 10.1242/dev.074047. 10.1242/dev.074047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kraus WE, Bhapkar M, Huffman KM, et al. 2 years of calorie restriction and cardiometabolic risk (CALERIE): exploratory outcomes of a multicentre, phase 2, randomised controlled trial. Lancet Diabetes Endocrinol. 2019;7:673–83. 10.1016/S2213-8587(19)30151-2. 10.1016/S2213-8587(19)30151-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krittika S, Yadav P. An overview of two decades of diet restriction studies using Drosophila. Biogerontology. 2019;20:723–40. 10.1007/s10522-019-09827-0. 10.1007/s10522-019-09827-0 [DOI] [PubMed] [Google Scholar]
- 105.Kumar S, Egan BM, Kocsisova Z, et al. Lifespan extension in C. elegans caused by bacterial colonization of the intestine and subsequent activation of an innate immune response. Dev Cell. 2019;49:100-117.e6. 10.1016/j.devcel.2019.03.010. 10.1016/j.devcel.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Labbadia J, Morimoto RI. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol Cell. 2015;59:639–50. 10.1016/j.molcel.2015.06.027. 10.1016/j.molcel.2015.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lahoz EG, Xu L, Schreiber-Agus N, DePinho RA. Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1. Proc Natl Acad Sci U S A. 1994;91:5503–7. 10.1073/pnas.91.12.5503. 10.1073/pnas.91.12.5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–6. 10.1073/pnas.95.22.13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lapierre LR, De Magalhaes Filho CD, McQuary PR, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:1–8. 10.1038/ncomms3267. 10.1038/ncomms3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lapierre LR, Kumsta C, Sandri M, et al. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11:867–80. 10.1080/15548627.2015.1034410. 10.1080/15548627.2015.1034410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lazaro-Pena MI, Cornwell AB, Diaz-Balzac CA, et al. Homeodomain-interacting protein kinase maintains neuronal homeostasis during normal Caenorhabditis elegans aging and systemically regulates longevity from serotonergic and GABAergic neurons. Elife. 2023;12:e85792. 10.7554/eLife.85792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lazaro-Pena MI, Ward ZC, Yang S, et al. HSF-1: guardian of the proteome through integration of longevity signals to the proteostatic network. Front Aging. 2022;3: 861686. 10.3389/fragi.2022.861686. 10.3389/fragi.2022.861686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20:2131–6. 10.1016/j.cub.2010.10.057. 10.1016/j.cub.2010.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leroux MR, Melki R, Gordon B, et al. Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides. J Biol Chem. 1997;272:24646–56. 10.1074/jbc.272.39.24646. 10.1074/jbc.272.39.24646 [DOI] [PubMed] [Google Scholar]
- 115.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. 10.1186/1471-2105-12-323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. 10.1093/bioinformatics/btt656. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 117.Lim YP, Go MK, Raida M, et al. (2018) Synthetic enzymology and the fountain of youth: repurposing biology for longevity. ACS Omega 3. 10.1021/acsomega.8b01620 [DOI] [PMC free article] [PubMed]
- 118.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Sci. 1997;278:1319–22. 10.1126/science.278.5341.1319. 10.1126/science.278.5341.1319 [DOI] [PubMed] [Google Scholar]
- 119.Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Curr Opin Cell Biol. 2016;39:61–8. 10.1016/j.ceb.2016.02.010. 10.1016/j.ceb.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. 10.1186/s13059-014-0550-8. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Luo S, Shaw WM, Ashraf J, Murphy CT (2009) TGF-ß Sma/Mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genet 5. 10.1371/journal.pgen.1000789 [DOI] [PMC free article] [PubMed]
- 122.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–54. 10.1146/annurev.biochem.77.061206.171059. 10.1146/annurev.biochem.77.061206.171059 [DOI] [PubMed] [Google Scholar]
- 123.Mair W, Panowski SH, Shaw RJ, Dillin A (2009) Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS ONE 4. 10.1371/journal.pone.0004535 [DOI] [PMC free article] [PubMed]
