Abstract
DNA-PK (DNA-dependent protein kinase) is a double-strand break sensor involved in DNA repair and signal transduction. In the present study, we constructed site-directed cross-linking probes to explore the range of DNA discontinuities that are recognized by DNA-PKCS (DNA-PK catalytic subunit). A comparison between different substrate architectures showed that DNA-PKCS associates preferentially with the crossover region of synthetic Holliday junctions. This interaction with four-way junctions was preserved when biotin–streptavidin complexes were assembled at the termini to exclude the binding of Ku proteins. The association of DNA-PKCS with Holliday junctions was salt-labile even in the presence of Ku proteins, but this interaction could be stabilized when the DNA probes were incubated with the endogenous enzyme in nuclear extracts of human cells. Cross-linking of the endogenous enzyme in cellular extracts also demonstrated that DNA-PKCS binds to DNA ends and four-way junctions with similar affinities in the context of a nuclear protein environment. Kinase assays using p53 proteins as a substrate showed that, in association with four-way structures, DNA-PKCS adopts an active conformation different from that in the complex with linear DNA. Our results are consistent with a structure-specific, but Ku- and DNA end-independent, recruitment of DNA-PKCS to Holliday junction intermediates. This observation suggests an unexpected functional link between the two main pathways that are responsible for the repair of DNA double-strand breaks in mammalian cells.
Keywords: DNA-dependent protein kinase (DNA-PK), DNA strand break, Holliday junction, homologous recombination, Ku, non-homologous end joining, replication arrest
Abbreviations: DNA-PK, DNA-dependent protein kinase; DNA-PKCS, DNA-PK catalytic subunit; HR, homologous recombination; NHEJ, non-homologous end-joining
INTRODUCTION
DNA-PK (DNA-dependent protein kinase) is a nuclear serine/threonine kinase that is activated by double-strand breaks in DNA [1]. Biochemical studies have shown that DNA-PK is composed of a 470 kDa catalytic subunit (DNA-PKCS) and the Ku70/Ku80 regulatory component [2,3]. In mammalian cells, both DNA-PKCS and the Ku heterodimer are required for the repair of double-strand breaks and for V(D)J rearrangements in developing lymphocytes [4–6]. The loss of DNA-PKCS activity results in severe combined immunodeficiency, hypersensitivity to ionizing radiation and radiomimetic chemicals, as well as cancer predisposition in mouse models [7–9]. In addition to its function in DNA double-strand break repair and V(D)J recombination, DNA-PKCS is also involved in p53-dependent signalling pathways that regulate cell death [10]. After genotoxic stress, DNA-PKCS phosphorylates RPA, a potential downstream effector of replication arrest during the S phase [11]. DNA-PKCS may also contribute to a checkpoint function in response to DNA damage through phosphorylation of the transcription factor E2F-1 (E2 promoter binding factor 1) [12]. Additional roles for DNA-PKCS involve telomer maintenance [13] and regulation of transcription [14,15].
Despite its extraordinary size of 4127 amino acids, DNA-PKCS is a highly abundant protein in primate cells. Its C-terminus comprises a catalytic domain of 380 amino acids, which shares homology with members of the phosphoinositide 3-kinase family [2]. DNA-PKCS associates with the Ku heterodimer through a domain (amino acids 3002–3850) that is adjacent to the kinase motif [16], but the function of the remaining large portion of the molecule is poorly understood. In the amino acid region 1503–1602, DNA-PKCS contains a leucine zipper motif that mediates the interaction with C1D, a nuclear matrix-associated DNA-binding protein. In contrast with the Ku subunits, which assemble with DNA-PKCS in the presence of DNA termini, C1D recruits DNA-PKCS to covalently closed DNA molecules and is capable of activating its kinase activity in the absence of strand breaks [17]. These observations suggest that DNA-PKCS is recruited to different substrates via association with distinct regulatory partners.
A number of issues concerning the role and mechanism of action of DNA-PKCS remain unresolved. For example, DNA-PKCS co-ordinates the repair of double-strand breaks by NHEJ (non-homologous end-joining) and, as a consequence, competes with the alternative repair of broken chromosomes by HR (homologous recombination). Interestingly, DNA-PKCS suppresses spontaneous HR events during replication more efficiently compared with HR elicited by exogenously induced DNA cleavage, suggesting that DNA-PKCS may adopt an active role in regulating homologous strand exchanges [18]. This view is supported by the finding that at least one interaction partner of DNA-PKCS, the C1D protein, is also implicated in the HR process [19]. In the present study, we tested whether DNA-PKCS may recognize, besides other DNA discontinuities that interrupt the double helix, non-linear intermediates of HR. Therefore we took advantage of photoreactive probes, designed in a site-specific manner, to analyse the structure-specific DNA-binding properties of the individual DNA-PK subunits. This approach revealed that DNA-PKCS, unlike the Ku component, binds to the crossover region of Holliday junctions.
EXPERIMENTAL
Proteins
DNA-PKCS was purified from cultured HeLa cells as described in [20]. The human DNA-PK holoenzyme (270 units/μg) was obtained from Promega, recombinant Ku heterodimer from Trevigen and p53 from Santa Cruz Biotechnology. Nuclear extracts from HeLa cells were prepared by the method of Dignam et al. [21], yielding a protein content of 10–15 mg/ml and a final conductivity equivalent to 80 mM NaCl. HeLa and TK6 (human lymphoblasts) were obtained from A.T.C.C. (Rockville, MD, U.S.A.). SV40-transformed fibroblasts (GM00637) were obtained from Coriell Institute for Medical Research (Camden, NJ, U.S.A.).
Antibody and Western-blot analysis
A rabbit polyclonal antibody raised against DNA-PKCS was obtained from Abcam (Cambridge, U.K.). Western blots on PVDF hydrophobic membranes (0.45 μm pore size; Bio-Rad Laboratories) were performed at an antibody dilution of 1:2000 using 5% (w/v) fat-free milk powder as the blocking solution and goat secondary antibodies conjugated with horseradish peroxidase. The protein was visualized by chemoluminescent detection after incubation with the SuperSignal substrate (Pierce).
DNA probes
The photoreactive oligonucleotides ON1 (5′-ACCACCCTTCGATCGTGTC-3′, where the reactive base analogue is underlined), ON2 (5′-ACCACCCTTCGATCGTGTC-3′), ON3 (5′-CGACTGCAGACGTACCACCCTTCGAACCACACGAATTCTAGTG-3′) and ON4 (5′-CGACTGCAGACGTACCACCCTTCGAACCACACGAATTCTAGTG-3′) were obtained by azido modification of thymine via a six-carbon triethylene spacer (Genset, Evry, France). The photoreactive oligonucleotides were radiolabelled using [γ-32P]ATP and T4 polynucleotide kinase. Biotinylated sequences were synthesized by Microsynth (Switzerland). The oligonucleotides were incubated with streptavidin (Roche) at 25 °C for 30 min. To generate DNA duplexes or four-way DNA junctions, the oligonucleotides were annealed in 50 mM Tris/HCl (pH 8.0) and 10 mM MgCl2, incubated for 5 min at 85 °C, followed by slow cooling [22]. To prepare duplex 19-mers, ON1 or ON2 was annealed to ON5 (5′-GACACGATCGAAGGGTGGT-3′). Duplex 43-mers were obtained by annealing ON3 or ON4 with ON6 (5′-CACTAGAATTCGTGTGGTTCGAAGGGTGGTACGTCTGCAGTCG-3′). Partial duplex DNA was generated by annealing ON3 to ON7 (5′-AGGGTGGTACGTCTGCAGTCG-3′). The three-way junction was constructed by annealing ON3 with ON8 (5′-CGATACGTCCCCAATATCCCAAGGGTGGTACGTCTGCAGTCG-3′) and ON9 (5′-CACTAGAATTCGTGTGGTTCGGGGATATTGGGGACGTATCG-3′). Four-way junctions were obtained by annealing ON3 with ON8, ON10 (5′-CACTAGAATTCGTGTGGTTCGGTATCACGACTAGC-3′) and ON11 (5′-GCTAGTCGTGATACGGGATATTGGGGACGTATCG-3′). When the photoreactive probe was used, the non-labelled strands were slightly in excess (10%) to ensure that the modified strand would be completely incorporated into the final substrate. The homogeneity of each DNA construct was confirmed by native PAGE (7% gel). Duplexes of 76 bp were assembled by hybridization of ON2 with a 76-mer oligonucleotide containing the complementary sequence at the centre and subsequent ligation with two other oligonucleotides of 28 and 29 nt to obtain a double-stranded fragment. Similarly, duplexes of 161 bp were recovered from the ligation of ON2 with five partially overlapping oligonucleotide sequences. These ligation products were purified on 10% denaturing polyacrylamide gels.
