Abstract
Solute carrier family 11 member 1 (Slc11a1, formerly Nramp1) is a highly glycosylated, 12 transmembrane domain protein expressed in macrophages. It resides in the membrane of late endosomes and lysosomes, where it functions as a bivalent cation transporter. Mice susceptible to infection by various intracellular pathogens including Leishmania donovani and Salmonella typhimurium carry a glycine to aspartic acid substitution at position 169 (G169D, Gly169→Asp), within transmembrane domain 4 of Slc11a1. To investigate the molecular pathogenesis of infectious disease susceptibility, we compared the behaviour of heterologously and endogenously expressed wild-type and mutant Slc11a1 by immunofluorescence, immunoelectron microscopy and Western-blot analysis. We found occasional late endosome/lysosome staining of mutant protein using immunoelectron microscopy, but most of the mutant Slc11a1 was retained within the ER (endoplasmic reticulum). Using glycosylation as a marker for protein maturation in two independent heterologous expression systems, we found that most mutant Slc11a1 existed as an ER-dependent, partially glycosylated intermediate species. Correct endosomal targeting of wild-type Slc11a1 continued despite disruption of N-glycosylation sites, indicating that glycosylation did not influence folding or sorting. We propose that the G169D mutation causes localized misfolding of Slc11a1, resulting in its retention in the ER and manifestation of the loss of function phenotype.
Keywords: endoplasmic reticulum retention, glycosylation, intracellular targeting, Nramp1, Slc11a1, subcellular localization
Abbreviations: GFP, green fluorescent protein; EGFP, enhanced GFP; endo H, endoglycosidase H; ER, endoplasmic reticulum; LE, late endosomes; Lys, lysosomes; M6PR, mannose 6-phosphate receptor; Pdi, protein disulphide-isomerase; PNGase F, peptide N-glycosidase F; TMD, transmembrane domain
INTRODUCTION
Solute carrier family 11 member 1 (Slc11a1, formerly Nramp1) is an extensively glycosylated, integral membrane protein with 12 putative TMDs (transmembrane domains), which is highly conserved across species as diverse as human and bacteria. Variants in Slc11a1 are associated with susceptibility to infectious and autoimmune diseases [1]. Slc11a1 displays a restricted expression profile including cells of the myeloid lineage [2], a subset of neurons [3] and endocrine tissues, including the anterior pituitary, adrenal medulla and pancreatic islets of Langerhans ([4,5]; J. K. White, unpublished work). In macrophages, Slc11a1 resides in the membrane of LE (late endosomes), Lys (lysosomes) and phagolysosomes [6,7]. Although the direction of transport is controversial, it is accepted that Slc11a1 functions as a protein-coupled bivalent cation transporter regulating cytoplasmic/endocytic bivalent cation concentrations [5,8–11].
Slc11a1 was originally identified in mice for its role in conferring innate resistance to a subset of phylogenetically distinct intracellular pathogens including Leishmania donovani, Salmonella typhimurium and Mycobacterium bovis BCG [12–14]. A naturally occurring glycine (wild-type; Slc11a1wt) to aspartic acid (mutant; Slc11a1mut) missense mutation at amino acid 169, within TMD4 of Slc11a1, renders mice susceptible to these pathogens. In fact, Slc11a1mut mice are phenotypically indistinguishable from Slc11a1−/− targeted disruption mutants [15]. Furthermore, insertion of this bulky, negatively charged aspartic acid residue within TMD4 affects the pleiotropic effects that Slc11a1 exerts on macrophage activation in vitro (reviewed in [14]). The molecular mechanism by which the Slc11a1mut allele causes these profound phenotypic differences in vivo and in vitro remains unclear but has major implications with respect to functional moieties within the protein.
The fate of Slc11a1mut protein is controversial with conflicting observations reported. Vidal et al. [16] and Gruenheid et al. [6] failed to identify Slc11a1mut protein in mutant macrophages either by immunoprecipitation or by confocal microscopy. However, using an antibody raised against the N-terminal 82 amino acids of Slc11a1, Atkinson and Barton [17,18] detected a 45 kDa precursor polypeptide in extracts from Slc11a1mut parental RAW264.7 cells. Furthermore, low levels of Slc11a1 protein were detected in the membrane of LE and Lys of control and activated mutant macrophages using fluorescence immunocytochemistry and immunoelectron microscopy [7,17,18].
To address the molecular pathogenesis of infectious disease susceptibility, we posed the question of whether the G169D mutation within TMD4 prevents appropriate targeting of Slc11a1 or whether the mutant protein reaches the LE/Lys compartment but the charged residue blocks its function. To answer this, we introduced wild-type and mutant Slc11a1 into cells and compared their behaviour by immunofluorescence, immunoelectron microscopy and Western-blot analysis. The results obtained in the present study suggest that after synthesis, most of the Slc11a1mut protein exists as a partially glycosylated intermediate form that is retained within the ER (endoplasmic reticulum).
EXPERIMENTAL
Preparation of expression constructs
Wild-type (Slc11alwt, Gly169) and mutant (Slc11almut, Asp169) Slc11a1 full-length cDNAs were PCR-amplified from pBABE constructs [19] with primers (5′-TGAATTCCACCATGATTAGTGACAAGAGCCC-3′ and 5′-TGGATCCCCGGAACCCTGCACGC-3′) designed to introduce a Kozak consensus site directly upstream of the ATG and a 5′-EcoRI and inframe 3′-BamHI cloning sites. Products were cloned into the mammalian expression vector pEGFP-N1 (ClonTech Laboratories, Oxford, U.K.), so that they were expressed with EGFP (enhanced green fluorescent protein) at their C-terminus. The two N-linked glycosylation consensus sites in Slc11alwt-EGFP were simultaneously disrupted by site-directed mutagenesis. Megaprimer technology [20] was used to convert the distal serine in N321SS and the threonine in N335NT into alanine (S323A+T337A). pcDNA3 was modified in-house by insertion of an 84 bp EcoRI–NotI cassette encoding a 22-amino-acid 3×FLAG epitope [21] preceded by a start methionine and a Kozak consensus site. Wild-type and mutant Slc11a1 cDNAs were PCR-amplified with primers (5′-AAGCGGCCGCCATGATTAGTGACAAGAGCCC-3′ and 5′-TTGGGCCCTCACCCGGAACCCTGCAC-3′) designed to introduce an inframe NotI site adjacent to the endogenous ATG and an ApaI site after the stop codon. Products were cloned into the NotI–ApaI cut, modified pcDNA, so that they were expressed with 3×FLAG at their N-terminus. All constructs were sequence-verified with ABI BigDye terminator chemistry, using an ABI377 automated DNA sequencer (Applied Biosystems, Warrington, Cheshire, U.K.), and prepared in bulk using the EndoFree plasmid maxi kit (Qiagen, Crawley, West Sussex, U.K.).
