Abstract
In the present paper, we report that a Cdc6 (cell-division control)-like factor from the hyperthermophilic crenarchaeon Sulfolobus solfataricus (referred to as SsoCdc6-2) has a modular organization of its biological functions. A reliable model of the SsoCdc6-2 three-dimensional structure was built up, based on the significant sequence identity with the Pyrobaculum aerophylum Cdc6 (PaeCdc6), whose crystallographic structure is known. This allowed us to design two truncated forms of SsoCdc6-2: the ΔC (residues 1–297, molecular mass 35 kDa) and the ΔN (residues 298–400, molecular mass 11 kDa) proteins. The ΔC protein contains the nucleotide-binding Rossmann fold and the Sensor-2 motif (Domains I and II in the PaeCdc6 structure), and retains the ability to bind and hydrolyse ATP. On the other hand, the ΔN protein contains the C-terminal WH (winged helix)-fold (Domain III), and is able to bind DNA molecules and to inhibit the DNA helicase activity of the SsoMCM (mini-chromosome maintenance) complex, although with lesser efficiency with respect to the full-sized SsoCdc6-2. These results provide direct biochemical evidence that the Cdc6 WH-domain is responsible for DNA-binding and inhibition of MCM DNA helicase activity.
Keywords: Archaea, DNA helicase, DNA replication, helicaseloader, Sulfolobus solfataricus, thermophilic organism
Abbreviations: AAA+, ATPases that are associated with various cellular activities; Afu, Archeoglobus fulgidus; Cdc, cell-division control; ds, double-stranded; GST, glutathione S-transferase; IPTG, isopropyl β-D-thiogalactoside; LB, Luria–Bertani; MCM, mini-chromosome maintenance; Mth, Methanobacterium thermoautotrophicum; Ni-NTA, Ni2+-nitrilotriacetate; ORC, origin-recognition complex; Pae, Pyrobaculum aerophilum; pre-RC, pre-replication complex; ss, single-stranded; Sso, Sulfolobus solfataricus; TBE, Tris/borate/EDTA; WH, winged helix
INTRODUCTION
The initiation of chromosomal DNA replication is of fundamental importance for the inheritance of genetic material and for cell-cycle regulation [1]. In Eukarya, chromosomal replication origins are specifically bound by a heterohexameric assembly named the ORC (origin recognition complex) [2]. At the onset of mitosis, various additional initiation factors associate with ORC to form the pre-RC (pre-replication complex), including the MCM (mini-chromosome maintenance) DNA helicase, Cdc6 (cell-division control) and Cdt1 (for a review see [3]). Cdc6 is believed to play a critical role in the MCM complex loading on to DNA at the replication origins [4,5]. In addition, the release and activation of the MCM DNA helicase in an active form requires several additional regulatory factors, such as MCM10, geminin, Cdc45 and the cell-cycle-dependent kinases [3].
The MCM complex is composed of six different, but orthologous, proteins, which were demonstrated to be essential not only for the initiation, but also for the elongation, phase of DNA replication in Saccharomyces cerevisiae [6,7]. However, only a subcomplex consisting of the MCM 4/6/7 subunits was shown to possess DNA helicase activity in vitro, whereas the MCM heterohexamer was found to be inactive in DNA-unwinding assays [8,9].
The Cdc6 protein is a member of the AAA+ family (ATPases that are associated with various cellular activities) [10]. These factors contain a nucleotide-binding domain of the Rossmann fold family and can bind and hydrolyse ATP. The AAA+ family includes several eukaryotic and bacterial replication factors, such as the Orc1, Orc4, Orc5 subunits, the six MCM proteins, the Escherichia coli initiation factors DnaA and DnaC, the γ and δ' subunits of the E. coli γ complex and the five subunits of the eukaryotic RF-C (replication factor C). Nucleotide binding and/or hydrolysis by the AAA+ proteins usually triggers structural changes that are transmitted to the target protein(s) and modulate its (their) biological functions [10]. The results of genetic analyses carried out in S. cerevisiae indicate that the integrity of the Cdc6 Walker A and B motifs is essential for a correct assembly of the pre-RC and normal cell growth [4–5,11]. It was hypothesized that Cdc6 could be the loader of the MCM DNA helicase at the eukaryotic replication origins. This hypothesis was supported by the finding that Cdc6 shows significant sequence similarity with the sliding clamp-loaders that promote assembly of ring-shaped proteins (such as PCNA or the β subunit) on to a primed DNA substrate and their association with the cognate replicative DNA polymerase [5]. However, at present, the molecular mechanism by which Cdc6 accomplishes this critical task is not known.
Comparative genomics indicated that the archaeal replication machinery is a simplified version of the eukaryotic one, thus suggesting that replication mechanisms could be similar in Archaea and Eukarya [12]. In fact, Archaea have genes coding for putative homologues of several eukaryotic replication proteins in their genomes, including the initiation factors MCM and Cdc6. The crystallographic structure of the Cdc6 factor from the crenarchaeon Pyrobaculum aerophilum (PaeCdc6) was solved and revealed that this protein is composed of three domains: the two N-terminal domains (Domains I and II) form an AAA+-type nucleotide-binding fold, whereas the C-terminal domain (Domain III) adopts a WH (winged-helix)-fold, as found in other known DNA-binding proteins [13]. Recently, we characterized the homohexameric MCM-like DNA helicase (SsoMCM) [14] and one of the three Cdc6 factors (SsoCdc6-2, previously named SsoCdc6-1) [15] from the hyperthermophilic crenarchaeon Sulfolobus solfataricus [16]. We found that SsoCdc6-2 is a monomer in solution, binds ss (single-stranded) and ds (double-stranded) DNA molecules, undergoes autophosphorylation in vitro, possesses a weak ATPase activity and strongly inhibits the SsoMCM DNA-helicase activity [15]. In the present paper, we investigated the modular organization of SsoCdc6-2 and demonstrated that a C-terminally deleted form of this protein (named ΔC), which includes Domains I and II, retains the ability to bind and hydrolyse ATP, whereas the C-terminal WH-domain (Domain III), produced as a separate recombinant protein (named ΔN), is able to bind ssDNA and dsDNA and to inhibit the SsoMCM DNA-helicase activity.
EXPERIMENTAL
Materials
All chemicals were of reagent grade. Restriction and modification enzymes were from New England Biolabs. Radioactive nucleotides were purchased from Amersham Biosciences. Oligonucleotides were synthesized by Proligo (Paris, France). SsoCdc6-2 wild-type and K286A (Lys286→Ala) mutant [15], and SsoMCM [14] were purified as described previously.