Cross-linking
Binding reactions were initiated in microcentrifuge tubes by incubating the photoreactive probes (0.2 nM) at 25 °C in 25 mM Hepes (pH 7.5) and 10 mM MgCl2, with Ku proteins, DNA-PK holoenzyme or fractions of purified DNA-PKCS, and the indicated concentrations of KCl. The final volume of each reaction was 20 μl. Cross-linking was performed by exposure for 40 s to UV light (325 nm) using the UVG-11 source obtained from UVP (Upland, CA, U.S.A.). The reactants were then dissolved in gel-loading buffer [62.5 mM Tris/HCl, pH 6.8/5% (v/v) glycerol/2% (w/v) SDS/5% (v/v) 2-mercaptoethanol/1% Bromophenol Blue]. After boiling for 5 min, the cross-linked products were separated by SDS/PAGE (10% gel) and visualized by autoradiography. The relative cross-linking efficiency was quantified by laser densitometry of the X-ray films.
DNA-PK activity assays
Kinase assays were performed in 25 mM Hepes (pH 7.5), 0.25 mM of the synthetic p53-derived peptide PESQEAFADLWKK (Promega), 10 mM MgCl2, 10 μg/ml sonicated calf thymus DNA and 0.1 mM [γ-32P]ATP (NCI; 1000 d.p.m./mol) in a volume of 40 μl. The reactions were started by the addition of DNA-PK and incubations were performed at 30 °C for 10 min. The reactions were stopped and kinase activity was quantified using the SignaTECT kit obtained from Promega. One unit is the amount of enzyme that incorporates 1 pmol of phosphate into 38 nmol of peptide substrate in 1 min at 30 °C.
RESULTS
Azido-mediated cross-linking of DNA-PKCS
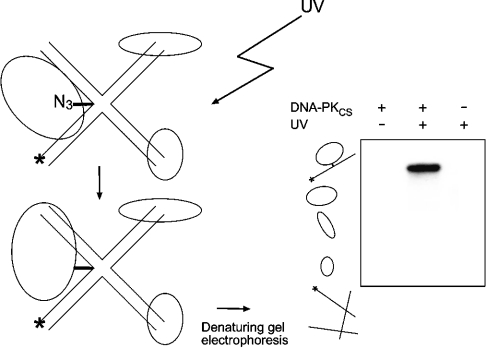
Previous UV cross-linking studies indicated that DNA-PKCS makes direct contacts with DNA substrates as short as 18 bp [1,23]. In the present study, we exploited the azido cross-linking chemistry [24,25] to analyse the ability of DNA-PKCS to interact with different DNA architectures. As outlined in Figure 1, this cross-linking technique involves the introduction of a photoreactive base analogue at specific positions of 32P-labelled DNA oligonucleotides. After irradiation with long-wave UV light, the azido group is converted into a nitrene intermediate that reacts with nucleophilic residues. This chemical process generates a covalent complex with the polypeptide that undergoes close contacts with the reactive site, e.g. the centre of a synthetic four-way junction. The radiolabelled cross-linked complexes are then separated by SDS/PAGE and visualized by autoradiography (Figure 1).
Figure 1. UV-induced cross-linking of proteins to oligonucleotide probes (2 nM) containing a site-directed N3 moiety.
The asterisk denotes the 32P-labelled residue at the 5′-end of each probe. Radiolabelled complexes were generated after UV irradiation, and the cross-linked protein was identified by analysis of the reaction products on SDS/polyacrylamide gels. The representative autoradiograph was obtained after a 2 min incubation with DNA-PKCS (0.1 μg). Lanes 1 and 3 (numbering from the left) show control reactions without UV or protein respectively. Free radioactive probes migrated ahead of the proteins into the electrophoresis buffer at the bottom of the gel.
Cross-linking of DNA-PKCS to linear DNA
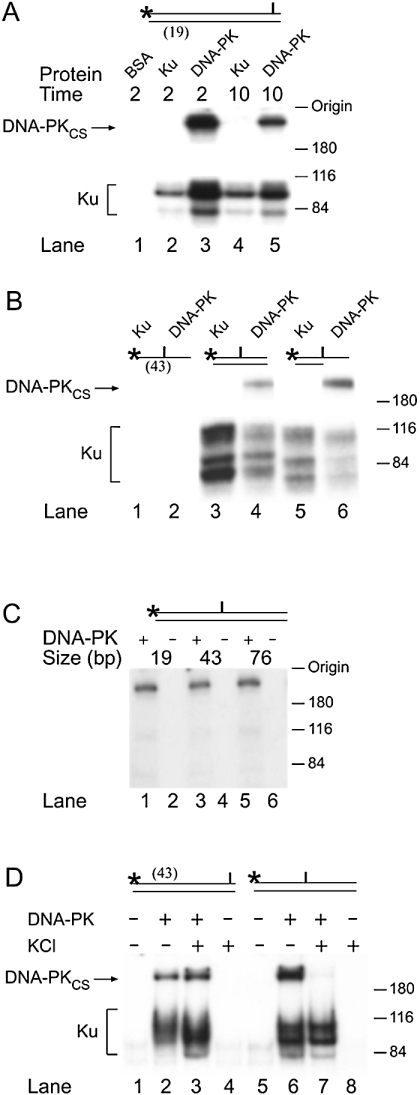
The advantage of site-directed cross-linking methods is that the target proteins can be immobilized at a unique position in the nucleic acid substrate. A photoreactive azido moiety was introduced in place of thymine 2 nt from the 3′-end of 19-mer DNA duplexes, whereas the 5′-end of the same strand was 32P-labelled. After reaction with the recombinant human Ku heterodimer (Ku70 and Ku80), this DNA probe generated the expected radiolabelled adducts that migrated faster than the 116 kDa molecular-mass standard on polyacrylamide gels (Figure 2A, lanes 2 and 4). In the presence of the DNA-PK holoenzyme, the same photoreactive probe was capable of cross-linking the two Ku proteins as well as the DNA-PKCS subunit. The Ku polypeptides appeared again as radiolabelled complexes ahead of the 116 kDa standard, whereas the catalytic subunit generated a radiolabelled product that migrated slower than the 180 kDa standard and remained near the gel origin (Figure 2A, lanes 3 and 5). The relative electrophoretic mobility of this larger protein was identical with that of the DNAPKCS subunit, and no radiolabelled product other than the expected cross-links with DNA-PKCS, Ku70 and Ku80, were detected. In time-course experiments, the highest cross-linking efficiency between DNA and DNA-PKCS was observed after a short incubation for 2 min before UV irradiation (Figure 2A, lane 3). At the same protein/DNA molar ratio (as with Ku proteins), BSA was not cross-linked to the DNA substrate (Figure 2A, lane 1). Preincubation of DNA-PK with ATP progressively decreased the cross-linking of the catalytic subunit (results not shown), in agreement with previous studies showing that DNA binding of DNA-PKCS is negatively regulated by autophosphorylation [26].
Figure 2. Binding of DNA-PKCS near the DNA ends and to internal positions of DNA fragments.
