Abstract
The binding characteristics of reduced hepatic membrane proteins for acetylated low-density lipoprotein (acetyl-LDL) and maleylated bovine serum albumin (Mal-BSA) have been examined. Two receptor activities were extracted from hepatic membranes in the presence of octyl beta-D-glucoside and beta-mercaptoethanol, and were separated by chromatography on Mal-BSA-Sepharose 4B. The receptors were revealed by ligand blotting. The active binding proteins had apparent molecular masses of 35 and 15 kDa in SDS/polyacrylamide gels. Equilibrium studies with protein-phosphatidylcholine complexes indicated that the reduced 35 kDa protein expresses two binding sites for Mal-BSA and one for acetyl-LDL, whereas the 15 kDa protein-phosphatidylcholine complex binds 131I-Mal-BSA and 131I-acetyl-LDL with a 4:1 stoichiometry. 131I-Mal-BSA binding was linear with both proteins, with a Kd of 4.8 nM at the 35 kDa protein and a Kd of 5.6 nM at the 15 kDa protein. The 35 kDa protein displayed saturable binding of 131I-acetyl-LDL with a Kd of 5 nM; the 15 kDa binding protein bound 131I-acetyl-LDL with a Kd of 2.3 nM. A 85 kDa protein was obtained by Mal-BSA-Sepharose chromatography when the hepatic membranes had been solubilized with Triton X-100 in presence of GSH/GSSG. This protein displayed saturable 131I-Mal-BSA binding with a Kd of 30 nM and 131I-acetyl-LDL binding with a Kd of 6.5 nM. The 131I-Mal-BSA binding capacity was four times higher than that of 131I-acetyl-LDL. Competition studies with the 35 kDa, 15 kDa and 85 kDa proteins binding Mal-BSA, acetyl-LDL, formylated albumin and polyanionic competitors provide evidence for the existence of more than one class of binding sites at the reduced binding proteins.
Full text
PDF



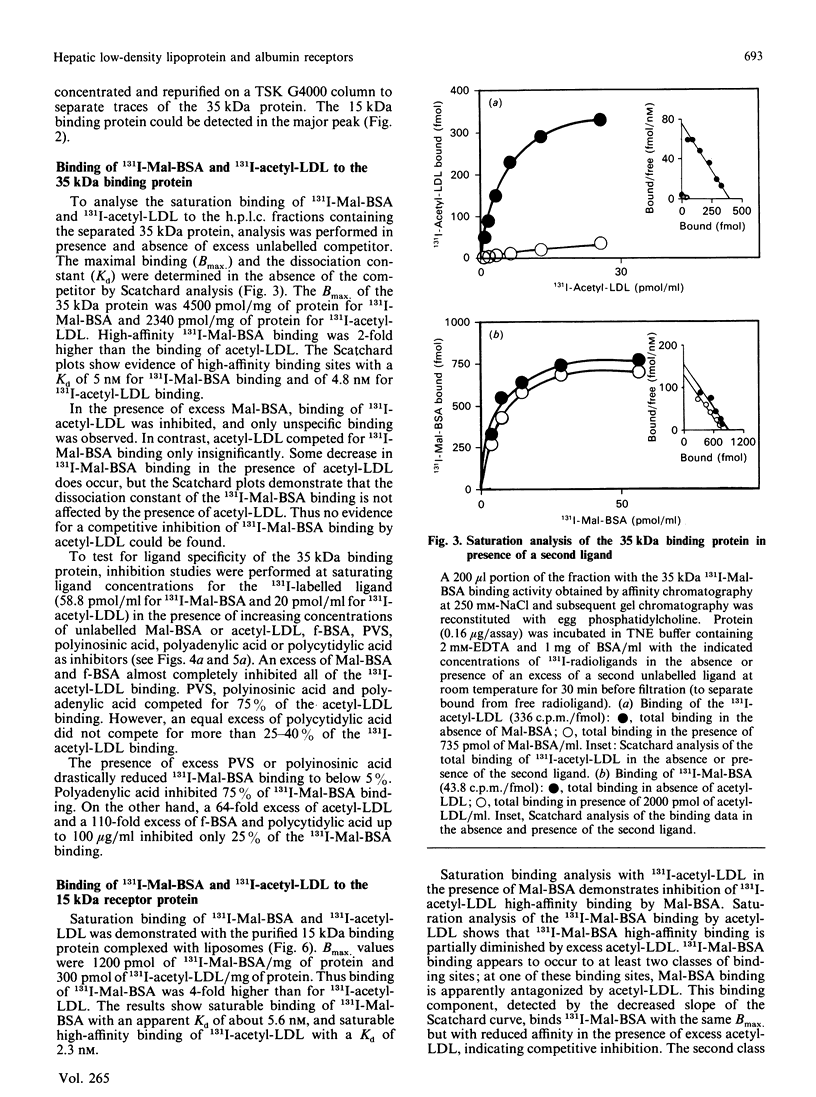



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basu S. K., Goldstein J. L., Anderson G. W., Brown M. S. Degradation of cationized low density lipoprotein and regulation of cholesterol metabolism in homozygous familial hypercholesterolemia fibroblasts. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3178–3182. doi: 10.1073/pnas.73.9.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomhoff R., Drevon C. A., Eskild W., Helgerud P., Norum K. R., Berg T. Clearance of acetyl low density lipoprotein by rat liver endothelial cells. Implications for hepatic cholesterol metabolism. J Biol Chem. 1984 Jul 25;259(14):8898–8903. [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Lipoprotein metabolism in the macrophage: implications for cholesterol deposition in atherosclerosis. Annu Rev Biochem. 1983;52:223–261. doi: 10.1146/annurev.bi.52.070183.001255. [DOI] [PubMed] [Google Scholar]
- Dresel H. A., Friedrich E., Via D. P., Schettler G., Sinn H. Characterization of binding sites for acetylated low density lipoprotein in the rat liver in vivo and in vitro. EMBO J. 1985 May;4(5):1157–1162. doi: 10.1002/j.1460-2075.1985.tb03754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresel H. A., Friedrich E., Via D. P., Sinn H., Ziegler R., Schettler G. Binding of acetylated low density lipoprotein and maleylated bovine serum albumin to the rat liver: one or two receptors? EMBO J. 1987 Feb;6(2):319–326. doi: 10.1002/j.1460-2075.1987.tb04757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresel H. A., Schettler G., Via D. P. Solubilization and purification of an acetyl-LDL binding membrane protein from human leukocytes. Agents Actions Suppl. 1984;16:153–161. doi: 10.1007/978-3-0348-7235-5_18. [DOI] [PubMed] [Google Scholar]