Homology modelling
Molecular modelling was performed on a Silicon Graphics O2+ workstation using the commercial software Insight II (version 2000.1, Accelrys, San Diego, CA, U.S.A.). The high-resolution X-ray crystal structure of PaeCdc6 [Protein Data Bank (PDB) code 1FNN] was used as a template structure [13]. The sequence alignment was obtained by exploiting the pair-wise sequence alignment tool of the package HOMOLOGY of Insight II with the option identity and default parameters. The alignment was manually corrected to avoid breakage of secondary structural elements. The final sequence identity was 30.6%. The correspondence of secondary-structural elements was verified by subjecting the SsoCdc6-2 sequence to a secondary-structure-prediction analysis with the program PHD (http://www.embl-heidelberg.de/predictprotein/predictprotein.html). Several alignments were tried and several three-dimensional models were constructed by using the MODELLER module [17] within Insight II. The final model was also opportunely minimized using the Discover3 Module. The resulting models were verified using the on-line software WHATH IF and ERRAT at the UCLA-DOE Structure Analysis and Verification Server (http://www.doe-mbi.ucla.edu/Services/SV/). Most of the analysed parameters had statistical values that were in the range of those expected for a naturally folded protein.
Construction of the SsoCdc6-2 truncated forms
The SsoCdc6-2 ΔC was generated by PCR carried out on the pProEX-Hta-SsoCdc6-2 construct [15] with the oligonucleotide Cdc6-2-for (5′-TTGGGAATTCTCATCTGTATTGATAATTAAACATAAGGAC-3′) as the forward primer (containing an EcoRI site, which is underlined) and the oligonucleotide 6-1ΔC (5′-TTGGCTGCAGTCACTCCGGATTTATTGTAGAATT-3′) as the reverse primer (containing a translational stop after codon 297, followed by a PstI site). The PCR product was cloned into EcoRI/PstI-linearized E. coli expression vector pProEX-Hta (Gibco) and sequenced. The SsoCdc6-2 ΔN was generated by PCR carried out on the pProEX-Hta-SsoCdc6-2 construct [15] with the oligonucleotide 6-1ΔNEco (5′-GGTTGAATTCAAGAGATTATAGATAG-3′) as the forward primer (containing an EcoRI site immediately upstream of codon 298) and the oligonucleotide Cdc6-1-rev (5′-TTGGCTGCAGTTAAGCTCTCGATTTTAACCTCACCATTAT-3′) as the reverse primer (containing a PstI site immediately downstream of the stop codon). The PCR product was cloned into the EcoRI/PstI-linearized E. coli expression vector pProEX-Hta and was sequenced. From this construct, the insert was then recovered by digestion with EcoRI and XhoI and subcloned into the EcoRI/XhoI linearized pGEX-4T-2 expression vector (Amersham Biosciences).
Expression and purification of recombinant proteins
E. coli BL21-CodonPlus(DE3)-RIL cells (Stratagene) transformed with the plasmid pProEX-Hta-SsoCdc6-2 ΔC were grown at 37 °C in 0.5 litre of LB (Luria–Bertani) medium containing 30 μg/ml chloramphenicol and 100 μg/ml ampicillin. When the culture reached a D600 of 0.7, protein expression was induced by addition of IPTG (isopropyl β-D-thiogalactoside) to 0.2 mM. The bacterial culture was incubated at 37 °C for an additional 2 h. Then cells were harvested by centrifugation for 30 min at 30000 rev./min (Beckman 60 Ti rotor) at 10 °C, and the pellet was stored at −20 °C until use. The pellet was thawed and resuspended in 20 ml of buffer A [25 mM Tris/HCl, pH 8.0, 2.5 mM MgCl2, 100 mM NaCl and 15% (v/v) glycerol] supplemented with protease inhibitors (50 μg/ml PMSF, 0.2 μg/ml benzamidine and 1 μg/ml aprotinin). Cells were broken by two consecutive passages through a French pressure cell apparatus (Aminco, Silver Spring, MD, U.S.A.) at 1500 p.s.i. (1 p.s.i.=6.9 kPa). The resulting cell extract was centrifuged for 30 min at 30000 rev./min (Beckman 60 Ti rotor) at 10 °C. The supernatant was subjected to heat treatment at 70 °C for 5 min, then incubated in ice for 10 min. The thermoprecipitated proteins were removed by centrifugation for 30 min at 30000 rev./min (Beckman 60 Ti rotor) at 10 °C. The supernatant was passed through a 0.22 μm pore-size filter (Millipore) and loaded on to a 0.5 ml Ni-NTA (Ni2+-nitrilotriacetate) Superflow agarose column (Qiagen) pre-equilibrated in buffer A. After a washing step with buffer A, the elution was carried out with 12 ml of an imidazole step gradient (50–500 mM) in buffer A. Fractions of 1.5 ml were collected and analysed by SDS/PAGE. Fractions containing the recombinant protein were pooled (10 ml) and dialysed overnight against buffer A. The dialysed sample was divided into 20 μl aliquots and stored at −80 °C. The final yield of the recombinant protein after this purification procedure was approx. 20 mg.
E. coli BL21-CodonPlus(DE3)-RIL cells (Stratagene) transformed with the plasmid pGEX-4T-2-SsoCdc6-2 ΔN were grown at 37 °C in 0.5 litre of LB medium containing 30 μg/ml chloramphenicol and 100 μg/ml ampicillin. When the culture reached a D600nm of 0.6, protein expression was induced by the addition of IPTG to a concentration of 0.5 mM. The bacterial culture was incubated at 37 °C for an additional 2 h. Then cells were harvested by centrifugation for 30 min at 30000 rev./min (Beckman 60 Ti rotor) at 10 °C and the pellet was stored at −20 °C until use. The pellet was thawed and resuspended in 20 ml of buffer PBS-T [100 mM sodium phosphate buffer, pH 7.5, 100 mM NaCl and 1% (v/v) Triton X-100] supplemented with protease inhibitors (50 μg/ml PMSF, 0.2 μg/ml benzamidine and 1 μg/ml aprotinin). Cells were broken by two consecutive passages through a French pressure cell apparatus at 1500 p.s.i. The resulting cell extract was centrifuged for 30 min at 30000 rev./min (Beckman 60 Ti rotor) at 10 °C. The supernatant was passed through a 0.22 μm pore-size filter and mixed with 3 ml of glutathione–Sepharose 4B resin (Amersham Biosciences) pre-equilibrated in PBS. The mixture was incubated at room temperature (22 °C) for 1 h with gentle shaking. The resin was then removed by centrifugation for 30 min at 30000 rev./min (Beckman 60 Ti rotor) at 10 °C and washed three times with 15 ml of PBS. The resin was then resuspended in 2 ml of PBS containing 100 units of thrombin protease (Amersham Biosciences) and incubated at 4 °C overnight. The resin was then removed by centrifugation for 30 min at 30000 rev./min (Beckman 60 Ti rotor) at 10 °C, and the supernatant (supernatant fraction I) was stored. Three washing steps were carried out using 3 ml aliquots of PBS. After each washing step, the resin was removed by centrifugation for 30 min at 30000 rev./min (Beckman 60 Ti rotor) at 10 °C, and the supernatant was recovered. The three supernatants were added to the supernatant fraction I and this pool (15 ml volume) was concentrated on a Centricon YM3 membrane (Amicon) to 0.8 ml. This sample was then dialysed overnight against a buffer containing 50 mM Tris/HCl, pH 8.0, 50 mM NaCl and 2.5 mM MgCl2. The dialysed sample was divided into 20 μl aliquots and stored at −20 °C. The final yield of the recombinant protein after this purification procedure was approx. 0.5 mg.