(A) Cross-linking of DNA-PK subunits to a 19-mer duplex probe containing the photoreactive residue 2 nt from the 3′-end. The incubation mixtures contained 1 μg of BSA (lane 1), 1 μg of Ku heterodimer (lanes 2 and 4) or 0.5 μg of DNA-PK. The samples were preincubated for 2 and 10 min before UV exposure and analysed by denaturing PAGE. (B) Cross-linking of DNA-PK subunits to 43-mer DNA substrates of variable topology containing the photoreactive site at the centre. The samples contained 1 μg of Ku heterodimer (lanes 1, 3 and 5) or 0.5 μg of DNA-PK (lanes 2, 4 and 6). (C) Cross-linking of DNA-PKCS (0.1 μg) to DNA duplexes of various lengths. The photoreactive residue was placed at the centre of each substrate. (D) Differential stability of the complexes between DNA-PKCS and 43-mer duplexes. The photoreactive residue was located 2 nt away from the DNA end (lanes 1–4) or at the centre of the substrate (lanes 5–8). The reaction mixtures contained DNA-PK (0.5 μg) and KCl (100 mM) as indicated.
DNA-PKCS and Ku can associate independently with DNA termini; however, if the nucleic acid substrate is long enough to accommodate both subunits (if it consists of at least 32 bp), Ku increases the affinity of DNA-PKCS for DNA termini [27,28]. Under these conditions, the Ku subunits slide to internal positions of the substrate [24]. Consistent with this translocation model, we observed that the Ku subunits could still be cross-linked when the photoreactive analogue was positioned exactly at the centre of a 43-mer DNA duplex (Figure 2B, lane 3). The appearance of multiple radiolabelled bands has already been described previously and reflects the formation of geometric isomers representing cross-links with different amino acid residues of the two Ku subunits [24].
Unexpectedly, when the experiment was repeated with the DNA-PK holoenzyme, not only Ku but also DNA-PKCS was crosslinked to the central residue of the 43-mer substrate (Figure 2B, lane 4). DNA-PK could also be immobilized to the central region of an overhang substrate (Figure 2B, lanes 5 and 6), which was assembled by annealing the photoreactive 43-mer oligonucleotide with a complementary 22-mer sequence. As with the double-stranded 43-mer fragment, both Ku and DNA-PKCS were immobilized to this partial duplex substrate. However, neither Ku nor DNA-PKCS could be cross-linked to single-stranded 19-mer (results not shown) or single-stranded 43-mer oligonucleotides (Figure 2B, lanes 1 and 2).
To confirm the direct interaction of DNA-PKCS with internal DNA sites, we employed a purified DNA-PKCS fraction, prepared by the method of Dvir et al. [20], which contained only trace amounts of the Ku-regulatory partners. After UV irradiation, the catalytic subunit was again susceptible to cross-linking to the centre of the 19- and 43-mer substrates; however, in this case, the intensity of the faster migrating bands representing cross-linked Ku was decreased (Figure 2C, lanes 1 and 3). In addition, we generated longer substrates of 76 and 161 bp containing the photoreactive group at the centre, i.e. at a distance of approx. 40 and 80 nt from each DNA terminus. As with the previously used duplexes, these longer probes were radiolabelled at one 5′-end before incubation (2 min) with the DNA-PKCS fraction. Surprisingly, the catalytic subunit was again cross-linked to the central region of the 76-mer probe (Figure 2C, lane 5). We also observed similar levels of cross-linking of DNA-PKCS to the centre of the 161-mer probe (gel not shown). Taken together, these results indicate that DNA-PKCS displays an intriguing ability to interact with double-stranded DNA at a considerable distance from the termini.
Characterization of the DNA-PKCS interactions with internal DNA sites
The binding of purified DNA-PKCS to intact portions of DNA is in conflict with previous studies, where the localization of the catalytic subunit was mapped to the vicinity of DNA ends with minimal inward translocation [23,24]. To characterize this unexpected interaction of DNA-PKCS with internal DNA segments, we tested the stability of nucleoprotein complexes at different ionic strengths. DNA-PK was cross-linked to the central residue of the 43-mer duplex in the presence of 100 mM KCl. Under these conditions, the formation of complexes with DNA-PKCS was completely disrupted, whereas cross-linking with the Ku subunits was not affected by the increased ionic strength (Figure 2D, lanes 6 and 7). Importantly, the cross-linking efficiency involving DNA-PKCS was not changed by the addition of 100 mM KCl when the photoreactive group was located only two residues away from the DNA terminus (Figure 2D, lanes 2 and 3). This finding is consistent with the known ability of the Ku subunits to recruit DNA-PKCS to DNA ends at physiological salt concentrations [27]. Instead, the interaction of DNA-PKCS with the more interior site of DNA duplexes is not stabilized by its association with Ku and, hence, can be detected only at low ionic strength.
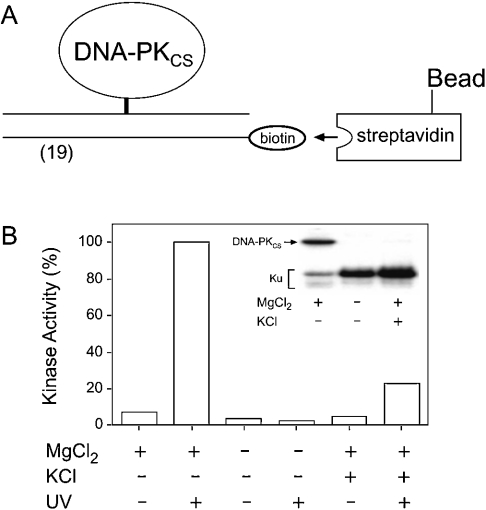
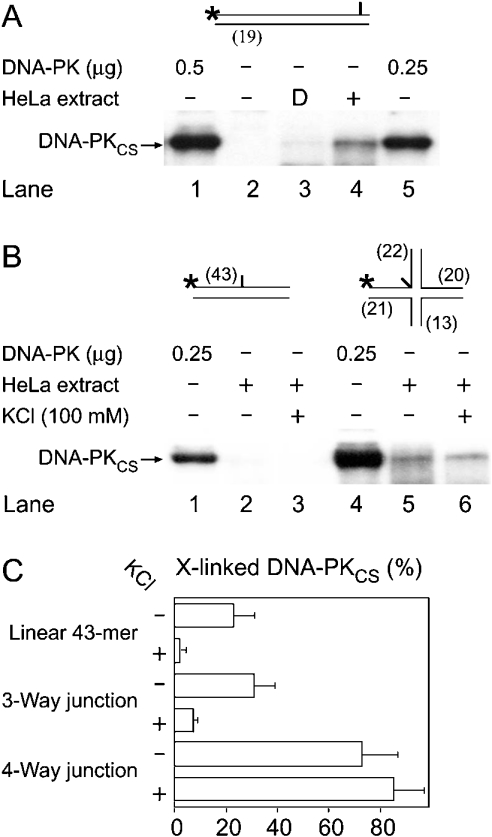
We wished to rule out the possibility that the results obtained so far could be due to a minor proportion of incorrectly folded and inactive DNA-PK molecules, which aggregate on DNA and become cross-linked in an unspecific manner. The strong interaction between biotin and streptavidin was exploited to isolate and characterize the cross-linked complexes. DNA duplexes were assembled as mentioned above, except that the unmodified complementary strand was supplemented with a biotin tag at its 3′-end (Figure 3A). DNA-PK was incubated with these biotinylated probes and, after 2 min, exposed to UV light for cross-linking. The resulting complexes were mixed with streptavidin-conjugated paramagnetic beads, pulled down and washed extensively with high-salt (1 M NaCl) buffer to remove any free protein that was not cross-linked to the biotinylated substrate. To determine the activity of the immobilized enzyme, the paramagnetic beads were incubated for 10 min with 32P-ATP and a p53 peptide substrate. These assays revealed a significant kinase activity associated with the immobilized enzyme, but only when the cross-linking reaction was performed under conditions that allow binding of DNA-PKCS to the central region of the substrate, i.e. in the presence of MgCl2 but without KCl (Figure 3B). Additional controls, where UV irradiation was omitted, demonstrate that the observed kinase activity results from cross-linked DNA-PKCS with little contamination of free enzyme (Figure 3B). Alternatively, the washed paramagnetic beads containing cross-linked enzyme were incubated with 32P-ATP in the absence of peptide substrate. Analysis of the reaction mixtures on denaturing polyacrylamide gels showed that the cross-linked DNA-PKCS molecules in the pellet were also susceptible to autophosphorylation (results not shown).