- 124.Matai L, Sarkar GC, Chamoli M, et al. Dietary restriction improves proteostasis and increases life span through endoplasmic reticulum hormesis. Proc Natl Acad Sci U S A. 2019;116:17383–92. 10.1073/pnas.1900055116. 10.1073/pnas.1900055116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mattison JA, Colman RJ, Beasley TM, et al. Caloric restriction improves health and survival of rhesus monkeys. Nat Commun. 2017;8:14063. 10.1038/ncomms14063. 10.1038/ncomms14063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McFerrin LG, Atchley WR. Evolution of the max and MlX networks in animals. Genome Biol Evol. 2011;3:915–37. 10.1093/gbe/evr082. 10.1093/gbe/evr082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mcferrin LG, Atchley WR (2012) A novel N-terminal domain may dictate the glucose response of Mondo proteins. 7:1–16.10.1371/journal.pone.0034803 [DOI] [PMC free article] [PubMed]
- 128.Mckay JP, Raizen DM, Gottschalk A, et al. eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genet. 2004;166:161–9. 10.1534/genetics.166.1.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McQuary PR, Liao C-Y, Chang JT, et al. C. elegans S6K mutants require a creatine-kinase-like effector for lifespan extension. Cell Rep. 2016;14:2059–67. 10.1016/j.celrep.2016.02.012. 10.1016/j.celrep.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meléndez A, Levine B (2009) Autophagy in C. elegans. WormBook: the online review of C elegans biology 1–26 [DOI] [PubMed]
- 131.Mesbahi H, Pho KB, Tench AJ, et al. Cuticle collagen expression is regulated in response to environmental stimuli by the GATA transcription factor ELT-3 in Caenorhabditis elegans. Genet. 2020;215:483–95. 10.1534/genetics.120.303125. 10.1534/genetics.120.303125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miller HA, Dean ES, Pletcher SD, Leiser SF. Cell non-autonomous regulation of health and longevity. eLife. 2020;9:e62659. 10.7554/eLife.62659. 10.7554/eLife.62659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mouchiroud L, Molin L, Mergoud-dit-lamarche A. Metabolomics analysis uncovers that dietary restriction buffers metabolic changes associated with aging in Caenorhabditis elegans. J Proteome Res. 2015;13:2910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mukhopadhyay A, Oh SW, Tissenbaum HA. Worming pathways to and from DAF-16/FOXO. Exp Gerontol. 2006;41:928–34. 10.1016/j.exger.2006.05.020. 10.1016/j.exger.2006.05.020 [DOI] [PubMed] [Google Scholar]
- 135.Murphy CT, McCarroll S, Bargmann CI, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–83. 10.1038/nature01789. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- 136.Nakagawa S, Lagisz M, Hector KL, Spencer HG. Comparative and meta-analytic insights into life extension via dietary restriction. Aging Cell. 2012;11:401–9. 10.1111/j.1474-9726.2012.00798.x. 10.1111/j.1474-9726.2012.00798.x [DOI] [PubMed] [Google Scholar]
- 137.Nakamura S, Karalay Ö, Jäger PS, et al. Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nat Commun. 2016;7:10944. 10.1038/ncomms10944. 10.1038/ncomms10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ogg S, Paradis S, Gottlieb S, et al. The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–9. 10.1038/40194. 10.1038/40194 [DOI] [PubMed] [Google Scholar]
- 139.Olsen A, Vantipalli MC, Lithgow GJ. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 2006;312:1381–5. 10.1126/science.1124981. 10.1126/science.1124981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.O’Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013;15:668–76. 10.1038/ncb2741. 10.1038/ncb2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pandit A, Jain V, Kumar N, Mukhopadhyay A. PHA-4/FOXA-regulated microRNA feed forward loops during Caenorhabditis elegans dietary restriction. Aging. 2014;6:835–55. 10.18632/aging.100697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pang S, Lynn DA, Lo JY, et al. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat Commun. 2014;5:5048. 10.1038/ncomms6048. 10.1038/ncomms6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Panowski SH, Wolff S, Aguilaniu H, et al. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–5. 10.1038/nature05837. 10.1038/nature05837 [DOI] [PubMed] [Google Scholar]