DNA band-shift assays
A 51-mer oligonucleotide (51nt-4U; 5′-CCCAGTCACGACGTTGTAAAACGACGGCCAGTGCGAGGCGCGCGAAGACCG-3′) was labelled with [γ-32P]ATP and T4 polynucleotide kinase, purified using Quantum Prep PCR Kleen Spin columns (Bio-Rad Laboratories), and used in ssDNA band-shift assays. To prepare dsDNA molecules, a 51-mer oligonucleotide complementary to 51nt-4U (51nt-4Urev; 5′-CGGTCTTCGCGCGCCTCGCACTGGCCGTCGTTTTACAACGTCGTGACTGGG-3′) was mixed with equal molar amounts of 51nt-4U; the mixture was incubated for 5 min at 95 °C and then slowly cooled to room temperature. These molecules were labelled with [γ-32P]ATP, purified as described above, and then used in dsDNA band-shift assays.
For the ssDNA and dsDNA band-shift assays, 10 μl mixtures were prepared containing 200 fmol of 32P-labelled ssDNA or dsDNA in 20 mM Tris/HCl, pH 7.5, 100 mM NaCl, 2 mM EDTA, 0.5 mM MgCl2, 0.7 mM 2-mercaptoethanol and the indicated amounts of SsoCdc6-2. These samples were incubated at room temperature for 10 min and then subjected to electrophoresis in a 5% (w/v) polyacrylamide/bis (29:1) gel in 0.5× TBE (Tris/borate/EDTA) buffer at a constant voltage of 100 V. The gel was then dried, and the radioactive bands were detected using a PhosphorImager (Molecular Dynamics).
ATPase assay
The standard ATPase assay reaction mixture (10 μl) contained 25 mM Tris/HCl, pH 8.0, 2.5 mM 2-mercaptoethanol, 50 mM sodium acetate, 5 mM MgCl2 and 100 μM [γ-32P]ATP (0.5–1 μCi). Incubations were performed for 30 min at 60 °C in a heated-top PCR machine to prevent evaporation and were stopped on ice. A 1 μl aliquot of each mixture was spotted on to a poly(ethyleneimine)–cellulose thin-layer plate (Merck) and run in 0.5 M LiCl and 1 M methanoic acid. The amounts of [γ-32P]ATP hydrolysed to [32P]orthophosphate were quantified using a PhosphorImager. The rate of ATP hydrolysis was determined in the linear range of reaction time and protein concentration dependence. The amount of spontaneously hydrolysed ATP was determined using blank reactions without enzyme and subtracted from the reaction rate values calculated as above.
In vitro autophosphorylation of SsoCdc6-2
Samples of the purified SsoCdc6-2 wild-type, KA mutant, ΔN, and ΔC proteins were incubated for 30 min at 70 °C in a reaction mixture (20 μl volume) containing 1.66 pmol of [γ-32P]ATP in 25 mM Tris/HCl, pH 8.0, 2.5 mM 2-mercaptoethanol, 50 mM sodium acetate and 5 mM MgCl2, in the absence or presence of 0.5 μg of M13 ssDNA or dsDNA (Amersham Biosciences). The proteins were then separated by SDS/15% PAGE and 32P-labelled bands were detected using a PhosphorImager.
DNA-helicase activity assay
A 85-mer oligonucleotide was used for the preparation of the DNA helicase substrate. This oligonucleotide (5′-TTGAACCACCCCCTTGTTAAATCACTTCTACTTGCATGCCTGCAGGTCGACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCG-3′) was complementary to the M13mp18(+) strand except for a 30-nt 5′-tail (which is underlined). The oligonucleotide was labelled with [γ-32P]ATP and T4 polynucleotide kinase, and purified as previously described for the 51nt-4U oligonucleotide. To prepare partial duplexes, mixtures containing equimolar amounts of the labelled oligonucleotide and M13mp18(+) strand were incubated for 5 min at 95 °C and then slowly cooled to room temperature. Helicase-assay reaction mixtures (20 μl) contained 50 fmol of 32P-labelled substrate (approx. 1×103 c.p.m./fmol) in 25 mM Tris/HCl, pH 8.0, 2.5 mM 2-mercaptoethanol, 50 mM sodium acetate and 5 mM MgCl2. The reaction mixtures were incubated for 30 min at 70 °C in a heated-top PCR machine to prevent evaporation and stopped by addition of 5 μl of 5× stop solution [0.5% (w/v) SDS, 40 mM EDTA, 0.5 mg/ml proteinase K, 20% (v/v) glycerol and 0.1% (w/v) Bromophenol Blue], then run on an 8% (w/v) polyacrylamide gel in TBE containing 0.1% (w/v) SDS at a constant voltage of 150 V. After electrophoresis, the gel was soaked in 20% trichloroacetic acid and analysed using a PhosphorImager. The reaction products were quantified, and any free oligonucleotide in the absence of enzyme was subtracted.