Figure 3. Characterization of cross-linked DNA-PKCS.
(A) The photoreactive probe was tagged with a biotin residue to isolate the cross-linked complexes. (B) Phosphorylation of an heterologous acceptor. DNA-PK (0.5 μg) was cross-linked to the biotinylated 19-mer probe under different conditions. MgCl2 and KCl were added at a concentration of 10 and 100 mM respectively. The resulting complexes (immobilized on streptavidin beads) were separated from the population of free enzymes and assayed for kinase activity in 10 min incubations with 32P-ATP (100 μM) and a peptide substrate. The autoradiograph shown in the inset demonstrates the different extents of DNA-PKCS cross-linking.
Binding of DNA-PKCS to the junction region of four-way intermediates
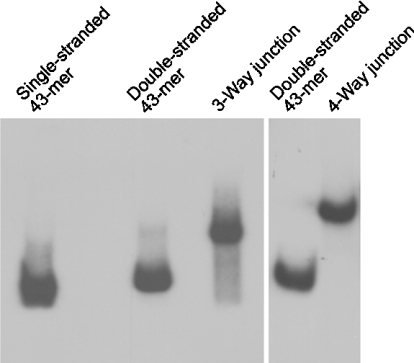
The efficient cross-linking of DNA-PKCS to internal substrate positions, located at a considerable distance from the DNA ends, prompted us to test whether DNA-PKCS is capable of interacting with architecturally more complex DNA structures. In particular, we explored its interaction with a Holliday junction molecule that constitutes the universal HR intermediate and is also generated during replication [29]. A synthetic Holliday junction with four double-helical stems of 13, 20, 21 and 22 bp was constructed by hybridization of the photoreactive 43-mer probe with three other oligonucleotides. For comparison, we also generated a three-way junction molecule (consisting of helical stems of 20, 21 and 22 bp) by annealing three different oligonucleotides. These DNA substrates contained a single photoreactive group in the central junction region and, in addition, the photoreactive 43-mer oligonucleotide was 32P-labelled at its 5′-end. Subsequent analysis on a native polyacrylamide gel demonstrated that the resulting hybridization products migrated as a single band, indicating homogeneous populations of four- and three-way DNA structures (Figure 4).
Figure 4. DNA preparations containing three- and four-way junctions.
The annealed oligonucleotides were analysed by native PAGE. The autoradiograph shows a 7% polyacrylamide gel demonstrating the purity of the DNA junction structures.
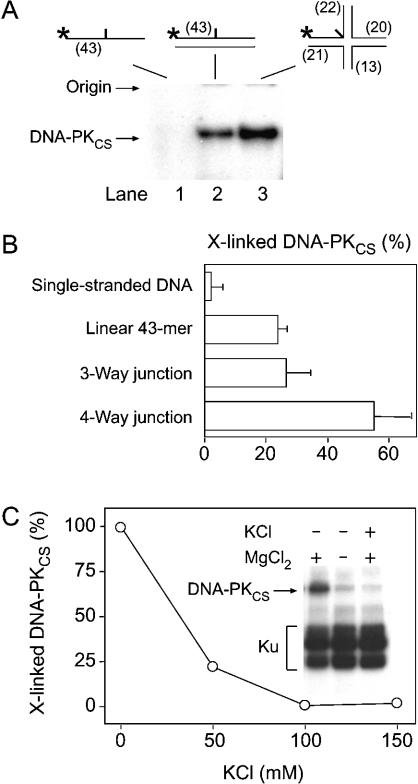
The photoreactive four-way DNA substrate, or the 43-mer linear control, was incubated with the purified fraction of DNA-PKCS. After 2 min, the samples were UV-irradiated and the amount of cross-links with the four-way junction was compared with that induced in the presence of linear DNA. Both types of substrates yielded nucleoprotein complexes of identical size, migrating near the origin of denaturing gels, which consisted of DNA-PKCS covalently linked to the radiolabelled single-stranded 43-mer oligonucleotide (Figure 5A). Unexpectedly, we found that DNA-PKCS is cross-linked slightly more efficiently to the synthetic four-way junction compared with the 43-mer linear control (lanes 2 and 3). In similar cross-linking experiments, RPA and XPC proteins, two factors that share a high affinity for single-stranded DNA, displayed no preference for four-way junctions (results not shown). The present study also showed that DNA-PKCS is cross-linked to the junction region of three-way DNA molecules with a similar efficiency as to the central photoreactive residue of the linear control (Figure 5B).
Figure 5. Preferential binding of DNA-PKCS to Holliday junctions.
(A) Interaction of the catalytic subunit (0.1 μg) with the central site of architecturally different substrates. The cross-linking products were resolved by denaturing gel electrophoresis. Only a single band at the position of DNA-PKCS was visible in the autoradiograph. (B) Mean values of three independent cross-linking experiments. The helical stems of the three-way junction consisted of 20, 21 and 22 bp. All values are expressed as percentages of complexes obtained with the 19-mer duplex probe. (C) Stability of the interaction between DNA-PKCS and four-way junctions. The diagram shows the amount of cross-linked DNA-PKCS after incubation of the holoenzyme (0.5 μg) with photoreactive Holliday junctions at different KCl concentrations (mean values of two experiments). A representative autoradiograph of cross-linked complexes is illustrated in the inset. The MgCl2 and KCl concentrations were 10 and 100 mM respectively.
The affinity for four-way junctions was confirmed in experiments performed with the holoenzyme fraction instead of just the catalytic subunit, indicating that the binding preference of DNA-PKCS for four-way junctions is maintained in the presence of Ku subunits (Figure 5C, inset). To determine the stability of the complex generated by DNA-PKCS with four-way junctions, DNA-PK was mixed with photoreactive four-way probes in the presence of increasing KCl concentrations. The formation of nucleoprotein complexes between DNA-PKCS and the four-way junction region was significantly decreased at a KCl concentration of 50 mM, and these interactions were completely suppressed with 100 mM KCl (Figure 5C). Under the same conditions, however, the Ku subunits were still efficiently cross-linked. These results indicate that the binding of DNA-PKCS to the central region of Holliday junctions is salt-sensitive and is not stabilized by the Ku heterodimeric partner. Finally, dose-dependent experiments show that the cross-linking efficiency increased in a linear manner when DNA-PK was incubated with the photoreactive probe at concentrations ranging from 5 to 50 ng/μl (results not shown).
Mechanism of binding to the four-way junction molecules
Although the Ku subunits are not capable of stabilizing the binding of DNA-PKCS to four-way junctions, it was still possible that the loading of DNA-PKCS to four-way junctions could be transiently mediated by Ku. The results shown in Figure 5(A), where the purified DNA-PKCS subunit was used, could be attributed to the presence of a small amount of Ku contaminating the enzyme preparation and, possibly, facilitating the binding of DNA-PKCS to four-way substrates. To solve this issue, we took advantage of the tight interaction between biotin and streptavidin. In the following experiments, the end of each double-helical stem of the four-way junction structure was supplemented with a biotin residue by modifying the 3′-terminus of each oligonucleotide (Figure 6A). A biotin moiety was also placed on each end of the 43-mer linear control fragment by modifying both the 3′- and 5′-terminal nucleotides of the bottom strand. The ends of these specially designed probes can now be blocked by the addition of streptavidin, which consists of a tetramer of four 15 kDa subunits, forcing the DNA-PK components to bind directly to the internal substrate region.