- 144.Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:169–73. 10.1093/nar/gkt393. 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pickett CL, Breen KT, Ayer DE. A C. elegans Myc-like network cooperates with semaphorin and Wnt signaling pathways to control cell migration. Dev Biol. 2007;310:226–39. 10.1016/j.ydbio.2007.07.034. 10.1016/j.ydbio.2007.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.R Core Team R. R: A language and environment for statistical computing. 2013. [Google Scholar]
- 147.Rahimi M, Sohrabi S, Murphy CT. Novel elasticity measurements reveal C. elegans cuticle stiffens with age and in a long-lived mutant. Biophys J. 2022;121:515–24. 10.1016/j.bpj.2022.01.013. 10.1016/j.bpj.2022.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Reinke V, Smith HE, Nance J, et al. (2000) A global profile of germline gene expression in C. elegans. Molec Cell 6. 10.1016/S1097-2765(00)00059-9 [DOI] [PubMed]
- 149.Richards P, Ourabah S, Montagne J, et al (2017) MondoA/ChREBP: the usual suspects of transcriptional glucose sensing; implication in pathophysiology. Metabol: Clin Experiment 70:133–151. 10.1016/j.metabol.2017.01.033 [DOI] [PubMed]
- 150.Rodríguez-Palero MJ, López-Díaz A, Marsac R, et al. An automated method for the analysis of food intake behaviour in Caenorhabditis elegans. Sci Rep. 2018;8:1–10. 10.1038/s41598-018-21964-z. 10.1038/s41598-018-21964-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saito TL, Hashimoto SI, Gu SG, et al. The transcription start site landscape of C. elegans. Genome Res. 2013;23:1348–61. 10.1101/gr.151571.112. 10.1101/gr.151571.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Salminen A, Kaarniranta K. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res Rev. 2012;11:230–41. 10.1016/j.arr.2011.12.005. 10.1016/j.arr.2011.12.005 [DOI] [PubMed] [Google Scholar]
- 153.Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–94. 10.1101/gad.1588907. 10.1101/gad.1588907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sanborn AL, Yeh BT, Feigerle JT, et al. Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to mediator. eLife. 2021;10:1–42. 10.7554/ELIFE.68068. 10.7554/ELIFE.68068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sandhu A, Badal D, Sheokand R, et al. Specific collagens maintain the cuticle permeability barrier in Caenorhabditis elegans. Genet. 2021;217. 10.1093/GENETICS/IYAA047. [DOI] [PMC free article] [PubMed]
- 156.Scharf A, Pohl F, Egan BM, et al. Reproductive aging in Caenorhabditis elegans: from molecules to ecology. Front Cell Dev Biol. 2021;9:718522. 10.3389/fcell.2021.718522 [DOI] [PMC free article] [PubMed]
- 157.Schreiber-Agus N, Chin L, Chen K, et al. Evolutionary relationships and functional conservation among vertebrate Max-associated proteins: the zebra fish homolog of Mxi1. Oncogene. 1994;9:3167–77. [PubMed] [Google Scholar]
- 158.Seo K, Choi E, Lee D, et al. Heat shock factor 1 mediates the longevity conferred by inhibition of TOR and insulin/IGF-1 signaling pathways in C. elegans. Aging Cell. 2013;12:1073–81. 10.1111/acel.12140. 10.1111/acel.12140 [DOI] [PubMed] [Google Scholar]
- 159.Sharples AP, Hughes DC, Deane CS, et al. Longevity and skeletal muscle mass: the role of IGF signalling, the sirtuins, dietary restriction and protein intake. Aging Cell. 2015;14:511–23. 10.1111/acel.12342. 10.1111/acel.12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shaw WM, Luo S, Landis J, et al. The C. elegans TGF-B Dauer pathway regulates longevity via insulin signaling. Curr Biol. 2007;17:1635–45. 10.1016/j.cub.2007.08.058. 10.1016/j.cub.2007.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sheaffer KL, Updike DL, Mango SE. The target of rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–64. 10.1016/j.cub.2008.07.097. 10.1016/j.cub.2008.07.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shih HM, Liu Z, Towle HC. Two CACGTG motifs with proper spacing dictate the carbohydrate regulation of hepatic gene transcription. J Biol Chem. 1995;270:21991–7. 10.1074/jbc.270.37.21991. 10.1074/jbc.270.37.21991 [DOI] [PubMed] [Google Scholar]
- 163.Shimabukuro K, Roberts TM. Major sperm protein and sperm locomotion. In: Lennarz WJ, Lane MD, editors. Encyclopedia of biological chemistry. Academic Press; 2013. [Google Scholar]