Limited proteolysis analyses
Samples of purified SsoCdc6-2 (3 μg) were incubated at 37 °C for 30 min in mixtures (20 μl volume) containing 5 mM Tris/HCl, pH 8.0, 0.5 mM MgCl2, 20 mM NaCl, 5% (v/v) glycerol, with (or without) 5 mM ATP, and the following amounts of trypsin (Worthington Biochemicals Corporation, Lakewood, NJ, U.S.A.): 0 ng, 125 ng, 250 ng, 500 ng and 1000 ng. Reactions were stopped by adding 10 μl of 3 × SDS/PAGE stop solution [1.5 M Tris/HCl, pH 6.8, 600 mM 2-mercaptoethanol, 30% (v/v) glycerol, 2% (w/v) SDS and 0.06% (w/v) Bromophenol Blue) to each mixture. Samples were then incubated at 95 °C for 5 min and run on an SDS/12.5% polyacrylamide gel. Protein bands were detected by staining with Coomassie Brilliant Blue G-250 (Serva).
RESULTS
Homology modelling
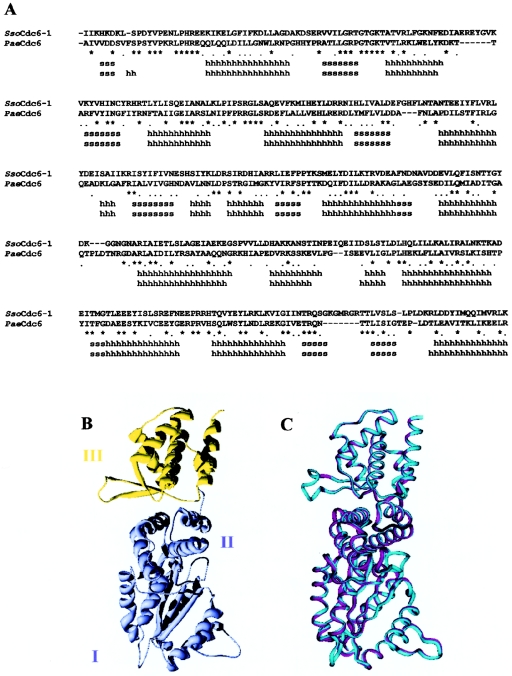
In a recent report, we demonstrated that SsoCdc6-2 is able to hydrolyse ATP, bind ssDNA and dsDNA, and strongly inhibit the DNA-helicase activity of SsoMCM on DNA substrates consisting of oligonucleotides annealed to M13 ssDNA [15]. To address further the structure–function relationships in SsoCdc6-2, a model of its three-dimensional structure was built up. For our modelling purposes, we used as a reference the PaeCdc6 three-dimensional structure, that was solved at 2.0 Å (1 Å=0.1 nm) [13]. A sequence alignment was generated between SsoCdc6-2 and PaeCdc6 (Figure 1A) and submitted to the modelling procedure described in the Experimental section. Residues 18–410 of SsoCdc6-2 were modelled on to residues 1–388 of PaeCdc6 (numbering according to the structure). Because residues 353–361 were missing in the PaeCdc6 structure, this region was modelled as a loop. Figure 1(B) shows the general folding pattern of the SsoCdc6-2 model with the Domains I and II (in pale blue) forming an AAA+ module and the Domain III (in yellow), that was proposed to contain a WH-fold [13]. The superimposed backbone traces of SsoCdc6-2 and PaeCdc6 (Figure 1C) displayed a 0.44 Å RMS (root mean square) deviation on 1456 superposed backbone atoms, by using a 2.0 Å cut-off distance. All the secondary-structural elements observed in PaeCdc6 were observed also in SsoCdc6-2, except for the short helix α1. Most of the residues that in PaeCdc6 are involved in the MgADP binding are fully conserved in SsoCdc6-2 and placed at corresponding positions in the SsoCdc6-2 model (G. Manco, M. De Felice and F. M. Pisani, unpublished work). An important feature of the PaeCdc6 crystallographic structure is the presence of a WH-fold. It belongs to the large family of helix–turn–helix DNA-binding domains, and is found in proteins from various sources and with different biological functions, such as transcription factors, the histone H5 and LexA (reviewed in [19]). All the structural elements of the PaeCdc6 WH-domain (the αβααββ fold) are conserved in our model of the SsoCdc6-2 three-dimensional structure.
Figure 1. Molecular modelling of SsoCdc6-2.
(A) Structural sequence alignment between SsoCdc6-2 model and the PaeCdc6 structure. Identical or similar residues are marked with an asterisk or a dot respectively. Stretches of ‘s’ or ‘h’ indicate β-strands or α-helices respectively. Superposition and alignment were performed with the Swiss-PDB viewer program [29]. (B) SsoCdc6-2 structural model with a ribbon representation. Domains I and II are coloured pale blue; Domain III (WH) is coloured yellow. (C) Superposition of the SsoCdc6-2 model (cyan) and PaeCdc6 structure (magenta). Pictures were generated with the Swiss-PDB viewer program [29] and rendered with POV (Persistance of Vision) ray for Windows v. 3.1.
Production of SsoCdc6-2 truncated forms
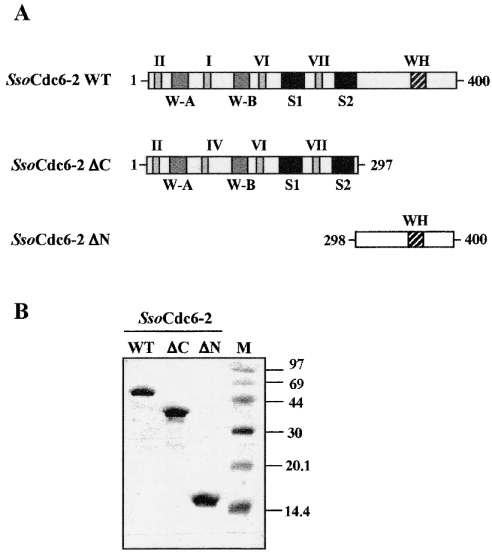
In order to investigate whether or not SsoCdc6-2 had a modular organization of its biological functions, as suggested by the structural analysis described above, we produced deleted forms of this protein and investigated their biochemical properties. Figure 2 shows a schematic representation of the polypeptide chain of SsoCdc6-2 and its C- and N-terminally deleted derivatives, named ΔC and ΔN respectively. The ΔC protein (residues 1–297; molecular mass 35 kDa) contains the Walker A and Walker B motifs, and the Sensor 1 and 2 boxes, whereas the ΔN protein (residues 298–400; molecular mass 11 kDa) includes only the C-terminal third of the SsoCdc6-2 polypeptide chain, which contains the WH-domain.
Figure 2. Production of the SsoCdc6-2 truncated forms.