Cell culture and treatment
COS7 cells were routinely maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal calf serum, 50 units/ml penicillin, 50 μg/ml streptomycin, 2 mM L-glutamine and 1 mM sodium pyruvate. The Slc11a1 mutant BALB/c-derived macrophage cell line RAW 264.7 [22] was grown in RPMI 1640 supplemented as above omitting sodium pyruvate. For glycosylation analysis, increasing doses of tunicamycin (0.5, 0.75 and 1.0 μg/ml) were added to subconfluent cultures of cells for 48 h before harvesting. Bone marrow-derived macrophages from C57BL10/ScSn (Slc11almut) or congenic N20 B10.L-Lshr (Slc11alwt) mice [23] were prepared as described in [24] using L-cell conditioned media. At day 10, primary macrophages were stimulated for 24 h with 10 units/ml recombinant mouse interferon γ and 1 ng/ml lipopolysaccharide before harvesting.
Antibodies
The rabbit polyclonal antibody to Slc11a1 (1:1000), prepared [17] against recombinant fusion protein corresponding to amino acids 1–82 of murine Slc11a1, was a gift from Dr C. H. Barton (University of Southampton, Southampton, U.K.). The rat monoclonal antibody to KDEL (1:500) was provided by Dr S. Munro (Laboratory for Molecular Biology, Cambridge, U.K.) and the mouse monoclonal antibody to the C-terminus of human Pdi (protein disulphide-isomerase; clone 1D3, 1:5) was a gift from Dr D. Vaux (Dunn School of Pathology, Oxford, U.K.). The rabbit polyclonal anti-bovine M6PR (mannose 6-phosphate receptor) antibody, diluted to 1:50 for immunoelectron microscopy, was kindly provided by Dr S. Pfeffer (Stanford University, Stanford, CA, U.S.A.) [25]. Commercially available antibodies used in the present study were two rabbit polyclonal antibodies to GFP [green fluorescent protein; Molecular Probes, Leiden, The Netherlands (1:200 for fluorescence immunocytochemistry) and Abcam, Cambridge, U.K. (1:5000 for Western blotting and 1:50 for immunoelectron microscopy)], anti-FLAG (M2) mouse monoclonal antibody (1:400; Sigma, Poole, Dorset, U.K.), anti-EEA1 (N-19) goat polyclonal antibody (1:100; Santa Cruz, Heidelberg, Germany) and anti-Lamp1 (1D4B) rat monoclonal antibody (1:100; Developmental Studies Hybridoma Bank, Iowa City, IA, U.S.A.).
Secondary antibodies used for immunofluorescence were Alexa™594-coupled donkey anti-goat, goat anti-rat and donkey anti-mouse IgG (1:200) and Alexa™488-coupled goat anti-rabbit IgG (1:400; Molecular Probes). For Western-blot analysis, horseradish peroxidase-conjugated rabbit anti-mouse and goat anti-rabbit IgG were used at 1:8000. For immunoelectron microscopy, goat anti-rabbit IgG-labelled with 15 nm gold and goat anti-mouse IgG labelled with 10 nm gold (1:70) were purchased from Biocell (Cardiff, U.K.). Protein A labelled with 15 or 5 nm gold (1:70) was from the Department of Cell Biology (University of Utrecht, The Netherlands).
Transfection of cells
For transfection, RAW264.7 cells were grown in 100 mm dishes to approx. 70% confluence and transfected transiently or stably according to the manufacturer's instructions with 10 μg of pEGFP-N1 constructs, using Superfect (Qiagen). For transient transfection, cells were harvested 48 h post-transfection and sorted on the basis of GFP-positive signal using a Mo Flo cytometer (DakoCytomation, Glostrup, Denmark). Selected cells were plated on to 8-well chamber slides overnight, then fixed for immunofluorescence staining. For stable lines, cells were passaged 1:6 into a medium containing 400 μg/ml G418, 40 h post-transfection. Selectable media were replaced four times for 10–12 days. Colonies were visualized using an inverted microscope and picked into 24-well plates.
Transient transfection of COS7 cells is more efficient and was performed in approx. 70% confluent six-well plates according to the manufacturer's instruction with 1 μg of 3×FLAG-tagged constructs using LIPOFECTAMINE™Plus (Invitrogen Life Technologies, Paisley, U.K.). Protein was harvested for Western-blot analysis 48 h post-transfection as described below.
Immunofluorescence
For immunofluorescence, cells adhered to chamber slides were fixed for 20 min at 4 °C with 100% methanol, rinsed in PBS then blocked for 1 h at room temperature (21–25 °C) in 0.1% Triton X-100/PBS supplemented with 5% (w/v) skimmed milk [3% (v/v) normal donkey serum for anti-EEA1]. Where stated, cells were permeabilized with 0.05% saponin in PBS for 90 s and washed in PBS, before fixation. For co-localization studies, antibodies were incubated sequentially. Primary antibody, diluted in blocking buffer, was incubated for 1 h at room temperature. Slides were washed three times for 5 min in 0.1% Triton X-100/PBS. Secondary antibody, diluted in 0.1% Triton X-100, PBS supplemented with 1% skimmed milk (1% normal donkey serum for anti-EEA1), was incubated for 1 h at room temperature. Slides were washed as above, then blocked at room temperature for an additional 30 min before application of the second primary antibody. Finally, cells were rinsed in PBS and mounted in Prolong Antifade (Molecular Probes). Cells were viewed with a Nikon optiphot-2 epifluorescence microscope coupled with a Bio-Rad MRC 1000 confocal laser scanning attachment (Bio-Rad Laboratories, Hemel Hempstead, U.K.). Images were collected using Lasersharp 2000 software.
Labelling of macrophages with BSA gold
To load Lys, but not LE, cells grown in tissue culture flasks were incubated with BSA-5 nm gold for 4 h at 37 °C followed by a 20 h chase in conjugate-free medium [26]. BSA-5 nm gold was prepared using tannic acid and sodium citrate to reduce gold chloride as described in [27].
Immunoelectron microscopy
Cells were prepared for ultrastructure immunocytochemistry essentially as described in [28,29]. Cells were fixed with 4% (w/v) paraformaldehyde/0.1% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) at room temperature for 1 h and processed for immunocytochemistry. Frozen ultrathin (60 nm) sections were cut using a Reichert Ultracut T ultramicrotome equipped with a cryochamber attachment (Leica, Milton Keynes, U.K.). Single staining of GFP and dual staining of GFP and Pdi were performed using indirect labelling as described in [28,30]. Species-specific secondary antibodies conjugated to 15 nm (GFP) and 10 nm (Pdi) gold particles were used for detection. Alternatively, dual labelling of GFP and M6PR, which were both rabbit polyclonal antibodies, was accomplished using Protein A-gold (15 and 5 nm) respectively [31]. Finally, for both techniques, cells were washed in water and stained with 1.8% methyl cellulose/0.3% uranyl acetate [32]. Sections were observed on a Philips CM 100 transmission electron microscope (Philips Electron Optics, Cambridge, U.K.) at an accelerating voltage of 80 kV.