- 164.Shioda T, Takahashi I, Ikenaka K, et al. Neuronal MML-1/MXL-2 regulates systemic aging via glutamate transporter and cell nonautonomous autophagic and peroxidase activity. Proc Natl Acad Sci USA. 2023;120: e2221553120. 10.1073/pnas.2221553120. 10.1073/pnas.2221553120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shpigel N, Shemesh N, Kishner M, Ben-Zvi A. Dietary restriction and gonadal signaling differentially regulate post-development quality control functions in Caenorhabditis elegans. Aging Cell. 2019;18:e12891. 10.1111/acel.12891. [DOI] [PMC free article] [PubMed]
- 166.Singson A. Every sperm is sacred: fertilization in Caenorhabditis elegans. Dev Biol. 2001;230:101–9. 10.1006/dbio.2000.0118. 10.1006/dbio.2000.0118 [DOI] [PubMed] [Google Scholar]
- 167.Sloan EJ, Ayer DE. Myc, Mondo, and metabolism. Genes Cancer. 2010;1:587–96. 10.1177/1947601910377489. 10.1177/1947601910377489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. 10.1126/science.273.5271.59. 10.1126/science.273.5271.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Son HG, Altintas O, Kim EJE, et al. Age-dependent changes and biomarkers of aging in Caenorhabditis elegans. Aging Cell. 2019;18:1–11. 10.1111/acel.12853. 10.1111/acel.12853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Soo SK, Traa A, Rudich ZD, et al. (2023) Genetic basis of enhanced stress resistance in long‐lived mutants highlights key role of innate immunity in determining longevity. Aging Cell 22. 10.1111/acel.13740 [DOI] [PMC free article] [PubMed]
- 171.Soukas AA, Kane EA, Carr CE, et al. Rictor / TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. 10.1101/gad.1775409.2004. 10.1101/gad.1775409.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Soultoukis GA, Partridge L. Dietary protein, metabolism, and aging. Annu Rev Biochem. 2016;85:5–34. 10.1146/annurev-biochem-060815-014422. 10.1146/annurev-biochem-060815-014422 [DOI] [PubMed] [Google Scholar]
- 173.Souza Matos M, Platt B, Delibegovic M. Effects of dietary restriction on metabolic and cognitive health. Proc Nutr Soc. 2021;80:126–38. 10.1017/S0029665120007910. 10.1017/S0029665120007910 [DOI] [PubMed] [Google Scholar]
- 174.Steinkraus KA, Smith ED, Davis C, et al. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. 2008;7:394–404. 10.1111/j.1474-9726.2008.00385.x. 10.1111/j.1474-9726.2008.00385.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stekovic S, Hofer SJ, Tripolt N, et al (2019) Alternate day fasting improves physiological and molecular markers of aging in healthy, non-obese humans. Cell Metabolism 1–15. 10.1016/j.cmet.2019.07.016 [DOI] [PubMed]
- 176.Stroustrup N, Ulmschneider BE, Nash ZM, et al (2014) The C. elegans lifespan machine. 10:665–67010.1038/nmeth.2475.The [DOI] [PMC free article] [PubMed]
- 177.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56. 10.1016/0012-1606(77)90158-0. [DOI] [PubMed]
- 178.Tabrez SS, Sharma RD, Jain V, et al. Differential alternative splicing coupled to nonsense-mediated decay of mRNA ensures dietary restriction-induced longevity. Nat Commun. 2017;8:306. 10.1038/s41467-017-00370-5. 10.1038/s41467-017-00370-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tacutu R, Thornton D, Johnson E, et al. Human ageing genomic resources: new and updated databases. Nucleic Acids Res. 2018;46:D1083–90. 10.1093/nar/gkx1042. 10.1093/nar/gkx1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life span. Annu Rev Physiol. 2008;70:191–212. 10.1146/annurev.physiol.70.113006.100533. 10.1146/annurev.physiol.70.113006.100533 [DOI] [PubMed] [Google Scholar]
- 181.Templeman NM, Murphy CT. Regulation of reproduction and longevity by nutrient-sensing pathways. J Cell Biol. 2018;217:93–106. 10.1083/jcb.201707168. 10.1083/jcb.201707168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tepper RG, Ashraf J, Kaletsky R, et al. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–90. 10.1016/j.cell.2013.07.006. 10.1016/j.cell.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Timmons L, Fire A. Specific interference by ingested dsRNA [10]. Nature. 1998;395:854. 10.1038/27579. 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- 184.Tóth ML, Sigmond T, Borsos É, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;8627. 10.4161/auto.5618. [DOI] [PubMed]
- 185.Tullet JMA, Hertweck M, An JH, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–38. 10.1016/j.cell.2008.01.030. 10.1016/j.cell.2008.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Verdin E. NAD + in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–13. 10.1126/science.aac4854. 10.1126/science.aac4854 [DOI] [PubMed] [Google Scholar]