(A) Schematic representation of the polypeptide chain of SsoCdc6-2 and its C- (SsoCdc6-2 ΔC) and N-terminally (SsoCdc6-2 ΔN) deleted forms. W-A and W-B stand for Walker A and Walker B motifs respectively. S1 and S2 stand for Sensor 1 and Sensor 2 respectively. (B) SsoCdc6-2 and its truncated forms were produced as recombinant proteins and purified as described in the Experimental section. Samples of the homogeneous proteins (5 μg) were subjected to SDS/15% PAGE. Protein bands were detected by Coomassie-Blue-staining of the gel.
The C-terminally deleted form was overproduced in E. coli as an His6-tagged protein using the vector pProEx-Hta, as was SsoCdc6-2 [15]. The recombinant protein was found to be expressed at a high level in a soluble form, and was purified using a procedure that included a thermal treatment of the cell extracts and a chromatographic step on an Ni-NTA column (see Figure 2B). The ΔN protein was overproduced in E. coli cells as a GST (glutathione S-transferase) fusion protein using the expression vector pGEX-4T-2. This chimaeric protein was expressed at high level in a soluble form and was purified on a glutathione–Sepharose 4B column. The GST portion was then removed by thrombin digestion, as described in the Experimental section (Figure 2B). To assess the oligomeric state of the SsoCdc6-2 truncated forms, we carried out gel-filtration experiments on to a Superose 6 column (Amersham Biosciences), and found that these proteins behave as monomers in solution.
Autophosphorylation of SsoCdc6-2 truncated forms
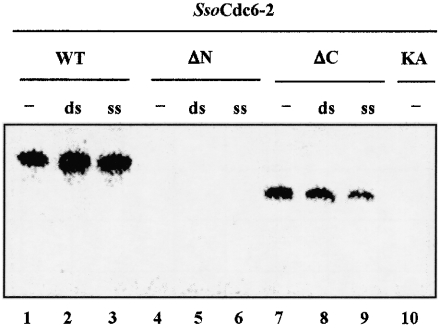
It was reported that the full-sized SsoCdc6-2 is able to autophosphorylate in vitro [15], as can the two Methanobacterium thermoautotrophicum (Mth) Cdc6 proteins [18]. This ability requires an integral Walker A motif, since the SsoCdc6-2 KA mutant protein did not undergo autophosphorylation, as also reported for the corresponding Walker A mutant of MthCdc6-2 [18]. We checked the ability of the ΔC and ΔN proteins to autophosphorylate in vitro when incubated with [γ-32P]ATP at 70 °C. As shown in Figure 3, whereas ΔC is still able to autophosphorylate under these conditions, ΔN does not possess this capability. In addition, autophosphorylation of ΔC is not inhibited in the presence of ssDNA or dsDNA (Figure 3).
Figure 3. Autophosphorylation of SsoCdc6-2 and its truncated forms.
Each protein was incubated at 70 °C for 30 min in a mixture (20 μl) containing [γ-32P]ATP in the absence (−) or in the presence of 0.5 μg of ss or ds M13 DNA, as described in the Experimental section. Following incubation, the samples were subjected to SDS/10% PAGE. The radioactive bands were visualized using a PhosphorImager.
ATPase activity of SsoCdc6-2 and its truncated forms
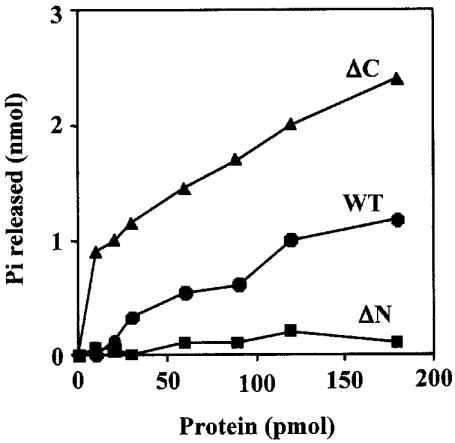
The primary structure of SsoCdc6-2 and ΔC contains the Walker A and B motifs, as shown in Figure 2. These sequence boxes are typically found in proteins endowed with ATP-hydrolytic activity. Therefore we tested the ATP-hydrolysis capability of SsoCdc6-2, ΔC and ΔN. The reaction mixtures were incubated at 60 °C and not at the optimal growth temperature for S. solfataricus (87 °C) to limit the thermally-induced autohydrolysis of ATP. Release of [γ-32P]orthophosphate was measured by TLC, as described in the Experimental section. We first assayed the SsoCdc6-2 ATPase activity at various ATP concentrations, and found that the maximal activity was observed at an ATP concentration of 5 mM (M. De Felice, L. Esposito and F. M. Pisani, unpublished work). Therefore we used this concentration in all the ATPase assays. As shown in Figure 4, 180 pmol of purified SsoCdc6-2 (10 μl reaction volume) was able to hydrolyse approx. 1.2 nmol of ATP in 60 min at 60 °C. The ΔC protein retains ATPase activity and is even more active than SsoCdc6-2: 180 pmol of purified ΔC was able to hydrolyse 2.4 nmol of ATP in the same conditions used for the full-sized protein. In contrast, ΔN was found to be completely devoid of ATPase activity even in assays where 180 pmol of purified protein was used.
Figure 4. ATPase activity of the SsoCdc6-2 and its truncated forms.
ATPase activity assays were carried out at 60 °C for 30 min using increasing amounts of SsoCdc6-2, ΔC or ΔN in the presence of radiolabelled [γ-32P]ATP and 5 mM ATP as described in the Experimental section. The orthophosphate (Pi) released during the hydrolysis reaction was plotted against the amount of protein used. Results are mean values of at least three independent experiments.