Western blotting
Protein was extracted from cell pellets by incubation in 10 vol. of RIPA buffer (150 mM NaCl, 1% Nonidet P40, 12 mM deoxycholic acid, 0.1% SDS and 50 mM Tris/HCl, pH 8), supplemented with protease inhibitors (1 mM dithiothreitol, 1 mM PMSF, 1 μg/ml leupeptin and 1 μg/ml aprotinin) for 30 min at 4 °C followed by centrifugation (15000 g, 20 min, 4 °C) to remove debris. For deglycosylation, 10 μg of protein was treated with 500 units of PNGase F (peptide N-glycosidase F; New England Biolabs, Hitchin, Herts., U.K.) for 4 h at 37 °C according to the manufacturer's instructions, except that the initial denaturation step was performed at 70 °C for 2 min. Western-blot analysis was performed following standard procedures [33]. Before loading, an equal volume of 2× loading buffer [60 mM dithiothreitol, 6 M urea, 4% (w/v) SDS, 20% (v/v) glycerol, 0.02% Bromophenol Blue and 125 mM Tris/HCl, pH 6.8] was added and the samples were incubated at room temperature for 30 min. Immunoblotting was performed as described in [33] and detection was achieved using enhanced chemiluminescence (ECL®; Amersham Biosciences, Chalfont St. Giles, U.K.).
RESULTS
Subcellular localization of heterologously expressed Slc11a1
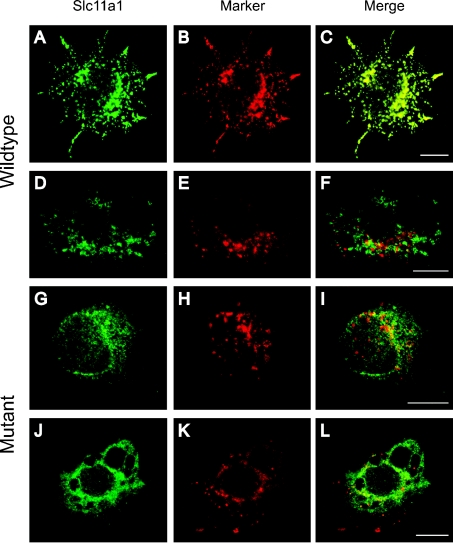
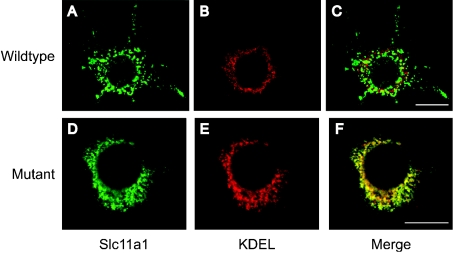
Expression of EGFP-tagged wild-type and mutant Slc11a1 was analysed approx. 65 h after transient transfection into RAW264.7 cells using fluorescence immunocytochemistry. The presence of the EGFP tag provided an alternative to using antibodies against Slc11a1, which have yielded conflicting results in the literature. However, EGFP is a large moiety (27 kDa) and the effect of adding it to the C-terminus of Slc11a1 was tested. RAW264.7 cells transfected with Slc11a1wt-EGFP were double-labelled with anti-GFP and an antibody against either the LE/Lys marker Lamp1 (Figures 1A–1C) or the EE (early endosome) marker EEA1 (Figures 1D–1F). Consistent with previous findings, Slc11a1wt-EGFP localized in vesicular compartments that co-stained with Lamp1, indicating that normal Slc11a1 subcellular targeting was unaffected by the EGFP tag. In contrast, Slc11a1mut-EGFP gave a fine, reticular distribution with some perinuclear focusing and failed to co-localize with either Lamp1 (Figures 1G–1I) or EEA1 (Figures 1J–1L). The pattern observed for mutant Slc11a1 was characteristic of the ER. To address this possibility, transiently transfected macrophages were co-stained for the defined ER marker KDEL and GFP (Figure 2). Whereas Slc11a1wt-EGFP failed to co-localize with this marker, most of the mutant Slc11a1 resided in the KDEL-positive ER compartment (Figure 2F).
Figure 1. Fluorescence localization of EGFP-tagged Slc11a1.
Immunofluorescence analysis of the subcellular localization of EGFP-tagged wild-type (A, D) and mutant (G, J) Slc11a1 protein (anti-GFP, green) following transient transfection into RAW264.7 cells. Cells were co-stained (red) either for the LE/Lys marker Lamp1 (B, H) or the early endosome marker EEA1 (E, K). For (D–L), cells were permeabilized with saponin before staining. In the merged panels (C, F, I and L), yellow (C only) indicates co-localization of signal. Scale bar, 10 μm.
Figure 2. Localization of mutant Slc11a1 to the ER.
Representative examples of immunofluorescence patterns observed for EGFP-tagged wild-type (A) or mutant (D) Slc11a1 protein (anti-GFP, green) in transiently transfected RAW264.7 cells. Cells were co-stained (red) for the ER marker KDEL (B, E). In the merged panels (C, F), yellow (F only) indicates co-localization of signal. Scale bar, 10 μm.
Cell lines stably expressing wild-type or mutant, EGFP-tagged Slc11a1 were generated to confirm the above observation and facilitate further studies. G418-resistant clones were genotyped by PCR and sequenced to establish the presence of the transgene (results not shown). Positive clones were evaluated for transgene expression using FACS analysis and immunofluorescence (results not shown). From an initial series of 21 and 19 independent wild-type and mutant clones respectively, two lines of each were selected as stable expressers and found to yield comparable results. WT3 and MUT12 are representative examples used in the present study. Consistent with the previous immunofluorescence results, wild-type Slc11a1 localized to the LE/Lys compartment of WT3, whereas the bulk of mutant Slc11a1 was retained within the ER of MUT12. Immunofluorescence alone was insufficient to determine if any mutant protein progressed beyond the ER. This was clarified using immunoelectron microscopy.
Immunoelectron microscopy detects low levels of Slc11a1mut protein in LE/Lys
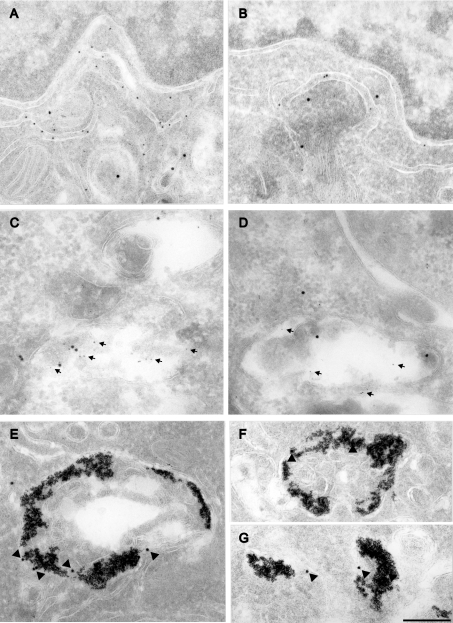
To investigate the localization of EGFP-tagged wild-type and mutant Slc11a1 further, WT3 and MUT12 cells were processed for immunoelectron microscopy. Dual-labelling cells confirmed the ER retention of Slc11a1mut, observed by immunofluorescence, for GFP and the defined ER marker Pdi (Figures 3A and 3B). Slc11a1mut was readily detected within the Pdi-positive ER of MUT12 (Figure 3B). In contrast, wild-type protein was not apparent in WT3 ER but was seen in membrane-bound vesicular structures on the same micrograph (Figure 3A).