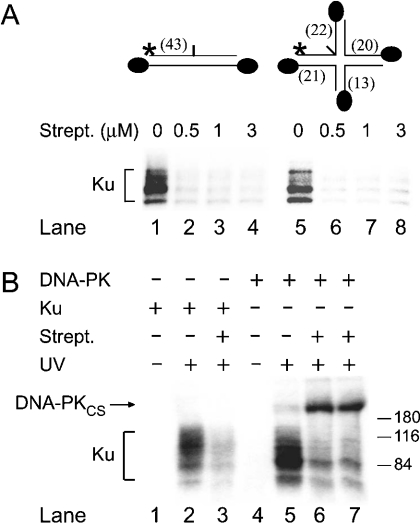
Figure 6. Direct interaction of DNA-PKCS with the four-way junction region.
Each double-strand end was modified with biotin, as illustrated, and accessibility to the substrates was blocked by preincubation with streptavidin (0.5 μM). (A) Binding of the Ku heterodimer to internal sites is suppressed in the absence of free DNA ends. After 2 min of incubation with Ku proteins (1 μg), the samples were exposed to UV light to induce the cross-linking reaction. (B) Binding of DNA-PKCS to four-way junctions is stimulated in the absence of free DNA ends. The substrates of lanes 3, 6 and 7 were obstructed by preincubation with streptavidin. The four-junction molecules were then incubated for another 2 min with 1 μg of Ku proteins (lanes 1–3) or 0.5 μg of DNA-PK holoenzyme (lanes 4–7) before cross-linking by UV irradiation.
The biotinylated probes were first incubated with recombinant human Ku proteins and each sample was exposed to UV light after 2 min. The presence of biotin alone did not interfere with the binding capacity of Ku, neither on the linear duplex (Figure 6A, lane 1) nor on the four-way junction (lane 5). Subsequently, the photoreactive probe was mixed for 30 min with streptavidin and then incubated with Ku proteins for 2 min, followed by exposure to UV light to induce the cross-linking reaction. In this case, the addition of streptavidin suppressed the binding of Ku to both linear DNA (Figure 6A, lanes 2–4) and four-way junction DNA (lanes 6–8).
When these cross-linking experiments were repeated with the DNA-PK holoenzyme, complex formation with Ku was again decreased by the addition of streptavidin (Figure 6B, lanes 5–7). However, after obstruction of the DNA ends, the four-way junction became the preferential locus of interaction with DNA-PKCS (duplicate, lanes 6 and 7). No decrease in Ku cross-links and no increase in DNA-PKCS cross-links were observed when streptavidin was added in the absence of biotin modifications (results not shown), indicating that the change in the binding pattern observed in Figure 6(B) requires streptavidin–DNA binding. Thus capping the DNA by biotin–streptavidin complexes favours the recognition of four-way junctions by the catalytic subunit, whereas binding of the Ku components to the same site is inhibited.
Recognition of Holliday junctions by DNA-PKCS in nuclear extracts of human cells
We next examined whether DNA-PKCS is capable of binding to four-way junctions in the context of the nuclear protein environment of human cells. DNA-PK constitutes the most abundant DNA end-binding activity in crude nuclear extracts [30] and is also the major target for nucleoprotein cross-links when short DNA duplexes are incubated in cellular extracts [1]. Thus we took advantage of nuclear cell extracts to compare the affinity of DNA-PKCS for DNA ends and four-way junctions.
A control experiment was performed by incubating the radiolabelled DNA duplex of 19 bp, which is used to monitor the DNA end-binding activity of DNA-PK, in a standard HeLa cell nuclear preparation. After 2 min, the mixture was exposed to UV light to induce the cross-linking reaction. After electrophoretic analysis, the cross-linked DNA-PKCS subunit was identified as a specific radiolabelled product (Figure 7A, lane 4), which migrated near the gel origin with exactly the same mobility as the corresponding cross-link generated, in parallel samples, by the 19-mer probe using purified DNA-PKCS enzyme (lanes 1 and 5). As expected, the formation of cross-linked complexes depended completely on the presence of nuclear extract proteins (lane 2), and these complexes were also absent (lane 3) when the reaction was performed with an extract that was depleted of DNA-PKCS by ammonium sulphate fractionation [31].
Figure 7. Stable binding of endogenous DNA-PKCS to Holliday junctions.
(A) Cross-linking of the 470 kDa subunit to 19-mer duplexes. The probe was incubated with purified holoenzyme (lanes 1 and 5), without any protein (lane 2) with a HeLa cell extract depleted of DNA-PKCS (10 μg of proteins; lane 3) or with HeLa nuclear extract (10 μg; lane 4). (B) Comparison between linear 43-mer duplexes and four-way junctions containing a photoreactive residue at the centre. Lanes 1 and 4: reactions with purified DNA-PK. Lanes 2 and 5: reactions in HeLa nuclear extract (10 μg). Lanes 3 and 6: reactions in HeLa nuclear cell extract, supplemented with 100 mM KCl. (C) Summary of three independent experiments in HeLa nuclear cell extract (10 μg), showing the relative amount of cross-linked DNA-PKCS. The assays were performed in low ionic strength buffer or in the presence of 100 mM KCl. Results are expressed as percentages of complexes obtained with the 19-mer duplex probe.
The cross-linking procedure was subsequently applied to monitor the interaction of DNA-PKCS in cell extracts with the four-way junction and the corresponding 43-mer linear control. A photoreactive residue was again placed at the centre of each DNA probe as indicated in Figure 7(B). The preference for the four-way substrate over linear DNA was confirmed by a parallel experiment with the purified holoenzyme (Figure 7B, lanes 1 and 4). Moreover, a striking binding preference for the four-way junction substrate was also observed when the two different DNA substrates were incubated in HeLa nuclear cell extracts (cf. lanes 2 and 5). In contrast with the results obtained for the 19-mer probe (Figure 7A), a cross-linked product between DNA-PKCS and the linear 43-mer substrate was barely detectable, indicating that other cellular proteins occupy the centre of this larger DNA molecule (Figure 7B, lane 2). However, the reaction with the Holliday junction generated a cross-linked product (lane 5), which displayed the same electrophoretic mobility as the complex generated by the purified enzyme with both linear and four-way DNA probes (lanes 1 and 4). Thus the endogenous enzyme in nuclear extracts exhibits the same architectural bias that had already been observed previously for the isolated holoenzyme or the purified catalytic subunit. Surprisingly, unlike the interaction of purified DNA-PK in the absence of other cellular factors, the binding of the endogenous enzyme to four-way junctions was more stable, such that it could be detected even in the presence of 100 mM KCl (Figure 7B, lane 6). The same experiment was extended to a variety of human cell lines. Significant cross-linking of DNA-PKCS to four-way junctions was observed in nuclear extracts from both human fibroblasts and lymphoblasts, indicating that the preference of DNA-PKCS for four-way junctions is a general phenomenon (results not shown).
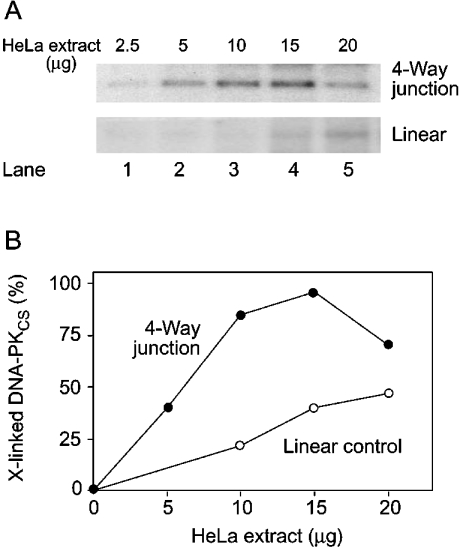
The quantitative evaluation of three independent cross-linking experiments performed in HeLa nuclear extracts showed that DNA-PKCS is cross-linked to four-way junctions with nearly the same efficiency as to the 19-mers that are used as a probe for its DNA end-binding activity (Figure 7C). These results indicate that the affinities of DNA-PKCS for DNA ends and four-way junctions are comparable. Also, the quantification of triplicate experiments confirmed that the binding of the endogenous enzyme to four-way junctions was not diminished by the addition of 100 mM KCl (Figure 7C). However, the interaction with linear 43-mer duplexes or with three-way junctions was significantly decreased in the buffer having higher ionic strength. Dose–response experiments were performed to compare the relative affinity of the endogenous DNA-PKCS enzyme for Holliday junctions and linear DNA (Figure 8). Interestingly, at high concentrations of HeLa nuclear extract, the radioactive band indicative of the covalent complex of DNA-PKCS with four-way DNA junctions was diminished (Figure 8A, lane 5). This observation suggests the presence of competing factors, displaying a low affinity for four-way junctions, which can bind to the reactive probe and inhibit the interaction of DNA-PKCS with these sites only when present at a higher concentration in the reaction mixture.