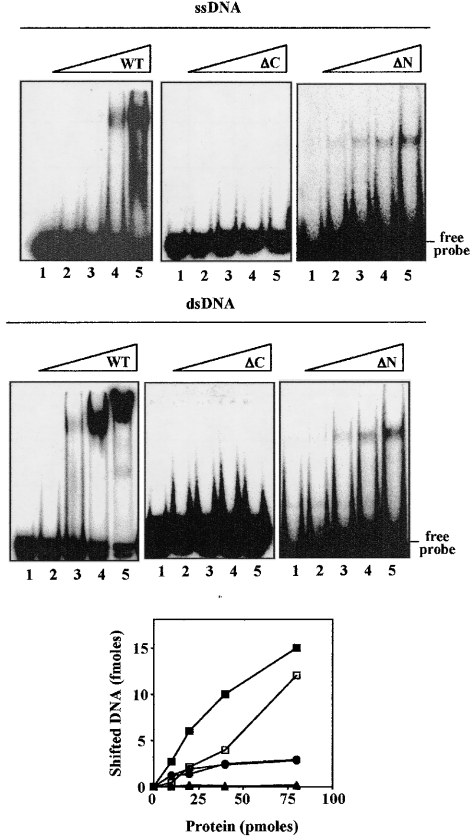
Binding of SsoCdc6-2 and its truncated forms to ssDNA or dsDNA
To determine whether the purified ΔC and ΔN proteins could bind ssDNA and/or dsDNA, electrophoretic mobility-shift assays were carried out using a γ-32P-labelled 51-mer synthetic oligonucleotide in ss and ds form. As shown in Figure 5, the ΔC protein was unable to bind either ssDNA or dsDNA. In contrast, ΔN was able to bind both ssDNA and dsDNA. However, a quantitative analysis of the shifted DNA indicated that the ΔN protein binds DNA with 5-fold reduced efficiency with respect to the full-sized SsoCdc6-2. Electrophoretic mobility-shift assays were carried out using mixtures of the SsoCdc6-2 ΔN and ΔC truncated forms in order to address the possibility that the N-terminal module could indirectly modulate DNA binding. The results of these experiments indicated that addition of the ΔC protein did not exert any significant effect on the DNA-binding capability of the ΔN truncated form (M. De Felice, L. Esposito and F. M. Pisani, unpublished work).
Figure 5. DNA-binding activity of full-sized SsoCdc6-2 and its truncated forms.
The ability of the proteins to bind a 32P-labelled 51-mer synthetic oligonucleotide in ss and ds form was analysed by gel-shift assays, as indicated in the Experimental section. In these experiments, increasing amounts of SsoCdc6-2, ΔC or ΔN were used (10, 20, 40 or 80 pmol). Detection and quantification of the radioactive bands was carried out using a PhosphorImager. A plot of shifted DNA (ss form, open symbols; ds form, closed symbols) against the amount of protein used (full-sized SsoCdc6-2, squares; ΔN, circles; ΔC, triangles) is shown in the bottom panel. Results are mean values of at least three independent experiments.
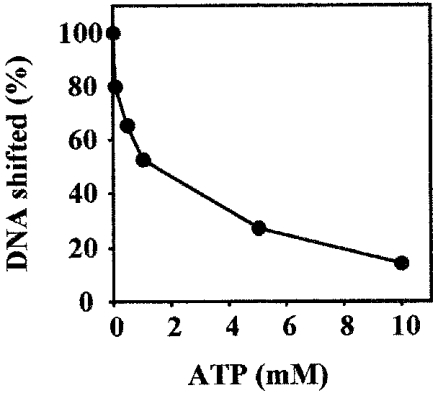
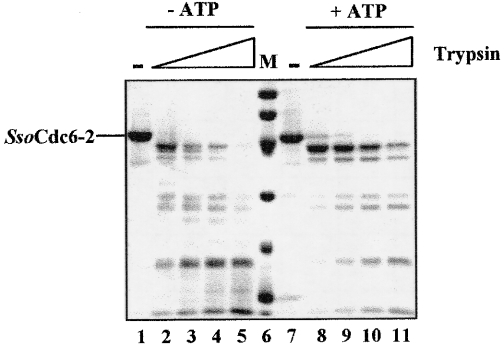
ATP effects on DNA binding and limited tryptic digestion of SsoCdc6-2
To test whether ATP had any effect on SsoCdc6-2 DNA-binding activity, electrophoretic mobility-shift assays were carried out using a ds γ-32P-labelled 51-mer synthetic oligonucleotide, 20 pmol of purified SsoCdc6-2 and increasing amounts of ATP. As shown in Figure 6, ATP inhibited SsoCdc6-2 DNA-binding activity on this substrate. The extent of this inhibition was proportional to the ATP concentration used (up to 10 mM): a 14% residual DNA-binding activity was detected in the presence of 10 mM ATP. In contrast, no effect of ATP on SsoCdc6-2 DNA-binding activity was observed when the 51-mer ssDNA was used (M. De Felice, L. Esposito and F. M. Pisani, unpublished work). These results suggest that upon ATP binding, SsoCdc6-2 undergoes structural changes that affect its ability to interact with dsDNA. In order to investigate this issue further, purified SsoCdc6-2 was subjected to partial trypsin proteolysis. In these experiments, 5.8 pmol of purified SsoCdc6-2 was incubated with increasing amounts of trypsin in the absence or presence of ATP (at 5 mM; see Figure 7). Comparison of the two digestion patterns by densitometric analysis and N-terminal sequencing of the proteolytic products indicated that ATP protects SsoCdc6-2 from trypsin proteolysis and suggested that the protein undergoes a conformational change upon ATP binding.
Figure 6. Effects of ATP on the SsoCdc6-2 DNA-binding activity.
The ability of SsoCdc6-2 to bind a 32P-labelled 51-mer synthetic oligonucleotide in a ds form was analysed by gel-shift assays in the presence of increasing concentrations of ATP. Detection and quantification of the radioactive bands was carried out using a PhosphorImager. A plot of the shifted DNA against the amount of protein used is shown. Results are mean values of at least three independent experiments.
Figure 7. Partial trypsin proteolysis of SsoCdc6-2.
Aliquots of homogeneous SsoCdc6-2 (3 μg) were subjected to partial trypsin digestion as described in the Experimental section. Reactions (20 μl volume) were carried out in the absence (lanes 2–5) or in the presence (lanes 8–11) of 5 mM ATP with increasing amounts of protease (0–100 ng, lanes 2–5 and 8–11). Samples were analysed by SDS/12.5% PAGE and Coomassie-Blue-staining.