Figure 3. EM-gold localization of EGFP-tagged Slc11a1.
Representative micrographs showing immunoelectron microscopic localization of EGFP-tagged wild-type (A, C and E) and mutant (B, D, F and G) Slc11a1 in the stable macrophage cell lines WT3 and MUT12 respectively. Cryosections were labelled with a rabbit polyclonal antibody to GFP (15 nm gold; A–D), and either a mouse monoclonal to Pdi (10 nm gold; A, B) or a rabbit polyclonal to M6PR (5 nm gold; C, D; arrows). (E–G) Lys were detected by preloading cells with BSA-5 nm gold and labelled for Slc11a1 using the anti-GFP antibody (15 nm gold; arrow heads). Two examples (F, G) are presented for MUT12 because of the low level of staining. Scale bar, 250 nm.
Anti-M6PR staining was used to define LE. Furthermore, a pulse–chase strategy using BSA-5 nm gold was used to mark predominantly Lys, and not LE, in WT3 and MUT12. Consistent with the immunofluorescence results from the present study, and previous observations [6,7], Slc11a1wt-EGFP protein, detected using an anti-GFP antibody, was abundant in the M6PR-labelled LE compartment (Figure 3C) and BSA-5 nm gold-loaded Lys (Figure 3E) of WT3. Interestingly, in MUT12, occasional co-localization of Slc11a1mut-EGFP protein with both M6PR-positive LE (Figure 3D) and BSA-5 nm gold-loaded Lys (Figures 3F and 3G) was detectable, indicating that, at a decreased level, mutant protein did traffic to its target organelles.
Altered glycosylation state correlates with ER retention
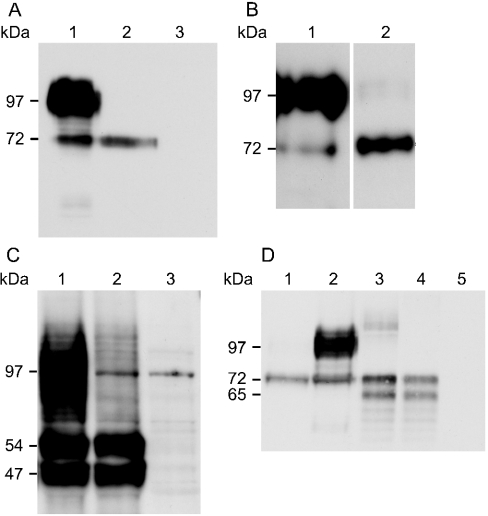
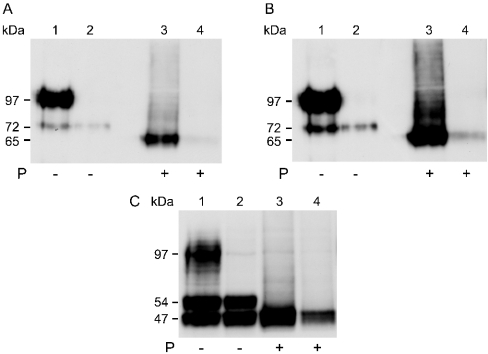
Protein extracts from WT3 and MUT12 were immunoblotted with an antibody to GFP (Figures 4A and 4B). WT3 yielded a smear flanking the 97 kDa standard and a 72 kDa band presumed to correspond to the fully N-glycosylated protein and the unglycosylated polypeptide respectively (Figure 4A, lane 1). In contrast, the predominant species in MUT12 extracts was the 72 kDa form (Figure 4A, lane 2). Interestingly, a long exposure of excess MUT12 extract revealed some extensively glycosylated, 90–100 kDa mutant protein (Figure 4B, lane 2). Immunoreactive bands of 90–100 kDa fully N-glycosylated protein and 45 kDa unglycosylated polypeptide were reported previously using Slc11a1 antiserum [17]. We propose that the 45 kDa band corresponded to our 72 kDa product. The shift in mobility is consistent with the additional 27 kDa contributed by the EGFP tag. However, this shift was less apparent in the fully N-glycosylated smear, which clearly flanked the 97 kDa standard. Abnormal migration of mature Slc11a1 may be caused by the extensive carbohydrate moieties present. To minimize the influence of the tag on protein migration, an alternative heterologous expression system was used. The 22-amino-acid, 3×FLAG epitope was engineered in frame, directly 5′ of the wild-type and mutant Slc11a1 start codon and the resulting expression constructs transiently transfected into COS7 cells. Protein was extracted 48 h post-transfection and immunoblotted using an antibody to FLAG (Figure 4C). A broad smear of mature protein was detected for wild-type and, to a lesser extent, mutant protein, migrating smaller than the EGFP-tagged protein and falling within the 90–100 kDa range previously observed [17] for endogenous Slc11a1wt. Two additional lower molecular-mass bands (54 and 47 kDa) of unknown glycosylation status were detected in similar abundance for both the wild-type and mutant Slc11a1 extracts.
Figure 4. Western-blot analysis of Slc11a1 glycosylation patterns.
(A) Extracts (10 μg) from the stable macrophage cell lines WT3 (lane 1) and MUT12 (lane 2) expressing EGFP-tagged wild-type and mutant Slc11a1 respectively, and from untransfected RAW264.7 cells (lane 3) were separated by SDS/PAGE (10% gel). Immunoblotting was performed using an anti-GFP rabbit polyclonal antibody (1:5000) and visualized following a 30 s exposure. Positions of the 72 kDa precursor species and the 97 kDa standard that is flanked by a broad smear of fully glycosylated mature wild-type Slc11a1 are shown on the left. (B) 5 μg of WT3 (lane 1) and 15 μg of MUT12 (lane 2) extracts were immunoblotted as in (A) and visualized following a 10 min exposure to facilitate detection of low levels of mature mutant protein. No staining of untransfected RAW264.7 cells was apparent at this exposure. (C) COS7 cells were transiently transfected with 3×FLAG-tagged wild-type (lane 1) and mutant (lane 2) Slc11a1 and non-recombinant vector (lane 3). Protein was extracted and 10 μg resolved by SDS/PAGE (10% gel), 48 h post-transfection. Immunoblotting was performed using an anti-FLAG monoclonal antibody (1:400). (D) Comparison of EGFP-tagged Slc11a1 expression in RAW and COS7 cells. Extracts (10 μg) from MUT12 (lane 1), WT3 (lane 2) and COS7 cells transiently transfected for 24 h with Slc11a1wt-EGFP (lane 3), Slc11a1mut-EGFP (lane 4) and non-recombinant pEGFP-N1 (lane 5) were resolved by SDS/PAGE (10% gel). Immunoblotting was performed as in (A). Positions of the 47 and 54 kDa (C) and 65 and 72 kDa (D) precursor species and the 97 kDa standard band are shown on the left.