Figure 8. Titration of the cross-linking reaction in HeLa cell extract.
The photoreactive probes (four-way junctions or linear DNA) were incubated with increasing concentrations of HeLa nuclear extract and subjected to UV cross-linking. (A) Representative autoradiograph showing the various amounts of cross-linked DNA-PKCS. (B) Quantification of two independent experiments demonstrating the dose-dependent cross-linking of endogenous DNA-PKCS after incubation of the nuclear extract with four-way DNA junctions. Results are expressed as percentages of complexes obtained with the 19-mer duplex probe.
Identification of DNA-PKCS in the covalent complex with four-way DNA junctions
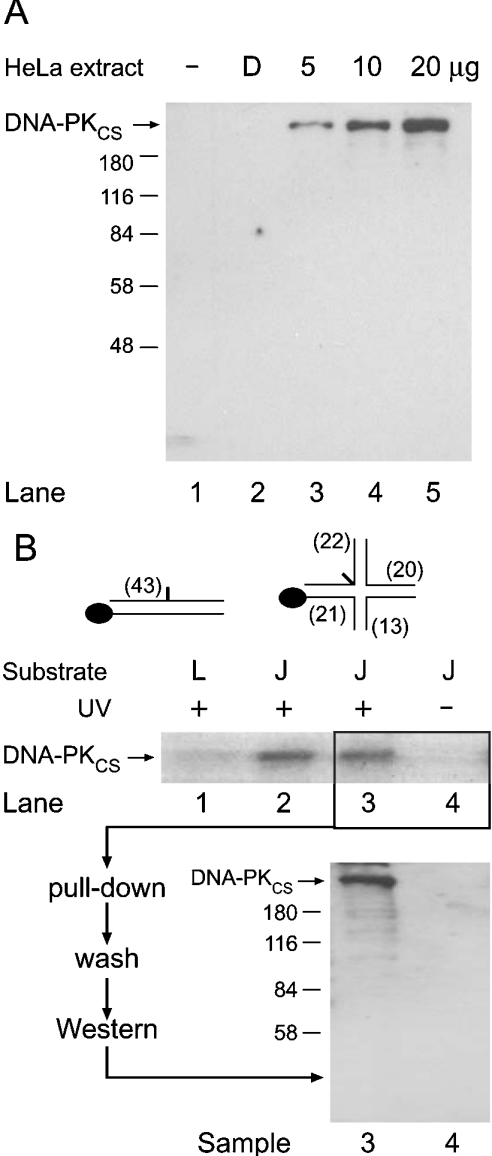
A specific polyclonal antibody was used to prove that the radiolabelled band co-migrating with DNA-PKCS represents covalent complexes of this enzyme with DNA. This particular antibody, directed against DNA-PKCS, generated a single band when tested in Western blots of HeLa nuclear extracts (Figure 9A). A pull-down experiment was performed by biotinylation of one end of the photoreactive four-way DNA junction as indicated in Figure 9(B). After incubation in HeLa nuclear extract, the presence of such a biotin residue did not change the preference of DNA-PKCS for the four-way probe relative to the linear control [cf. the amount of cross-linked products in Figure 9(B, upper panel, lanes 1–3)]. No cross-linked complexes were observed when UV irradiation was omitted (Figure 9B, upper panel, lane 4). A representative aliquot of the reaction of lane 3, incubated in HeLa cell extract and cross-linked under UV light, was mixed with streptavidin-conjugated paramagnetic beads. A control sample, identical with the reaction of lane 4, was subjected to the same treatment but without exposure to UV light. The complexes were pulled down and washed extensively with high salt (1 M NaCl) to remove free proteins not cross-linked to the biotinylated substrate. The resulting fractions of paramagnetic beads were resolved by SDS/PAGE, followed by Western-blot analysis using the specific anti-DNA-PKCS antibody. These control experiments confirmed that DNA-PKCS is indeed cross-linked to the four-way junctions (Figure 9B, lower panel). No specific antibody signal was detected when the UV cross-linking was omitted (Figure 9B, sample 4) or when the same probes were tested with a corresponding preimmune serum (results not shown). These results confirm that the radiolabelled complexes co-migrating with DNA-PKCS are indeed the product of a direct interaction of this enzyme with four-way DNA junctions.
Figure 9. Immunological identification of DNA-PKCS in the covalent complexes with four-way DNA junctions.
(A) Western blot of HeLa nuclear extracts demonstrating the selectivity of the anti-DNA-PKCS antibody. (B) Visualization of cross-linked DNA-PKCS. Biotinylated four-way DNA junctions (0.2 nM) were cross-linked in HeLa nuclear extract (10 μg). After separation on streptavidin-conjugated beads, the immobilized complexes were analysed by SDS/PAGE. Sample 3, analysis of cross-linked complexes; sample 4, control experiment without UV irradiation. L, linear; J, four-way junction.
Functional consequence of structure-specific nucleoprotein complexes
Incubation of DNA-PK with the 43-mer fragments or the four-way junction revealed a similar level of kinase activity. In fact, we observed that both types of DNA molecules induced the autophosphorylation reaction as well as the phosphorylation of a p53 peptide. Nevertheless, it is probable that the DNA-PKCS subunit adopts distinctly different conformations when assembled with either linear DNA or the architecturally more complex four-way junction. To test this hypothesis, we generated a panel of differentially biotinylated substrates.
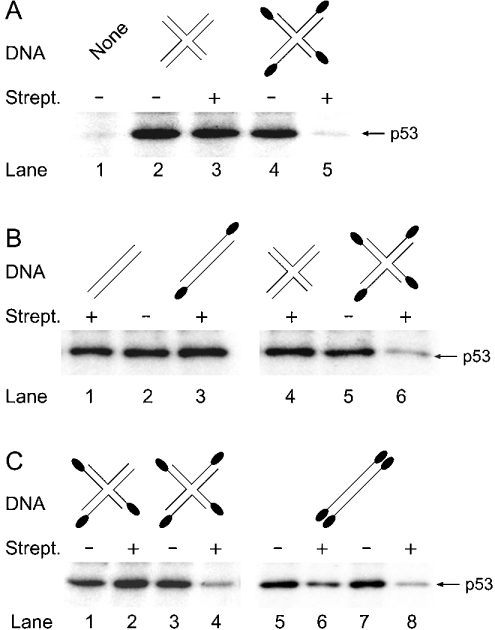
Previous reports showed that, to become activated, DNA-PK must be capable of interacting with at least one end of duplex substrates [32]. To investigate the structural requirements for activation by four-way structures, we constructed a substrate in which all the four ends were modified by the introduction of a biotin residue on the 3′-terminal nucleotide. The stimulation of kinase activity was then assessed by monitoring the phosphorylation of the p53 protein during 10 min reactions with 32P-ATP. The resulting incubation mixtures were analysed on denaturing polyacrylamide gels to determine the level of p53 phosphorylation. The autoradiograph of Figure 10(A) shows that biotin modification of the four-way junction molecule by itself, in the absence of streptavidin, had little or no effect on the ability of the DNA to activate the kinase activity (lane 4). However, addition of streptavidin to the substrate in which the DNA strands ended with biotin residues resulted in nearly complete suppression of kinase activity (lane 5). Inhibition of DNA-PKCS activity required streptavidin–DNA binding since no decrease in p53 phosphorylation was observed when streptavidin was added to DNA lacking biotin modifications (lane 3). Thus the obstruction of DNA ends by the formation of biotin–streptavidin complexes interfered with enzyme activation, but not with its binding to the substrate, since the same streptavidin-capped four-way junction provided a preferential site for the recruitment of DNA-PKCS (cf. Figure 6B). In agreement with the results obtained by Hammarsten et al. [32], we found that p53 phosphorylation was not decreased when only the 3′-terminus of each complementary oligonucleotide in the 43-mer duplex was biotin-labelled, such that the linear fragment ended with only one biotin–streptavidin complex on each side (Figure 10B, lane 3). In contrast, as confirmed in lane 6, DNA-PKCS activity was severely decreased in the presence of one obstructing complex at each 3′-end of the four-way substrate.