- 187.Vieira AFC, Xatse MA, Tifeki H, et al. Monomethyl branched-chain fatty acids are critical for Caenorhabitis elegans survival in elevated glucose conditions. J Biol Chem. 2022;298:101444. 10.1016/j.jbc.2021.101444 [DOI] [PMC free article] [PubMed]
- 188.von Frieling J, Roeder T. Factors that affect the translation of dietary restriction into a longer life. IUBMB Life. 2020;72:814–24. 10.1002/iub.2224. 10.1002/iub.2224 [DOI] [PubMed] [Google Scholar]
- 189.Vos MJ, Carra S, Kanon B, et al. Specific protein homeostatic functions of small heat-shock proteins increase lifespan. Aging Cell. 2016;15:217–26. 10.1111/acel.12422. 10.1111/acel.12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Weir HJ, Yao P, Huynh FK, et al. Dietary restriction and AMPK increase lifespan via mitochondrial network and peroxisome remodeling. Cell Metab. 2017;26:884-896.e5. 10.1016/j.cmet.2017.09.024. 10.1016/j.cmet.2017.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–92. 10.1073/pnas.0801030105. 10.1073/pnas.0801030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang W, Dierking K, Schulenburg H. WormExp: a web-based application for a Caenorhabditis elegans-specific gene expression enrichment analysis. Bioinformatics (Oxford, England). 2016;32:943–5. 10.1093/bioinformatics/btv667. 10.1093/bioinformatics/btv667 [DOI] [PubMed] [Google Scholar]
- 193.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. 10.1186/gb-2010-11-2-r14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yuan J, Tirabassi RS, Bush AB, Cole MD. The C. elegans MDL-1 and MXL-1 proteins can functionally substitute for vertebrate MAD and MAX. Oncogene. 1998;17:1109–18. 10.1038/sj.onc.1202036. 10.1038/sj.onc.1202036 [DOI] [PubMed] [Google Scholar]
- 195.Yuan Y, Kadiyala CS, Ching T, et al. Enhanced energy metabolism contributes to the extended life span of calorie-restricted Caenorhabditis elegans. J Biol Chem. 2012;287:31414–26. 10.1074/jbc.M112.377275. 10.1074/jbc.M112.377275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–41. 10.1101/gad.176826.111. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. 10.1016/j.gde.2015.12.003. 10.1016/j.gde.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zarse K, Schmeisser S, Groth M, et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–65. 10.1016/j.cmet.2012.02.013. 10.1016/j.cmet.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zečić A, Dhondt I, Braeckman BP. The nutritional requirements of Caenorhabditis elegans. Genes Nutr. 2019;14. 10.1186/s12263-019-0637-7. [DOI] [PMC free article] [PubMed]
- 200.Zhang Y, Shao Z, Zhai Z, et al. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS ONE. 2009;4:e6348. 10.1371/journal.pone.0006348. 10.1371/journal.pone.0006348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhao Y, Wang H, Poole RJ, Gems D. A fln-2 mutation affects lethal pathology and lifespan in C. elegans. Nat Commun. 2019;10:1–10. 10.1038/s41467-019-13062-z. 10.1038/s41467-019-13062-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhong M, Niu W, Lu ZJ, et al. In: Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4 / FOXA in development and environmental response, vol. 6. 2010. 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. RNA-Seq biological replicate correlations are strong, and variability is not associated with sample batches. (A) Pearson correlations for biological replicate sample are strong, and not lower than 0.847 in any case. Sample groups are organized by strain as indicated on the left side and RNAi condition as indicated on the top. All sample groups have at least two biological replicates. Histograms indicate the density of the points in the correlation dot plots. All 17,907 genes considered as expressed in any sample in the dataset are included. All correlations were significant. (B) Due to the size of the experiment, animals were prepared in six batches, indicated by the shape of the points in the PCA scatterplots. Variability between samples is not associated with these batches. All RNA-Seq libraries were sequenced at the same time to eliminate a common source of technical variation. Supplementary figure 1 (PDF 6403 KB)
