Abstract
DGK (diacylglycerol kinase) regulates the concentration of two bioactive lipids, diacylglycerol and phosphatidic acid. DGKδ1 or its PH (pleckstrin homology) domain alone has been shown to be translocated to the plasma membrane from the cytoplasm in PMA-treated cells. In the present study, we identified Ser-22 and Ser-26 within the PH domain as the PMA- and epidermal-growth-factor-dependent phosphorylation sites of DGKδ1. Experiments in vitro and with intact cells suggested that the cPKC (conventional protein kinase C) phosphorylated these Ser residues directly. Puzzlingly, alanine/asparagine mutants at Ser-22 and Ser-26 of DGKδ1 and its PH domain are still persistently translocated by PMA treatment, suggesting that the PH domain phosphorylation is not responsible for the enzyme translocation and that the translocation was caused by a PMA-dependent, but cPKC-independent, process yet to be identified. Interestingly, the aspartate mutation, which mimics phosphoserine, at Ser-22 or Ser-26, inhibited the translocation of full-length DGKδ1 and the PH domain markedly, suggesting that the phosphorylation regulates negatively the enzyme translocation. Our results provide evidence of the phosphorylation of the DGKδ1 PH domain by cPKC, and suggest that the phosphorylation is involved in the control of subcellular localization of DGKδ1.
Keywords: diacylglycerol kinase, pleckstrin homology domain, phorbol ester, protein kinase C, translocation
Abbreviations: cPKC, conventional protein kinase C; DG, diacylglycerol; DGK, DG kinase; DMEM, Dulbecco's modified Eagle's medium; EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; GFP, green fluorescent protein; GST, glutathione S-transferase; HEK, human embryonic kidney; MEK, mitogen-activated protein kinase/ERK kinase; nPKC, novel protein kinase C; NTR, N-terminal region; PH, pleckstrin homology; PKC, protein kinase C; SAM, sterile α motif; YFP, yellow fluorescent protein
INTRODUCTION
Upon cell activation by various hormones, neurotransmitters, growth factors and other agonists, DG (diacylglycerol) is liberated from phosphatidylinositols and other phospholipids [1,2]. DGK (DG kinase) phosphorylates DG to produce phosphatidic acid [3]. The roles of DG and phosphatidic acid as lipid second messengers have been attracting much attention. DG is known to be an activator of cPKC and nPKC (conventional and novel protein kinase C respectively), Unc-13 and Ras guanine nucleotide-releasing protein [4–7]. Phosphatidic acid has been reported in a number of studies to modulate phosphatidylinositol-4-phosphate kinase, Raf-1 kinase, atypical PKC and many other important enzymes [8,9]. Because the cellular concentration of these signalling lipids must be strictly regulated, DGK is thought to be one of the key enzymes involved in the cellular signal transduction.
To date, nine mammalian DGK isoenzymes (α, β, γ, δ, ε, ζ, η, θ and ι), containing in common the zinc-finger structures and the catalytic region, have been identified and classified into five subgroups according to their structural features [10–13]. Interestingly, the occurrence of alternative splicing, which supplements further the complexity of the DGK gene family members, has recently been detected for five mammalian DGK genes (γ [14], ζ [15], β [16], δ [17] and η [18] isoforms). The structural diversity and the tissue- and cell-dependent expression patterns observed distinctively for these isoforms [10–13] suggest that each member is regulated by a distinct mechanism, performing differentiated functions in particular types of cells.
We previously cloned a cDNA encoding DGKδ1 (type II isoenzyme), which contains a PH (pleckstrin homology) domain at the N-terminus and a SAM (sterile α motif) domain at the C-terminus [19]. The alternative splicing product (DGKδ2) contains the 52 residues extended N-terminally [17]. The SAM domain, which is an evolutionally conserved protein–protein interaction motif, was found to occur in a wide range of proteins [20,21]. We demonstrated by gel filtration and co-immunoprecipitation analyses that DGKδ1 formed homo-oligomers [22] as well as hetero-oligomers with other type II DGK members, DGKδ2 and DGKη2, through their common SAM domains [17,18]. The PH domain is a small protein module of 100–120 amino acids that is found in many proteins involved in cell signalling, cytoskeletal rearrangement and other processes [23,24]. We reported that DGKδ1 was translocated from the cytoplasm to the plasma membrane in phorbol-ester-stimulated cells [22], and that the PH domain was critically involved in the translocation [17]. Interestingly, the subcellular localization of DGKδ2 was not changed even after phorbol ester stimulation [17].