This alternative steady-state glycosylation profile could have been cell-type-specific, expression-level-dependent or related to the nature or position of the epitope tag. To address this, the wild-type and mutant Slc11a1-EGFP expression constructs were transiently transfected into COS7 cells. Three species were apparent in both wild-type and mutant transfected COS7 cells, 48 h post-transfection; a 90–100 kDa smear (faint for Slc11almut-EGFP) and two lower molecular-mass bands of unknown glycosylation status migrating at 72 and 65 kDa (results not shown). However, 24 h post-transfection, the 72 and 65 kDa species were dominant, whereas the 90–100 kDa smear was absent (Slc11a1mut) or at very low abundance (Slc11a1wt), indicating that steady-state levels of fully glycosylated, mature protein were yet to be attained (Figure 4D). From this, we concluded that the two lower molecular-mass proteins were normal, stable precursors in the COS7 cell glycosylation process, rather than deglycosylated degradation products, and that the difference between RAW and COS7 transfection results was cell-type-dependent. The anti-FLAG antibody was found to cross-react with endogenous RAW cell extracts, precluding the reciprocal experiment to test 3×FLAG-tagged constructs in RAW cells.
To remove carbohydrate moieties, protein extracts were treated exhaustively with PNGase F before Western-blot analysis. PNGase F cleaves the asparagine-N-acetyl glycosamine bond that joins N-linked oligosaccharides to a broad class of glycoproteins. Treatment caused the EGFP-tagged 90–100 and 72 kDa bands in WT3 (Figure 5A) and MUT12 (Figure 5B) extracts to disappear with the concomitant appearance of a 65 kDa immunoreactive species. Similarly, deglycosylation of extracts from transiently transfected COS7 cells caused all immunoreactive 3×FLAG-tagged protein in both wild-type and mutant transfectants to resolve as the lower, 47 kDa species (Figure 5C). These results demonstrate the presence of a partially glycosylated form of wild-type and mutant Slc11a1 in both stable macrophage cell lines (72 kDa) and in transiently transfected COS7 cells (54 kDa). In line with the immunofluorescence data presented, we hypothesized that the intermediates were products of ER-dependent glycosylation. As confirmation, WT3 and MUT12 extracts were treated with endo H (endoglycosidase H) before SDS/PAGE analysis (results not shown). Endo H cleaves oligosaccharides of the high mannose and hybrid (ER-dependent) form, but not complex carbohydrate structures processed in the Golgi apparatus. The 90–100 kDa, fully N-glycosylated protein was endo H resistant. In contrast, the 72 kDa form shifted to 65 kDa after treatment with endo H, indicating that it was ER-derived.
Figure 5. Sensitivity to treatment with the glycoamidase PNGase F.
(A, B) The protein extract (10 μg) from the stable cell lines WT3 (lanes 1 and 3) and MUT12 (lanes 2 and 4) was denatured and then incubated for 4 h in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 500 units of PNGase F. Samples were resolved by SDS/PAGE (10% gel). Immunoblotting was performed using an anti-GFP rabbit polyclonal antibody (1:5000). After chemiluminescent detection, the blot was exposed for 3 s (A) and 10 s (B). Positions of the 65 and 72 kDa species and the 97 kDa standard are shown on the left. Some degradation of the MUT12 sample is apparent. (C) The extracts (10 μg) from COS7 cells transiently transfected (48 h) with 3×FLAG-tagged wild-type (lanes 1 and 3) or mutant (lanes 2 and 4) Slc11a1 were treated in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of PNGase F, as described above. Immunoblotting was performed with an anti-FLAG monoclonal antibody (1:400). Positions of the 47 and 54 kDa species and the 97 kDa standard are shown on the left.
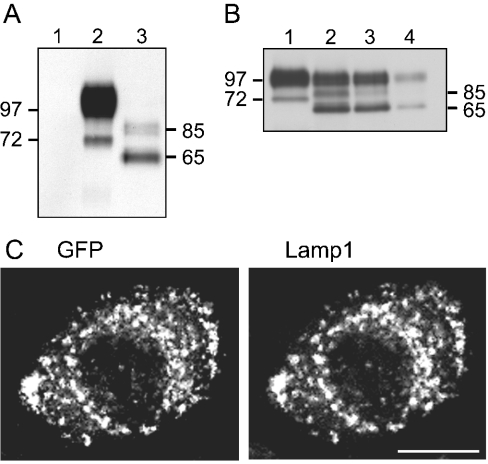
Further confirmation was obtained by disruption of the two highly conserved N-glycosylation sites situated in loop 7 of the EGFP-tagged Slc11alwt expression construct. Site-directed mutagenesis was used to substitute simultaneously the distal serine in N321SS and the threonine in N335NT with alanine, to create the N-glycosylation-deficient construct S323A+T337A. Stable RAW cell lines were generated and assessed for expression as before. Protein extracts from two stable expressers were analysed by SDS/PAGE. Results were comparable and are presented in Figure 6(A) for clone CHO-9. As predicted, removal of the consensus sites resulted in the absence of both mature (90–100 kDa) and partially glycosylated (72 kDa) forms of Slc11a1. The 65 kDa unmodified primary polypeptide was dominant, and a faint doublet of approx. 85 kDa was evident. This product profile was reproducible in transient transfections of S323A+T337A into RAW cells (results not shown) and in WT3 cells after treatment with the GlcNAc transferase inhibitor tunicamycin, which interferes with N-glycosylation in the ER. WT3 cells were cultured for 48 h in the presence of increasing concentrations of tunicamycin. Doses above 1.0 μg/ml caused extensive cell death. After treatment with doses ≤1.0 μg/ml, immunoblotting with an antibody to GFP revealed the absence of the 72 kDa partially glycosylated species in conjunction with the appearance of the 65 kDa unmodified precursor polypeptide and a faint 85 kDa product (Figure 6B). Some fully glycosylated mature protein was visible in extracts treated with tunicamycin. This may indicate incomplete inhibition of GlcNAc transferase; however, given the absence of the 72 kDa product, it probably represents persisting mature protein synthesized before tunicamycin treatment. The nature of the 85 kDa doublet remains unclear but may indicate additional post-translational modifications. O-glycosylation is unlikely since only two weak consensus sites are predicted in Slc11a1 and they both reside in the cytoplasmic N-terminal domain. Phosphorylation is possible since Slc11a1 is a known phosphoprotein [16] possessing eight candidate phosphorylation sites. Alternatively, this form may be targeted for degradation by ubiquitination, a mechanism for down-regulating proteins post-translationally. Slc11a1 has ten cytoplasmic lysine residues that could conjugate to ubiquitin. We conclude that the intermediates identified in our heterologous expression systems are ER-dependent glycosylated forms of Slc11a1. Interestingly, despite the absence of all N-linked glycosylation, Slc11a1wt continued to co-localize with the Lamp1-positive LE/Lys compartment in the S323A+T337A stable expresser CHO-9 (Figure 6C), and was not detectable anywhere else, including the Pdi-positive ER (results not shown). This indicated that glycosylation did not influence folding and sorting.