Figure S2. A subset of innate immune response-associated genes are upregulated in eat-2, but not specific to DR. We observed enrichment of innate immune response Gene Ontology (GO) terms in eat-2 animals (Figure 2C), and thus further investigated expression of innate immune response-associated genes in the other samples. (A) Venn diagram indicating the number of C. elegans genes associated to GO terms for innate immune response, from annotation in WormBase (WS282). 391 genes are represented in the union of the three innate immune response terms. (B) The 391 immune response genes from (A) were intersected with the genes significantly up-regulated in any of eat-2, mxl-2, or N2 pha-4 RNAi, yielding 85 genes. The heatmap represents foldchanges for the indicated sample group compared to wild-type (N2) for these 85 genes. Red blocks on the left bar indicate GO terms associated with each gene. A subset of these genes is similarly upregulated in eat-2, mxl-2, and N2 pha-4 RNAi, and is thus not specific to DR. Supplementary file 2 (PDF 51 KB)
Figure S3. Genes with demonstrated or predicted functions in phosphorylation or dephosphorylation were significantly enriched among transcripts downregulated in eat-2. (A) We found 76 de-phosphorylation genes and 111 phosphorylation genes significantly downregulated in eat-2 based on Gene Ontology annotation. (B, C) We looked at other functions and pathways associated with these genes and represented the relative frequency of the functional terms as a treemap each for the de-phosphorylation genes (B) and phosphorylation genes (C) downregulated in eat-2. The box size represents the number of genes associated with the term relative to all the associations for the group. Only terms for which more than two genes were associated were considered, and phosphorylation cycle terms used to select the genes in (A) were removed. (D) Tissue enrichment for the phosphorylation and de-phosphorylation genes down-regulated in eat-2 from (A). Somatic tissue gene sets are based on the union of tissuespecific and tissue-enriched gene lists from cell type specific bulk RNA-Seq in (Kaletsky et al. 2018). Sperm-specific genes are from (Reinke et al. 2000). Both phosphorylation and dephosphorylation genes are significantly enriched in sperm, rather than being distinctly regulated in different somatic tissues. Numbers in brackets indicate the size of the gene set or the number of genes in the intersection. Hypergeometric test adjusted p-value significance level: ‘***’ p < 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1. Supplementary file 3 (PDF 53 KB)
Figure S4. DR down-regulation of sperm and muscle genes is dependent on mxl-2. (A.) Tissue enrichment for significantly differentially expressed in genes in eat-2, mxl-2, and pha-4 RNAi all compared to wild-type (N2 EV). We found strong enrichment of muscle- and spermassociated genes in eat-2 but not loss of mxl-2 or pha-4. (B.) Tissue enrichment for eat-2 differentially expressed genes that maintained eat-2-like expression with mxl-2 loss (“mxl-2-independent”, bottom set of rows), and mxl-2-dependent genes (middle set of rows). 394 of 395 sperm-associated genes downregulated in eat-2 were dependent on mxl-2 to maintain reduced expression in DR. Rather than returning to WT-like-levels, a substantial fraction of sperm-related genes are significantly upregulated in eat-2;mxl-2 animals (top set of rows). Somatic tissue gene sets are based on the union of tissue-specific and tissue-enriched gene lists from cell-typespecific bulk RNA-Seq from (Kaletsky et al 2018). Sperm-specific genes are from (Reinke et al 2000). Numbers in brackets indicate the size of the gene set, or the number of genes in the intersection. Adjusted p-value significance level: ‘***’ p < 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1. Supplementary file 4 (PDF 66 KB)