Figure 10. Activity of complexes formed by DNA-PKCS with four-way junctions.
(A) Suppression of kinase activity by obstruction of each DNA end with one biotin–streptavidin complex. The streptavidin concentration was 0.5 μM. Each reaction contained four-way junction DNA molecules (0.2 nM), DNA-PK (0.5 μg), p53 protein (100 ng) and 32P-ATP (100 μM). After 10 min of incubation, the reaction products were resolved by denaturing gel electrophoresis to visualize the phosphorylated p53. Lane 1, control incubation without DNA. (B) Different requirements for activation by linear 43-mer duplexes (lanes 1–3) and four-way junctions (lanes 4–6). (C) Activation of DNA-PKCS by four-way junction molecules containing one free double-strand end (lanes 1–4) and inhibition of DNA-PKCS activity by complete obstruction of the ends of linear DNA (lanes 5–8).
To stimulate effectively the phosphorylation by DNA-PKCS, it was necessary to leave at least one double-strand end of the four helical arms completely devoid of streptavidin complexes (Figure 10C, lane 2). On the other hand, again in agreement with previous results [32], activation of the enzyme by linear fragments could be inhibited only when the DNA was capped by streptavidin complexes at all terminal nucleotides (lanes 5–8). These distinct substrate requirements observed with linear DNA and four-way junctions indicate a structural difference in the mechanism of enzyme activation, suggesting that DNA-PKCS adopts different allosteric conformations in the complexes with these two types of DNA substrates.
DISCUSSION
Failure to repair double-strand breaks can lead to chromosomal fragmentation, deletions or translocations, resulting in cell death or cancer. Two distinct mechanisms for double-strand break repair have been described, involving either NHEJ or HR. NHEJ connects DNA ends irrespective of their sequence and is supposed to be effective at all times in the cell cycle, whereas HR is functional in the S/G2 phase when the DNA breaks can be repaired using genetic information from a sister chromatid [33,34]. In the NHEJ pathway, DNA-PKCS occupies a strategic position near the DNA ends, where it acts as a bridging factor that mediates the synapsis between non-homologous break sites [35,36]. The present study indicates that DNA-PKCS is a more universal sensor of DNA discontinuities contrary to previous understanding, since this enzyme is also capable of recognizing the Holliday crossover intermediates that are formed between homologous DNA molecules.
In the present study, a cross-linking approach has been employed that had already been used previously to analyse the geometry of DNA-PK complexes associated with DNA [24,25]. Our results confirm that this azido-mediated cross-linking technique is a suitable method to characterize the interaction of DNA-PK with nucleic acid substrates. First, the catalytic subunit is cross-linked to double-stranded fragments, which activate the enzyme, but not to single-stranded oligonucleotides that fail to stimulate its kinase activity (Figure 2B; [32]). Secondly, the cross-linking of DNA-PKCS in a higher ionic strength buffer reflects the stabilizing effect exerted by the Ku subunits in the proximity of DNA ends (Figure 2D). Thirdly, cross-linking also faithfully reflects the autoregulatory properties of DNA-PKCS [26], since the binding efficiency was decreased by preincubation of the enzyme with ATP, supporting the notion that only the unphosphorylated fraction of DNA-PKCS can associate with DNA. Fourthly, we demonstrated that the cross-linked DNA-PKCS subunit retains its enzymic activity and is capable of phosphorylating itself as well as heterologous protein acceptors (Figure 3). Finally, the identification of DNA-PKCS bound to interior positions of DNA duplexes is not without precedent, as previous atomic force microscopy studies have shown trapping of the enzymic subunits both at the DNA ends and internally [37]. We noted, however, that this binding to internal positions of linear substrates is suppressed in the context of the nuclear protein environment (Figure 7B).
Consistent with DNA-PKCS and Ku functioning in concert with the NHEJ pathway, the absence of either component leads to defects in double-strand break repair and V(D)J rearrangements [2,4,6]. Thus DNA-PKCS and Ku are often considered to represent the individual subunits of the DNA-PK holoenzyme complex. However, DNA-PKCS and Ku associate weakly, if at all, in solution and the active complex is formed only in the presence of DNA [38], raising the possibility that alternative pathways of activation of the kinase subunit may occur in vivo through interactions with regulatory partners other than Ku. Homologues of Ku have been identified in a variety of organisms, from yeast to mammals, whereas DNA-PKCS seems to be restricted to higher eukaryotes [39]. The recent finding of the evolutionary nature of DNA-PKCS supports the view that this enzyme probably adopts other cellular functions in relation to DNA metabolism or signal transduction that are independent of the more ancient Ku subunits. An example for such an alternative mechanism has already been discovered, as the kinase function of DNA-PK can be activated by supercoiled DNA in the absence of double-strand breaks through association with the 18 kDa matrix protein C1D [17]. Unlike the Ku subunits, which are not involved in HR, C1D is an interaction partner of DNA-PKCS that operates in both NHEJ and HR [19].
The outstanding finding of the present study is that DNA-PKCS, but not the Ku heterodimer, binds directly to the Holliday junction region generated, e.g. as an intermediate of strand-exchange reactions during HR (Figures 5 and 6). Cross-linking experiments targeting the endogenous enzyme in cell extracts indicate that, in the context of a nuclear protein environment, DNA-PKCS is capable of binding to DNA ends and four-way junctions with similar affinities (Figures 7 and 8). DNA-PKCS was unequivocally identified in the covalent complexes with four-way junctions by immunological detection (Figure 9). In addition, we observed that the binding of DNA-PKCS to four-way junctions becomes resistant to higher ionic strength in the presence of nuclear protein extracts. In fact, isolated DNA-PKCS molecules could be cross-linked to Holliday junctions only at low ionic strength, even in the presence of Ku, indicating that the interaction of the catalytic subunit with four-way junctions is exquisitely salt-labile (Figure 5C). However, the cross-linking reaction between the 470 kDa catalytic subunit and Holliday junctions became insensitive to higher ionic strength conditions after incubation of the DNA probes with nuclear cell extracts, indicating that the interaction can be stabilized by cellular binding partners (Figure 7B). Finally, distinct molecular complexes formed by DNA-PKCS with linear DNA and four-way junctions could be distinguished at a functional level using a panel of differentially biotinylated substrates (Figure 10), indicating that DNA-PKCS adopts a specialized active conformation after interaction with four-way structures.
The recognition of Holliday junctions by DNA-PKCS bears on the question of how cells control the choice between the repair of double-strand breaks by NHEJ and HR. In fact, there might be passive and active modes of competition between the two diverging pathways. With passive competition, the repair outcome may simply depend on whether NHEJ or HR proteins bind first to broken DNA ends. For example, Ku proteins drive double-strand breaks into NHEJ and, in the absence of Ku in meiotic cells, the double-strand breaks become a substrate for HR [40]. With active competition, the proteins involved in NHEJ and HR may interact directly and influence each other's function. Such an active regulatory role of DNA-PKCS appears to be particularly important for the processing of stalled or blocked replication forks, as indicated by the observation that the spontaneous HR rates, which are dependent on DNA replication, are most efficiently suppressed by DNA-PKCS [18].