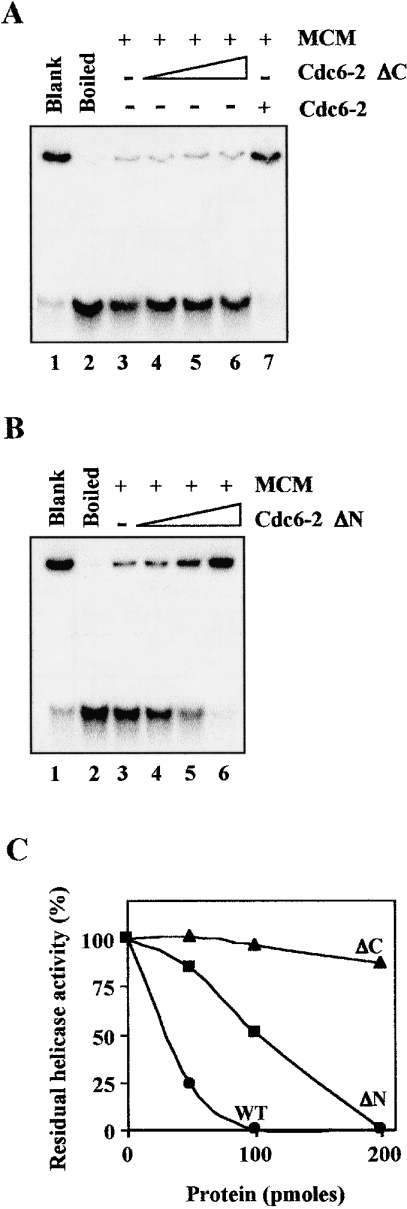
Effects of SsoCdc6-2 and its truncated forms on the SsoMCM DNA-helicase activity
In eukaryotic organisms, both Cdc6 and MCM are believed to play a critical role in DNA-replication initiation. Cdc6 is thought to act as the loader of the MCM DNA helicase on to DNA at the replication origins [3]. In a previous report, we showed that the SsoMCM DNA helicase activity was strongly inhibited by the full-sized SsoCdc6-2 [15]. In order to test the effects of the SsoCdc6-2 truncated forms on SsoMCM, the DNA-helicase activity of the SsoMCM complex (1.5 pmol as a hexamer) was assayed in the presence of increasing amounts of these proteins by means of a strand-displacement assay performed at 70 °C for 30 min (Figure 8). The substrate used was prepared by annealing a 32P-labelled synthetic 85-mer oligonucleotide to M13mp18 ssDNA, giving rise to partial duplexes having a 30-nt 5′-tail. As shown in Figure 8, the ΔN protein was able to inhibit the SsoMCM helicase activity, although with a lesser efficiency with respect to SsoCdc6-2: 200 pmol of the truncated protein was required for a total inhibition of the DNA helicase, whereas 100 pmol of the full-sized protein was enough to produce a similar effect. On the other hand, the C-terminally deleted form caused a negligible inhibition of the SsoMCM DNA-unwinding capability even in assays where 200 pmol of the purified protein was used.
Figure 8. Effects of the SsoCdc6-2 truncated forms on the SsoMCM DNA-helicase activity.
The DNA-unwinding activity of the SsoMCM (700 ng) was tested at 70 °C for 30 min in the presence of 50, 100 and 200 pmol of ΔC (A, lanes 4–6) or ΔN (B, lanes 4–6) proteins. The effects of 200 pmol of the full-sized SsoCdc6-2 on the SsoMCM DNA helicase activity was tested as control (A, lane 7). (C) Residual DNA-helicase activity of the SsoMCM was plotted against the amount of SsoCdc6-2, ΔC and ΔN protein present in each enzymic assay. The unwinding activity of SsoMCM in the absence of any other protein was taken as 100%. Results are mean values of at least three independent determinations. WT, wild-type.
DISCUSSION
In the present paper, we demonstrate that a Cdc6-like protein from the hyperthermophilic crenarchaeon S. solfataricus (referred to as SsoCdc6-2; previously named SsoCdc6-1 [15]) has a modular organization of its associated biological functions: the N-terminal AAA+ NTP-binding domain retains the ability to autophosphorylate in vitro and to hydrolyse ATP, whereas the C-terminal WH-domain is responsible for ssDNA- and dsDNA-binding activity and for the inhibition of the SsoMCM DNA helicase. In line with our biochemical data, a mutational analysis of Schizosaccharomyces pombe Cdc18 (the S. cerevisiae Cdc6 orthologue) showed that amino-acid changes in the WH-fold ‘recognition helix’ or in the nearby ‘wing’ region impair the in vivo functions of this protein, raising the possibility that these structural elements might contact DNA directly [13]. Furthermore, it was shown that in vitro autophosphorylation of the MthCdc6-2 factor is inhibited by dsDNA, and this effect seems to be mediated by the WH-domain because autophosphorylation of a C-terminally truncated form of this protein is not affected by dsDNA [18]. On the other hand, autophosphorylation of both the full-sized and the truncated MthCdc6-2 protein was inhibited by ssDNA, suggesting that the domain responsible for the interaction with ssDNA has a different location. In contrast, our results indicate that the C-terminal third of the SsoCdc6-2 polypeptide chain is able to bind either ssDNA or dsDNA, whereas the C-terminally deleted protein is completely devoid of DNA-binding capability.
The SsoCdc6-2 dsDNA-binding activity is modulated by ATP, since the affinity of the protein for dsDNA is noticeably decreased in the presence of this nucleotide, as determined by electrophoretic mobility-shift assays. This suggests that upon ATP-binding, the protein undergoes a conformational change that alters its ability to interact with DNA. The effect exerted by ATP is specific, because it was not observed when a SsoCdc6-2 Walker-A mutant, that is unable to bind ATP, was used in these DNA band-shift assays (M. De Felice, L. Esposito and F. M. Pisani, unpublished work). In addition, the results of partial proteolysis experiments indicate that in the presence of ATP, SsoCdc6-2 is protected against protease digestion, probably as a consequence of a structural change. Members of the AAA+ family involved in DNA replication were shown to adopt two conformational states (ATP- or ADP-bound form) with different functions, and the switching from one conformation to the other is triggered by ATP hydrolysis [20,21]. It was also shown by limited proteolysis experiments, that human Cdc6 undergoes structural changes promoted by nucleotide binding. However, these conformational changes were detected in the presence of ADP, but not ATP or ATPγS [22]. It is quite likely that Sensor 1 and 2 boxes, which are conserved in the AAA+ family, detect the status of the bound nucleotide, and may play a role in modulating the ATPase activity of these proteins and in triggering the ATP/ADP-induced structural change. Mutations of conserved amino acid residues in S. cerevisiae Cdc6 Sensor 1 and 2 boxes were reported to slow down cell growth due to inefficient MCM loading on to chromatin [23].