Figure 6. Targeting unaffected by glycosylation state.
Subcellular targeting of wild-type Slc11a1 to LE/Lys is unaffected by the glycosylation state. (A) The extracts (10 μg) from parental RAW cells (lane 1), WT3 (lane 2) and CHO-9, the RAW cell clone stably expressing N-glycosylation-deficient Slc11a1 (lane 3), were resolved by SDS/PAGE (10% gel). Immunoblotting was performed using an anti-GFP rabbit polyclonal antibody (1:5000). (B) WT3 cells were cultured for 48 h with tunicamycin: 0 (lane 1), 0.5 μg/ml (lane 2), 0.75 μg/ml (lane 3) and 1.0 μg/ml (lane 4). Protein extracts were prepared and analysed by Western blotting [SDS/PAGE (12% gel), 5–20 μg of protein depending on yield] with anti-GFP rabbit polyclonal antibody (1:5000). Positions of the 65, 72 and 85 kDa proteins and the 97 kDa standard are shown. (C) CHO-9 cells were methanol-fixed and stained for GFP combined with Lamp1. A representative example demonstrating the vesicular staining of N-glycosylation-deficient Slc11a1 clearly co-localizing with Lamp1 is shown. Scale bar, 10 μm.
Expression pattern of endogenous Slc11a1 confirms in vitro observations
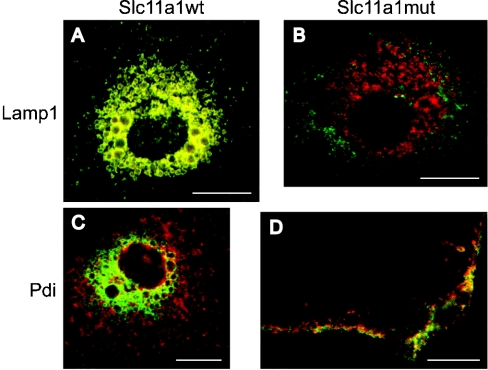
To establish the fate of endogenously expressed Slc11a1mut protein, bone marrow-derived macrophages from C57BL/10ScSn (Slc11a1mut) and congenic B10.L-Lshr (Slc11a1wt) mice were analysed by immunofluorescence. Wild-type (Figures 7A and 7C) and mutant (Figures 7B and 7D) macrophages were double-labelled with one of two antibodies against the N-terminus of Slc11a1, and either the LE/Lys marker Lamp1 (Figures 7A and 7B) or the ER marker Pdi (Figures 7C and 7D). Merged images are presented for the published Slc11a1 antibody [17]. The other Slc11a1 antibody, generated in-house, gave similar results. Consistent with previous findings, wild-type Slc11a1 stained a vesicular compartment that co-localized completely with Lamp1 (Figure 7A, yellow stain) and not with the ER marker Pdi (Figure 7C). Decreased levels of mutant Slc11a1 made imaging difficult. However, the staining that was detectable was reticular in nature, failed to co-localize with Lamp1 (Figure 7B) and did partially overlap with the ER marker Pdi (Figure 7D). These observations suggest that mutant Slc11a1 expressed endogenously in bone marrow-derived macrophages behaves in the same way as the recombinant proteins expressed in RAW264.7 and COS7 cells. That is, it is retained within the ER and subsequently degraded, resulting in a decrease in detectable levels of Slc11a1mut protein.
Figure 7. Localization of endogenous Slc11a1.
Localization of endogenous wild-type and mutant Slc11a1 in bone marrow-derived macrophages. Day 10 macrophages from Slc11a1 mutant C57BL/10ScSn versus congenic Slc11a1 wild-type N20, B10.L-Lshr mice were activated for 24 h with interferon γ/lipopolysaccharide. After methanol fixation, wild-type (A, C) and mutant (B, D) macrophages were stained with a polyclonal anti-N-terminal Slc11a1 antibody (green), combined with either anti-Lamp1 (A, B) or anti-Pdi (C, D), both shown in red. Merged images are presented with yellow identifying the co-localization of wild-type Slc11a1 with Lamp1 and partial co-localization of mutant Slc11a1 with Pdi. Scale bar, 10 μm.
DISCUSSION
In the present study, we addressed one of the controversies in Slc11a1 literature pertaining to the fate of mutant G169D Slc11a1 protein in mice. We found that the charged residue within TMD4 of Slc11a1mut resulted in retention of the protein in the ER, characterized using fluorescence immunocytochemistry as a fine, reticular-staining pattern with some perinuclear focusing. In two separate heterologous expression systems, Slc11a1mut protein exhibited an altered glycosylation pattern. A major decrease in the fully glycosylated 90–100 kDa form of Slc11a1 was observed concomitant with an increase in a partially glycosylated intermediate species. We propose that insertion of the charged residue at position 169 affects folding of the hydrophobic α-helix that constitutes TMD4. This localized misfolding results in the protein being retained by the ER quality control mechanism [34,35] and, subsequently, degraded. N-linked glycosylation begins in the lumen of the rough ER with the addition of a common, highmannose precursor oligosaccharide. This accounts for the partially glycosylated intermediate detected as the dominant species by SDS/PAGE analysis in mutant transfectants. These findings are consistent with current opinion on the fate of misfolded proteins within the ER [36]. They explain the susceptibility to infection by L. donovani, S. typhimurium and M. bovis BCG observed in mice carrying the mutation. This loss of function mutation adds to the list of disorders caused by defective protein folding/trafficking [37–39].
TMD4 is one of the five Slc11a1 membrane-spanning domains that carry a charged amino acid. Aspartate (Asp177) at position 13 of the predicted 20 residue TMD4 is conserved in Slc11 homologues from human to yeast. The G169D mutation observed in in-bred mice occurs at position 5. Periodicity analysis using a helical wheel projection [12] explains why one is tolerated, whereas the other causes the protein to misfold. Whereas Asp177 is predicted to fall within the hydrophilic face of the helix, G169D positions within the lipid-accessible region of TMD4. Hence, identical residues located eight amino acids apart can exert profoundly different effects on the fate of the protein.
In the experiments presented, Slc11a1wt is fully glycosylated, presumably in the Golgi apparatus, and then trafficked to its target organelle (LE/Lys). Mature EGFP-tagged Slc11a1wt resolved by SDS/PAGE at a molecular mass lower than predicted. This abnormal mobility may reflect the disproportionately large net negative charge added by the extensive N-linked glycosylation, masking the increase in molecular mass due to the EGFP tag. Interestingly, the subcellular localization of N-glycosylation-deficient Slc11a1 in cell lines stably expressing S323A+T337A was indistinguishable from that of the wild-type Slc11a1, indicating that glycosylation was not a prerequisite for effective trafficking of this protein. The same approach was taken to study the closely related family member Slc11a2 (alternatively designated Nramp2 or DMT1). Consistent with our findings, Tabuchi et al. [40] demonstrated that correct endosomal localization in non-polarized cells was observed despite disruption by site-directed mutagenesis of the two highly conserved N-glycosylation sites within human SLC11A2.