The newly discovered affinity of DNA-PKCS for the crossover structure of Holliday junctions provides an immediate mechanism by which this NHEJ subunit may regulate HR processes. It is thus conceivable that modulation of HR by DNA-PKCS can occur through recruitment of the enzyme to a subset of four-way intermediates, followed by phosphorylation of proteins involved in branch migration or Holliday junction resolution. A possible four-way DNA target of DNA-PKCS involves the chicken foot-like intermediate formed by backward migration of stalled replication forks [29]. These four-way intermediates are supposed to facilitate the replicative bypass of damaged replication templates before the replication fork is regenerated. The recognition of chicken foot intermediates by DNA-PKCS could prevent branch migration in the wrong direction or avoid the improper endonucleolytic resolution of this particular kind of Holliday junction. Also, tumour cells lacking DNA-PKCS are susceptible to gene amplification [41], suggesting that the special recombinogenic intermediates arising during such amplification events may constitute another target of DNA-PKCS.
Acknowledgments
We thank Dr G. Marra and Dr J. Jiricny for supplying us with the nuclear cell extracts. This research was supported by the Swiss National Science Foundation grant no. 3100A0-101747.
References
- 1.Gottlieb T. M., Jackson S. P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell (Cambridge, Mass.) 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 2.Smith G. C., Jackson S. P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 3.van Gent D. C., Hoeijmakers J. H., Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 4.Lieber M. R. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells. 1999;7:77–85. doi: 10.1046/j.1365-2443.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- 5.Chan D. W., Chen B. P.-C., Prithivirajsingh S., Kurimasa A., Story M. D., Quin J., Chen D. J. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 7.Schuler W., Weiler I. J., Schuler A., Phillips R. A., Rosenberg N., Mak T. W., Kearney J. F., Perry R. P., Bosma M. J. Rearrangement of antigen receptor gene is defective in mice with severe combined immune deficiency. Cell (Cambridge, Mass.) 1986;46:963–972. doi: 10.1016/0092-8674(86)90695-1. [DOI] [PubMed] [Google Scholar]
- 8.Fulop G. M., Phillips R. A. The scid mutation in mice causes a general defect in DNA repair. Nature (London) 1990;347:478–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 9.Taccioli G. E., Amatucci A. G., Beamish H. J., Gell D., Xiang X. H., Arzayus M. I., Priestly A., Jackson S. P., Rothstein A. M., Jeggo P. A., et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 10.Woo R. A., Jack M. T., Xu Y., Burma S., Chen D. J., Lee P. W. DNA damage-induced apoptosis requires the DNA-dependent protein kinase, and is mediated by the latent population of p53. EMBO J. 2002;21:3000–3008. doi: 10.1093/emboj/cdf307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao R. G., Cao C. X., Zhang H., Kohn K. W., Wold M. S., Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA–DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe F., Shinohara K., Teraoka H., Komatsu K., Tatsumi K., Suzuki F., Imai T., Sagara M., Tsuji H., Ogiu T. Involvement of DNA-dependent protein kinase in down-regulation of cell cycle progression. Int. J. Biochem. Cell Biol. 2003;35:432–440. doi: 10.1016/s1357-2725(02)00268-6. [DOI] [PubMed] [Google Scholar]
- 13.Gilley D., Tanaka H., Hande M. P., Kurimasa A., Li G. C., Oshimura M., Chen D. J. DNA-PKCS is critical for telomer capping. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15084–15088. doi: 10.1073/pnas.261574698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn A., Gottlieb T. M., Jackson S. P., Grummt I. DNA-dependent protein kinase: a potent inhibitor of transcription by RNA polymerase I. Genes Dev. 1995;9:193–203. doi: 10.1101/gad.9.2.193. [DOI] [PubMed] [Google Scholar]
- 15.Giffin W., Torrance H., Rodda D. J., Prefontaine G. G., Pope L., Hache R. J. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature (London) 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 16.Jin S., Kharbanda S., Mayer B., Kufe D., Weaver D. T. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J. Biol. Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 17.Yavuzer U., Smith G. C., Bliss T., Werner D., Jackson S. P. DNA end-independent activation of DNA-PK mediated via association with the DNA-binding protein C1D. Genes Dev. 1998;12:2188–2199. doi: 10.1101/gad.12.14.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen C., Kurimasa A., Brenneman M. A., Chen D. J., Nickoloff J. A. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erdemir T., Bilican B., Cagatay T., Goding C. R., Yavuzer U. Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol. Microbiol. 2002;43:947–957. doi: 10.1046/j.1365-2958.2002.03224.x. [DOI] [PubMed] [Google Scholar]
- 20.Dvir A., Stein L. Y., Calore B. L., Dynan W. S. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J. Biol. Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- 21.Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 22.Missura M., Buterin T., Hindges R., Hübscher U., Kasparkova J., Brabec V., Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaneva M., Kowalewski T., Lieber M. R. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo S., Dynan W. S. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKCS induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoo S., Kimzey A., Dynan W. S. Photocross-linking of an oriented DNA repair complex. J. Biol. Chem. 1999;274:20034–20039. doi: 10.1074/jbc.274.28.20034. [DOI] [PubMed] [Google Scholar]
- 26.Chan D. W., Lees-Miller S. P. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 27.Hammarsten O., Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc. Natl. Acad. Sci. U.S.A. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West R. B., Yaneva M., Lieber M. R. Productive and nonproductive complexes of Ku and DNA-dependent protein kinase at DNA termini. Mol. Cell. Biol. 1998;18:5908–5920. doi: 10.1128/mcb.18.10.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox M. M. The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 2002;510:107–120. doi: 10.1016/s0027-5107(02)00256-7. [DOI] [PubMed] [Google Scholar]
- 30.Getts R. C., Stamato T. D. Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J. Biol. Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- 31.Cheong N., Perrault A. R., Wang H., Wachsberger P., Mammen P., Jackson I., Iliakis G. DNA-PK-independent rejoining of DNA double-strand breaks in human cell extracts in vitro. Int. J. Radiat. Biol. 1999;75:67–81. doi: 10.1080/095530099140825. [DOI] [PubMed] [Google Scholar]
- 32.Hammarsten O., DeFazio L. G., Chu G. Activation of DNA-dependent protein kinase by single-stranded DNA ends. J. Biol. Chem. 2000;275:1541–1550. doi: 10.1074/jbc.275.3.1541. [DOI] [PubMed] [Google Scholar]
- 33.Jeggo P. A. DNA breakage and repair. Adv. Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 34.Takata M., Sasaki M. S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L., Trujillo K., Sung P., Tomkinson A. E. Interactions of the DNA ligase IV–Xrcc4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- 36.DeFazio L. G., Stansel R. M., Griffith J. D., Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cary R. B., Peterson S. R., Wang J., Bear D. G., Bradbury E. M., Chen D. J. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suwa A., Hirakata M., Takeda Y., Jesch S. A., Mimori T., Hardin J. A. DNA-dependent protein kinase (Ku protein–p350 complex) assembles on double-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimori A., Araki R., Fukumura R., Ohhata T., Takahashi H., Kawahara A., Tatsumi K., Abe M. Identification of four highly conserved regions in DNA-PKCS. Immunogenetics. 2000;51:965–973. doi: 10.1007/s002510000227. [DOI] [PubMed] [Google Scholar]
- 40.Goedecke W., Eijpe M., Offenberg H. H., van Aalderen M., Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat. Genet. 1999;23:194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- 41.Mondello C., Rebuzzini P., Dolzan M., Edmonson S., Taccioli G. E., Giulotti E. Increased gene amplification in immortal cells deficient for the DNA-dependent protein kinase catalytic subunit. Cancer Res. 2001;61:4520–4525. [PubMed] [Google Scholar]