We have found that, in addition to the translocation, endogenous or transiently expressed DGKδ1 is phosphorylated by endogenous protein kinase in response to phorbol ester stimulation in HepG2, COS-7 and HEK-293 (human embryonic kidney) cells [22]. However, the phosphorylation site(s) and the functional relationship between the enzyme phosphorylation and translocation remain to be explored. In the present study, we identified Ser-22 and Ser-26 within the PH domain as the phorbol-ester- and EGF (epidermal growth factor)-dependent phosphorylation sites. Several lines of evidence suggest that cPKC serves as a direct upstream kinase for these serine residues, and that, unexpectedly, the phosphorylation of one of these serine residues by cPKC regulates negatively, rather than positively, the translocation of DGKδ1.
EXPERIMENTAL
Plasmid constructs
The cDNA fragments of DGKδ1-NTR (amino acid residues 1–116), DGKδ1-ΔNTR (amino acid residues 109–1170) and DGKδ1-PH domain (amino acid residues 10–107) were generated from a human DGKδ1 cDNA clone [19] by PCR. Mutagenesis was carried out using the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.). For subsequent subcloning into expression vectors, primers were designed such that the resulting DGKδ1 cDNA fragments contained a 5′-EcoRI restriction site and a 3′-stop codon followed by a 3′-EcoRI site. The resulting DGKδ1 fragments were fused in-frame with YFP (yellow fluorescent protein) in the pEYFP-C1 vector (Clontech, Tokyo, Japan) and with 3×FLAG tag in the p3×FLAG–CMV-7.1 (Sigma-Aldrich, Tokyo, Japan). Plasmids encoding the DGKδ1-PH domain fused to GST (glutathione S-transferase) were obtained by inserting the PH domain cDNA fragments into the pGEX-6P-1 vector (Amersham Biosciences, Tokyo, Japan).
Cell culture and transfection
HEK-293 and COS-7 cells were maintained in DMEM (Dulbecco's modified Eagle's medium; Sigma-Aldrich) containing 10% (v/v) foetal bovine serum at 37 °C in an atmosphere containing 5% CO2. Cells were transiently transfected with cDNAs using Effectene transfection reagent according to the manufacturer's instruction (Qiagen, Tokyo, Japan). After 24–48 h, cells were used for further analysis.
Protein phosphorylation analysis in intact cells
COS-7 or HEK-293 cells expressing various 3×FLAG-tagged DGKδ1 constructs were used at 40 h post-transfection. The cells (35-mm-diameter dish) transfected were washed four times with Tris-buffered saline, incubated for 2 h in DMEM without sodium phosphate and sodium pyruvate (Invitrogen Life Technologies, Tokyo, Japan) containing 0.1% (w/v) BSA, and then labelled for 2 h with [32P]orthophosphate (0.1 mCi/ml; Amersham Biosciences) in phosphate-free DMEM/0.1% BSA. The labelled cells were subsequently incubated in phosphate-free DMEM/0.1% BSA containing 100 nM PMA or 0.1% DMSO for 30 min. When the effects of EGF stimulation were determined, cells were labelled for 4 h with 0.3 mCi/ml [32P]orthophosphate. Stimulated cells were then harvested in buffer 1 [50 mM Tris/HCl, pH 7.4, 1 mM EDTA, 1% (w/v) Nonidet P-40, 150 mM NaCl, 1 mM PMSF, Complete™ protease inhibitor mixture (Roche Molecular Biochemicals, Tokyo, Japan) and phosphatase inhibitor cocktail II (Sigma-Aldrich)]. The suspension was sonicated and then centrifuged at 10000 g for 20 min at 4 °C to give cell lysates. Cell lysates (300 μl) were pre-cleared with 10 μl of Protein A/G PLUS–agarose (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.). Anti-FLAG M2 monoclonal antibody (2 μg; Sigma-Aldrich) was added to pre-cleared lysates to immunoprecipitate 3×FLAG-tagged DGKδ1 proteins. After 1 h, 5 μl of Protein A/G PLUS–agarose was added, followed by a 1 h incubation at 4 °C. After washing the agarose beads five times with buffer 1, immunoprecipitated proteins were extracted with 50 μl of SDS sample buffer and then separated by SDS/PAGE. The radioactive signal in a dried gel was visualized by phosphorimaging using a BAS1800 Bio-Image Analyzer (Fuji Film, Tokyo, Japan).
Western blot analysis
Pre-cleared cell lysates and immunoprecipitates were separated by SDS/PAGE. The separated proteins were transferred on to a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany) and blocked with 10% Block Ace (Dainippon Pharmaceutical, Tokyo, Japan) as described previously [22]. The membrane was incubated with anti-FLAG M2 monoclonal antibody in Block Ace for 1 h. The immunoreactive bands were visualized using horse-radish-peroxidase-conjugated anti-mouse IgG antibody (Jackson Immunoresearch Laboratories, West Grove, PA, U.S.A.) and SuperSignal (Pierce, Rockford, IL, U.S.A.).