The SsoCdc6-2 WH-domain is also responsible for the inhibition of the SsoMCM DNA-helicase activity. These results are consistent with a recent report on the MthCdc6 factors showing that C-terminally truncated forms of these proteins were unable to inactivate the MthMCM DNA unwinding activity in vitro [24]. In addition, a two-hybrid analysis suggested that the WH-domain could be responsible for a physical interaction between MthCdc6 and MCM proteins [24]. ATP binding and hydrolysis by SsoCdc6-2 is not required for SsoMCM helicase inhibition, since the ΔN protein retains this inhibitory capability, although it is completely unable to bind and hydrolyse ATP. Furthermore, we previously demonstrated that a SsoCdc6-2 Walker A mutant inhibited the SsoMCM helicase with the same efficiency as the wild-type protein [15]. On the other hand, it was reported that ATP binding, but not hydrolysis, is needed for the MthMCM DNA-helicase inhibition by MthCdc6-2 [24]. At present, we are unable to explain the different behaviour of the Cdc6 factors from these archaeal species. In fact, the molecular mechanism by which Cdc6 inhibits the MCM DNA helicase is not known, and only speculative models can be envisaged. Cdc6 could directly interact with MCM, promoting a structural change that makes the helicase unable to translocate along DNA or destabilize its binding to DNA. A recent biochemical analysis of replication initiation factors from the euryarchaeon Archeoglobus fulgidus (Afu) showed that both Afu MCM and Cdc6 proteins are able to bind DNA in vitro, with a clear preference for molecules with a single-stranded bubble that mimic early replication intermediates [25]. The AfuCdc6 protein was found to displace a pre-bound MCM complex from these DNA molecules independently of the presence of ATP and Mg2+ ions [25]. The modulation of MCM function by Cdc6 ensures that the replicative DNA helicase is released in an active form only at appropriate times during the cell cycle. In E. coli, the helicase activity of DnaB is modulated by DnaC that acts as the helicase-loader, and inhibition of the DnaB unwinding activity is observed at high DnaC/DnaB molar ratios [26]. Very recently, it has been reported that the chromosome of S. solfataricus contains two active origins of replication, and that SsoCdc6-1 and SsoCdc6-2 bind to both of them at specific sites, as indicated by Dnase I footprinting analyses and chromatin-immunoprecipitation experiments [27]. It has been postulated that SsoCdc6-2 may act as a negative regulator of DNA-replication initiation, because its expression is greatly reduced in G1 and S phases [27]. Several archaeal species (in addition to S. solfataricus) possess multiple Cdc6-like factors [28]. The biochemical characterization of these factors and the analysis of their interplay with the other components of the DNA-replication machinery will help in understanding their biological role.
Acknowledgments
This work was supported by grants from the European Union (Contract No. QLK3-CT-2002-0207) and from MIUR/CNR (‘Biomolecole per la salute umana’ Legge 95/95 and Progetto Legge 449/97-DM 30/10/2000). The Istituto di Genetica e Biofisica-Consiglio Nazionale Ricerche/Tigem Sequencing core is acknowledged for excellent assistance in DNA sequencing.
References
- 1.Kornberg A., Baker T. A. DNA replication. 2nd edn. New York: W. H. Freeman & Co.; 1992. Replication mechanisms and operations; pp. 471–510. [Google Scholar]
- 2.Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multi-protein complex. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 3.Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 4.Donovan S., Harwood J., Drury L., Diffley J. F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perkins G., Diffley J. F. X. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 6.Tye B. K., Sawyer S. L. The hexameric eukaryotic MCM helicase: building symmetry from non-identical parts. J. Biol. Chem. 2000;275:34833–34836. doi: 10.1074/jbc.R000018200. [DOI] [PubMed] [Google Scholar]
- 7.Labib K., Tercero J. A., Diffley J. F. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 8.Lee J. K., Hurwitz J. Isolation and characterization of various complexes of the mini-chromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem. 2000;275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- 9.Ishimi Y. A. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 10.Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 11.Weinreich M., Liang C., Stillman B. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc. Natl. Acad. Sci. U.S.A. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tye B. K. Insights into DNA replication from the third domain of life. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2399–2401. doi: 10.1073/pnas.97.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., Smith C. L., DeRyckere D., DeAngelis K., Martin G. S., Berger J. M. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 14.Carpentieri F., De Felice M., De Falco M., Rossi M., Pisani F. M. Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 2002;277:12118–12127. doi: 10.1074/jbc.M200091200. [DOI] [PubMed] [Google Scholar]
- 15.De Felice M., Esposito L., Pucci B., Carpentieri F., De Falco M., Rossi M., Pisani F. M. Biochemical characterization of a Cdc6-like protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 2003;278:46424–46431. doi: 10.1074/jbc.M306075200. [DOI] [PubMed] [Google Scholar]
- 16.She Q., Singh R. K., Confalonieri F., Zivanovic Y., Allard G., Awayez M. J., Chan-Weiher C. C., Clausen I. G., Curtis B. A., De Moors A., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez R., Sali A. Evaluation of comparative protein structure modelling by MODELLER-3. Proteins. 1997;1(Suppl.):50–58. doi: 10.1002/(sici)1097-0134(1997)1+<50::aid-prot8>3.3.co;2-w. [DOI] [PubMed] [Google Scholar]
- 18.Gabrowski B., Kelman Z. Auto-phosphorylation of archaeal Cdc6 homologues is regulated by DNA. J. Bacteriol. 2001;183:5459–5464. doi: 10.1128/JB.183.18.5459-5464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gajiwala K. S., Burley S. K. Winged helix proteins. Curr. Opin. Struct. Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 20.Lee D. G., Bell S. P. ATPase switches controlling DNA replication initiation. Curr. Opin. Cell. Biol. 2000;12:280–285. doi: 10.1016/s0955-0674(00)00089-2. [DOI] [PubMed] [Google Scholar]
- 21.Davey M. J., Jeruzalmi D., Kuriyan J., O'Donnell M. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 2002;3:1–10. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- 22.Herbig U., Marlar C. A., Fanning E. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol. Biol. Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi T., Tsutsumi S., Tsuchiya T., Stillman B., Mizushima T. Functions of sensor 1 and sensor 2 regions of Saccharomyces cerevisiae Cdc6p in vivo and in vitro. J. Biol. Chem. 2002;277:16033–16040. doi: 10.1074/jbc.M108615200. [DOI] [PubMed] [Google Scholar]
- 24.Shin J.-H., Grabowski B., Kasiviswanathan R., Bell S. D., Kelman Z. Regulation of mini-chromosome maintenance helicase activity by Cdc6. J. Biol. Chem. 2003;278:38059–38067. doi: 10.1074/jbc.M305477200. [DOI] [PubMed] [Google Scholar]
- 25.Grainge I., Scaife S., Wigley D. B. Biochemical analysis of components of the pre-replication complex of Archeoglobus fulgidus. Nucleic Acids Res. 2003;31:4888–4898. doi: 10.1093/nar/gkg662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davey M. J., Fang L., McInerney P., Georgescu R. E., O'Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson N. P., Dionne I., Lundgren M., Marsh V. L., Bernander R., Bell S. D. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 28.Berquist B. R., DasSarma S. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 2003;185:5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PDB viewer: an environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]