Figure S5. Putative high-confidence trans-activation domains (TAD) are predicted on the amino-terminal side of MML-1 and the carboxy-terminal side of MDL-1. Trans-activation domains (TADs) in C. elegans Myc-family TFs (blue headers) and their human homologs (grey headers) were predicted with ADpred; the red horizontal line in each plot indicates an ADpred score of 0.8, which was suggested as a threshold of high-confidence TAD prediction in the ADpred manuscript (Erijman et al 2020). Scores are predicted along the protein sequence with black lines: raw scores per amino acid, and blue lines: smoothed to highlight broader trends. (A.) ADpred analysis identified a high-confidence TAD from 245-276 on the amino terminal side of MML-1 between the “Mondo-conserved” domains IV and V (Ceballos, Esse, Grishok 2021) that parallels similar predicted TADs in the human homologs MLXIP and MLXIPL. (B.) There are TAD regions of lower confidence on the C-terminal side of MXL-2, which also correspond to a region of similar confidence on the C-terminal side of human homolog MLX. (C.) C. elegans MDL-1, thought to function primarily as a transcriptional repressor, harbors a high confidence predicted TAD on the C-terminal end, indicating that MDL-1 may have roles in transcriptional activation in some contexts, unlike its human counterparts. (D.) Similar to mammalian MAX, there were no predicted TADs in MXL-1 but we found a predicted TAD of lower confidence on the C-terminal side of MXL-3. Protein sequences were from WormBase for C. elegans and UniProt for human: MLXIP (Q9HAP2), MXLIPL (Q9NP71), MLX (Q9UH92), MXI1 (P50539), MXD1/MAD (Q05195), MXD3/MAD3 (Q9BW11), MXD4/MAD4 (Q14582), MAX (P61244). Supplementary file5 (PDF 88 KB)
Figure S6. Plot for additional trial for brood size assay. Loss of MXL-2 in DR but not ad libitum animals compromises fecundity and embryo viability. Additional trial data for brood size assay, analogous to the representative trial in Figure 4. Brood size assays were performed to determine how reproduction-associated gene expression changes with mxl-2 loss in eat-2 affected reproductive fitness. (A) Viable progeny from each day over the course of the reproductive lifespan were counted for singled parent hermaphrodites for (i) N2 and mxl-2, and (ii) eat-2 and eat-2;mxl-2 animals. (B) Total brood size aggregated from across the observations in (A) for each strain. (C) Unhatched eggs were counted for each strain, but only found for eat-2;mxl-2 animals. Data is shown from a representative trial. See Table S4 for complete results and statistical analysis. Stars indicate FDR-corrected p-values: *< 0.05, **< 0.01, ***< 0.005. Supplementary file6 (PDF 68 KB)
Figure S7. Key autophagy-associated genes were not differentially expressed in eat-2 but are dysregulated in eat-2;mxl-2. Heatmap of log2 foldchanges for the indicated sample group compared to wild-type (N2) for key genes involved in autophagy, with their canonical role indicated by the labeled sidebar on the left. Autophagy-related gene expression was not up-regulated in eat-2 animals but was dysregulated by loss of mxl-2 in the eat-2 background. Columns are ordered by hierarchical clustering. Significantly differentially expressed genes are indicated by red stars, adjusted pvalue significance level: ‘***’ p < 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1. Supplementary file7 (PDF 41 KB)
Figure S8. Enrichment for predicted transcription factor binding in promoters of genes differentially expressed in eat-2, mxl-2, daf-16 (RNAi) or pha-4 (RNAi). Heatmaps representing enrichment of transcription factor motifs in promoters of genes (A) downregulated or (B) up-regulated compared to wild-type. (A) Consistent with the expected binding motif of MML-1::MXL-2, the most enriched motifs for genes down-regulated under DR correspond to canonical CACGTG E-box sites and the motif for MDL-1. DAF-16 motifs are most significantly enriched for genes down-regulated with RNAi for daf-16, but we do not observe enrichment for PHA-4 predicted sites in samples with RNAi for pha-4, or in eat-2 DR conditions. E-box sites or motifs are not enriched in the genes downregulated in mxl-2 mutants, however MML-1::MXL-2 activity is low under such conditions (Johnson et al 2014). (B). An invariant set of transcription factor motifs represent those most significantly enriched across all single-perturbation conditions for up-regulated genes, corresponding to GATA-binding factors. Numbers in brackets are the size of the gene set, or the size of the intersection. Stars indicate significant enrichment results. Adjusted p-value significance level: ‘***’ p < 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1. Supplementary file8 (PDF 55 KB)
Data Availability Statement
The RNA-Seq dataset is available from the NCBI Gene Expression Omnibus (GEO) under accession GSE240821.