- Eskild W., Henriksen T., Skretting G., Blomhoff R., Berg T. Endocytosis of acetylated low-density lipoprotein, endothelial cell-modified low-density lipoprotein, and formaldehyde-treated serum albumin by rat liver endothelial cells. Evidence of uptake via a common receptor. Scand J Gastroenterol. 1987 Dec;22(10):1263–1269. doi: 10.3109/00365528708996474. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Ho Y. K., Basu S. K., Brown M. S. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc Natl Acad Sci U S A. 1979 Jan;76(1):333–337. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAVEL R. J., EDER H. A., BRAGDON J. H. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J Clin Invest. 1955 Sep;34(9):1345–1353. doi: 10.1172/JCI103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi S., Takata K., Morino Y. Characterization of a membrane-associated receptor from rat sinusoidal liver cells that binds formaldehyde-treated serum albumin. J Biol Chem. 1985 Jan 10;260(1):475–481. [PubMed] [Google Scholar]
- Imber M. J., Pizzo S. V., Johnson W. J., Adams D. O. Selective diminution of the binding of mannose by murine macrophages in the late stages of activation. J Biol Chem. 1982 May 10;257(9):5129–5135. [PubMed] [Google Scholar]
- Kodama T., Reddy P., Kishimoto C., Krieger M. Purification and characterization of a bovine acetyl low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9238–9242. doi: 10.1073/pnas.85.23.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Nagelkerke J. F., Barto K. P., van Berkel T. J. In vivo and in vitro uptake and degradation of acetylated low density lipoprotein by rat liver endothelial, Kupffer, and parenchymal cells. J Biol Chem. 1983 Oct 25;258(20):12221–12227. [PubMed] [Google Scholar]
- Ottnad E., Via D. P., Sinn H., Friedrich E., Ziegler R., Dresel H. A. Evidence in liver for a disulphide-linked scavenger receptor containing a binding site for acetylated low-density lipoprotein and maleylated bovine serum albumin. Biochem J. 1988 Aug 1;253(3):835–838. doi: 10.1042/bj2530835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitas R. E., Boyles J., Mahley R. W., Bissell D. M. Uptake of chemically modified low density lipoproteins in vivo is mediated by specific endothelial cells. J Cell Biol. 1985 Jan;100(1):103–117. doi: 10.1083/jcb.100.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W. J., Goldstein J. L., Brown M. S. Partial purification and characterization of the low density lipoprotein receptor from bovine adrenal cortex. J Biol Chem. 1980 Dec 10;255(23):11442–11447. [PubMed] [Google Scholar]
- Sinn H. J., Schrenk H. H., Friedrich E. A., Via D. P., Dresel H. A. Radioiodination of proteins and lipoproteins using N-bromosuccinimide as oxidizing agent. Anal Biochem. 1988 Apr;170(1):186–192. doi: 10.1016/0003-2697(88)90107-8. [DOI] [PubMed] [Google Scholar]
- Via D. P., Dresel H. A., Cheng S. L., Gotto A. M., Jr Murine macrophage tumors are a source of a 260,000-dalton acetyl-low density lipoprotein receptor. J Biol Chem. 1985 Jun 25;260(12):7379–7386. [PubMed] [Google Scholar]