Consistent with previous observations [7,18] in the stable transfectant MUT12, a low level of fully glycosylated mutant protein was detected by Western-blot analysis and occasional staining of both LE and Lys was apparent by immunoelectron microscopy. It is unclear whether the small amount of Slc11a1mut protein that passes cellular quality control is functional. Substitution with a charged residue at the adjacent position within TMD4 of the highly conserved family member Slc11a2 is a natural mutation occurring in microcytic anaemia (mk) mice and Belgrade (b) rats [41]. This causes a substantial decrease in the amount of detectable Slc11a2 protein; however, some Slc11a2-driven bivalent cation transport activity is retained in mk and b animals [42]. Given the similarity of the mutation, it is conceivable that residual function is retained in Slc11a1mut mice. If so, it is insufficient to rescue the infectious disease susceptibility phenotype.
The cytomegalovirus promoter in the pEGFP-N1 expression vector used in the present study will drive constitutive expression of the transgene and, thus, yield high levels of Slc11a1 protein. This may result in the observed accumulation of mutant Slc11a1 within the ER. To confirm that the fate of endogenously expressed Slc11a1 was consistent with the results from our overexpression system, immunofluorescence was performed on bone marrow-derived macrophages prepared from Slc11a1 wildtype and mutant mice. Consistent with previous observations [7], only severely decreased levels of mutant protein were detectable. The staining that was seen was reticular in nature and localized predominantly to the ER, strongly suggesting that Slc11a1mut faces the same fate in vivo as the overexpression system highlighted. That is, it is retained by the ER quality control mechanism and subsequently degraded. This matches with results from Slc11a1mut BALB/c-derived parental RAW cell extracts, analysed by Western blot with the anti-Slc11a1 antibody used in the present study, where only the ER-dependent precursor polypeptide (45 kDa) was detectable [18]. In a previous study involving four antibodies against Slc11a1, three detected weak vesicular staining of endogenous Slc11a1mut in bone marrow-derived macrophages, whereas the fourth did not [7]. In the present study, no vesicular staining of endogenous Slc11a1mut was apparent. This discrepancy presumably reflects the different detection thresholds of the various anti-Slc11a1 antibodies as the overexpression system clearly demonstrated that it is possible, albeit at a very low level, for mutant Slc11a1 to evade the cellular quality control mechanisms and traffic to endocytic vesicles.
Protein misfolding leading to ER retention and degradation is an increasingly prevalent pathogenic mechanism [37–39]. Examples of diseases caused by misfolded integral membrane proteins include cystic fibrosis [43], retinitis pigmentosa [44], ocular albinism type 1 [45] and demyelinating or hypomyelinating disorders [46]. The murine G169D mutation described in the present study has not been identified in humans. However, it highlights a potentially important mechanism that must be considered in relation to human SLC11A1 disease association studies when characterizing putative functional polymorphisms.
In conclusion, we have used a combination of biochemistry and cell biology to shed light on the molecular basis of innate Slc11a1-mediated susceptibility to infectious disease. We have identified loss of function by defective subcellular targeting as the probable pathogenic mechanism of the infectious disease phenotype manifest in Slc11a1mut mice.
Acknowledgments
We thank A. Riddell for help with the Mo Flo cytometer. This work was supported by the Wellcome Trust. J.K.W. held a Beit Memorial Fellowship.
References
- 1.Blackwell J. M., Searle S., Mohamed H., White J. K. Divalent cation transport and susceptibility to infectious and autoimmune disease: continuation of the Ity/Lsh/Bcg/Nramp1/Slc11a1 gene story. Immunol. Lett. 2003;85:197–203. doi: 10.1016/s0165-2478(02)00231-6. [DOI] [PubMed] [Google Scholar]
- 2.Cellier M., Shustik C., Dalton W., Rich E., Hu J., Malo D., Schurr E., Gros P. Expression of the human NRAMP1 gene in professional primary phagocytes: studies in blood cells and in HL-60 promyelocytic leukemia. J. Leukoc. Biol. 1997;61:96–105. doi: 10.1002/jlb.61.1.96. [DOI] [PubMed] [Google Scholar]
- 3.Evans C. A. W., Harbuz M. S., Ostenfeld T., Norrish A., Blackwell J. M. Nramp1 is expressed in neurons and is associated with behavioural and immune responses to stress. Neurogenetics. 2001;3:69–78. doi: 10.1007/s100480100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clifford K. S., MacDonald M. J. Survey of mRNAs encoding zinc transporters and other metal complexing proteins in pancreatic islets of rats from birth to adulthood: similar patterns in the Sprague–Dawley and Wistar BB strains. Diabetes Res. Clin. Pract. 2000;49:77–85. doi: 10.1016/s0168-8227(00)00141-8. [DOI] [PubMed] [Google Scholar]
- 5.Blackwell J. M., Goswami T., Evans C. A. W., Sibthorpe D., Papo N., White J. K., Searle S., Miller E. N., Peacock C. S., Mohammed H., et al. SLC11A1 (formerly NRAMP1) and disease. Cell. Microbiol. 2001;3:773–784. doi: 10.1046/j.1462-5822.2001.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruenheid S., Pinner E., Desjardins M., Gros P. Natural resistance to infections with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J. Exp. Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Searle S., Bright N. A., Roach T. I. A., Atkinson P. G. P., Barton C. H., Meloen R. H., Blackwell J. M. Localisation of Nramp1 in macrophages: modulation with activation and infection. J. Cell Sci. 1998;111:2855–2866. doi: 10.1242/jcs.111.19.2855. [DOI] [PubMed] [Google Scholar]
- 8.Goswami T., Bhattacharjee A., Babal P., Searle S., Moore E., Li M., Blackwell J. M. Natural-resistance-associated macrophage protein 1 is an H+/bivalent cation antiporter. Biochem. J. 2001;354:511–519. doi: 10.1042/0264-6021:3540511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jabado N., Jankowski A., Dougaparsad S., Picard V., Grinstein S., Gros P. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 2000;192:1237–1248. doi: 10.1084/jem.192.9.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn D. E., Baker B. D., Lafuse W. P., Zwilling B. S. Differential iron transport into phagosomes isolated from the RAW264.7 macrophage cell lines transfected with Nramp1Gly169 or Nramp1Asp169. J. Leukoc. Biol. 1999;66:113–119. doi: 10.1002/jlb.66.1.113. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn D. E., Lafuse W. P., Zwilling B. S. Iron transport into mycobacterium avium-containing phagosomes from an Nramp1(Gly169)-transfected RAW264.7 macrophage cell line. J. Leukoc. Biol. 2001;69:43–49. [PubMed] [Google Scholar]
- 12.Vidal S. M., Malo D., Vogan K., Skamene E., Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell (Cambridge, Mass.) 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 13.Blackwell J. M., Searle S. Genetic regulation of macrophage activation: understanding the function of Nramp1 (=Ity/Lsh/Bcg) Immunol. Lett. 1999;65:73–80. doi: 10.1016/s0165-2478(98)00127-8. [DOI] [PubMed] [Google Scholar]
- 14.Blackwell J. M., Searle S., Goswami T., Miller E. N. Understanding the multiple functions of Nramp1. Microbes Infect. 2000;2:317–321. doi: 10.1016/s1286-4579(00)00295-1. [DOI] [PubMed] [Google Scholar]
- 15.Vidal S., Tremblay M. L., Govoni G., Gauthier S., Sebastiani G., Malo D., Skamene E., Olivier M., Jothy S., Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J. Exp. Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal S., Pinner E., Lepage P., Gauthier S., Gros P. Natural resistance to intracellular infections. Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1D169) mouse strains. J. Immunol. 1996;157:3559–3568. [PubMed] [Google Scholar]
- 17.Atkinson P. G. P., Barton C. H. Ectopic expression of Nramp1 in COS-1 cells modulates iron accumulation. FEBS Lett. 1998;425:239–242. doi: 10.1016/s0014-5793(98)00236-1. [DOI] [PubMed] [Google Scholar]
- 18.Atkinson P. G., Barton C. H. High level expression of Nramp1G169 in RAW264.7 cell transfectants: analysis of intracellular iron transport. Immunology. 1999;96:656–662. doi: 10.1046/j.1365-2567.1999.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barton C. H., Whitehead S. H., Blackwell J. M. Nramp transfection transfers Ity/Lsh/Bcg-related pleiotropic effects on macrophage activation: influence on oxidative burst and nitric oxide pathways. Mol. Med. 1995;1:267–279. [PMC free article] [PubMed] [Google Scholar]
- 20.Ke S. H., Madison E. L. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 1997;25:3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernan R., Heuermann K., Brizzard B. Multiple epitope tagging of expressed proteins for enhanced detection. Biotechniques. 2000;28:789–793. doi: 10.2144/00284pf01. [DOI] [PubMed] [Google Scholar]
- 22.Raschke W. C., Baird S., Ralph R., Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell (Cambridge, Mass.) 1978;15:261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 23.Blackwell J. M., Toole S., King M., Dawda P., Roach T. I., Cooper A. Analysis of Lsh gene expression in congenic B10.L-Lshr mice. Curr. Top. Microbiol. Immunol. 1988;137:301–309. doi: 10.1007/978-3-642-50059-6_45. [DOI] [PubMed] [Google Scholar]
- 24.Roach T. I. A., Barton C. H., Chatterjee D., Blackwell J. M. Macrophage activation: lipoarabinomannan (LAM) from avirulent and virulent strains of Mycobacterium tuberculosis differentially induces the early genes c-fos, KC, JE and TNF-alpha. J. Immunol. 1993;150:1886–1896. [PubMed] [Google Scholar]
- 25.Pfeffer S. R. The endosomal concentration of a mannose 6-phosphate receptor is unchanged in the absence of ligand synthesis. J. Cell Biol. 1987;105:229–234. doi: 10.1083/jcb.105.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths G., Hoflack B., Simons K., Mellman I., Kornfeld S. The mannose 6-phosphate receptor and the biogenesis of lysosomes. Cell (Cambridge, Mass.) 1988;52:329–341. doi: 10.1016/s0092-8674(88)80026-6. [DOI] [PubMed] [Google Scholar]
- 27.Slot J. W., Geuze H. J. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur. J. Cell Biol. 1985;38:87–93. [PubMed] [Google Scholar]
- 28.Bright N. A., Reaves B. J., Mullock B. M., Luzio J. P. Dense core lysosomes can fuse with late endosomes and are re-formed from the resultant hybrid organelles. J. Cell Sci. 1997;110:2027–2040. doi: 10.1242/jcs.110.17.2027. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths G. Berlin: Springer-Verlag; 1993. Fine Structure Immunocytochemistry; pp. 1–459. [Google Scholar]
- 30.Slot J. W., Geuze H. J., Gigengack S., Lienhard G. E., James D. E. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slot J. W., Geuze H. J. Immunohistochemistry. Chichester: John Wiley & Sons; 1983. The use of Protein A-colloidal gold (PAG) complexes as immunolabels in ultrathin frozen sections; pp. 323–346. [Google Scholar]
- 32.Tokuyasu K. T. A study of positive staining of ultrathin frozen sections. J. Ultrastruct. Res. 1978;63:287–307. doi: 10.1016/s0022-5320(78)80053-7. [DOI] [PubMed] [Google Scholar]
- 33.Harlow E., Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1988. Immunoblotting; pp. 267–310. [Google Scholar]
- 34.Klausner R. D., Sitia R. Protein degradation in the endoplasmic reticulum. Cell (Cambridge, Mass.) 1990;62:611–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- 35.Bonifacino J. S., Cosson P., Shah N., Klausner R. D. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell (Cambridge, Mass.) 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- 37.Aridor M., Balch W. E. Integration of endoplasmic reticulum signaling in health and disease. Nat. Med. (N.Y.) 1999;5:745–751. doi: 10.1038/10466. [DOI] [PubMed] [Google Scholar]
- 38.Aridor M., Hannan L. A. Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic. 2000;1:836–851. doi: 10.1034/j.1600-0854.2000.011104.x. [DOI] [PubMed] [Google Scholar]
- 39.Aridor M., Hannan L. A. Traffic jams II: an update of diseases of intracellular transport. Traffic. 2002;3:781–790. doi: 10.1034/j.1600-0854.2002.31103.x. [DOI] [PubMed] [Google Scholar]
- 40.Tabuchi M., Tanaka N., Nishida-Kitayama J., Ohno H., Kishi F. Alternative splicing regulates the subcellular localization of divalent metal transporter 1 isoforms. Mol. Biol. Cell. 2002;13:4371–4387. doi: 10.1091/mbc.E02-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleming M. D., Romano M. A., Su M. A., Garrick L. M., Garrick M. D., Andrews N. C. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su M. A., Trenor C. C., Fleming J. C., Fleming M. D., Andrews N. C. The G185R mutation disrupts function of the iron transporter Nramp2. Blood. 1998;92:2157–2163. [PubMed] [Google Scholar]
- 43.Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell (Cambridge, Mass.) 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 44.Sung C. H., Davenport C. M., Nathans J. Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J. Biol. Chem. 1993;268:26645–26649. [PubMed] [Google Scholar]
- 45.d'Addio M., Pizzigoni A., Bassi M. T., Baschirotto C., Valetti C., Incerti B., Clementi M., De Luca M., Ballabio A., Schiaffino M. V. Defective intracellular transport and processing of OA1 is a major cause of ocular albinism type 1. Hum. Mol. Genet. 2000;9:3011–3018. doi: 10.1093/hmg/9.20.3011. [DOI] [PubMed] [Google Scholar]
- 46.Gow A., Friedrich V. L., Jr, Lazzarini R. A. Many naturally occurring mutations of myelin proteolipid protein impair its intracellular transport. J. Neurosci. Res. 1994;37:574–583. doi: 10.1002/jnr.490370504. [DOI] [PubMed] [Google Scholar]