Phosphoamino acid analysis
3×FLAG-tagged DGKδ1-PH domain labelled with 32P was immunoprecipitated, separated by SDS/(16.5%) PAGE, and then transferred on to an Immobilon-PSQ membrane (Millipore, Tokyo, Japan). The transferred protein was visualized by autoradiography, excised from the membrane and hydrolysed in 6 M HCl at 110 °C for 90 min. The hydrolysate was dried under vacuum and redissolved in water containing unlabelled phosphoserine, phosphothreonine and phosphotyrosine standards. The hydrolysate was spotted on a cellulose TLC plate (Sigma-Aldrich). The electrophoresis was carried out in pH 3.5 buffer (5% ethanoic acid and 0.5% pyridine). After being dried, plates were sprayed with 0.25% (w/v) ninhydrin in acetone and heated at 65 °C to visualize the phosphoamino acid standards. The radioactive signal of phosphoamino acid was detected by phosphorimaging using a BAS1800 Bio-Image Analyzer.
Expression and purification of GST-fusion proteins
XL1-Blue cells (Stratagene) were transformed by various pGEX-6P-1 constructs, and GST or GST-fusion proteins were expressed and purified according to the procedure recommended by the manufacturer (Amersham Biosciences). In this case, the expression of fusion proteins was induced by 1 mM isopropyl β-D-thiogalactoside at 37 °C for 3 h. Cells were then lysed by sonication in PBS, and insoluble material was removed by centrifugation at 10000 g for 5 min. The supernatants were incubated in batches with glutathione–Sepharose 4B (Amersham Biosciences) for 2 h at 4 °C, and beads were then washed four times with PBS. The beads were finally washed once with the kinase buffer (see below) without ATP just before an in vitro protein kinase assay.
In vitro protein kinase assay
An in vitro protein kinase assay was carried out at 30 °C for 30 min in the kinase buffer (20 mM Tris/HCl, pH 7.4, 1 mM CaCl2, 1 mM dithiothreitol, 10 mM MgCl2, 200 μg/ml phosphatidylserine, 20 μg/ml diolein, 1 mM ATP and 2.5 μCi of [γ-32P]ATP). Phosphatidylserine and diolein in chloroform were dried under nitrogen and dispersed in the buffer by sonication for 30 s at 4 °C, before the addition of enzyme and radioactive ATP. The beads that bound GST or GST-fusion proteins (5 μg) were incubated with 15 m-units of purified rat PKCα (>90% pure; Sigma-Aldrich). Reactions were terminated by centrifugation at 10000 g for 5 min, and the beads were washed with the kinase buffer without ATP. GST or GST-fusion proteins were extracted with SDS sample buffer and then analysed by SDS/PAGE.
Fluorescence microscopy
HEK-293 cells were grown on poly(L-lysine)-coated glass coverslips and transiently transfected with cDNAs encoding various DGKδ1 constructs N-terminally fused with YFP. After 24 h, HEK-293 cells were washed four times with PBS and then cultured in DMEM/0.1% BSA. After 3 h, the medium was exchanged with DMEM/0.1% BSA containing 100 nM PMA or 0.1% DMSO. After 30 min, cells were fixed with 3.7% (w/v) formaldehyde in PBS for 10 min and then washed five times with PBS at room temperature (25 °C). The coverslips were mounted using Vectashield (Vector Laboratories, Burlingame, CA, U.S.A.). Cells were examined using an inverted confocal laser scanning microscope (Zeiss LSM 510).
RESULTS
Phosphorylation of the DGKδ1-PH domain by PMA stimulation
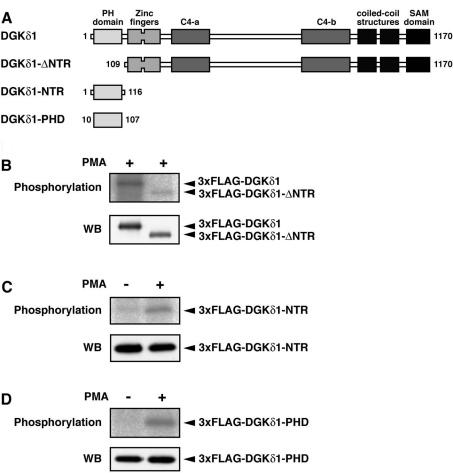
We previously described the phosphorylation of DGKδ1 in PMA-stimulated cells [22]. To identify the phosphorylation sites, we first assessed phosphorylation of wild-type DGKδ1 and its truncation mutant lacking the N-terminal region (DGKδ1-ΔNTR; Figure 1A). As shown in Figure 1(B), when compared with the wild-type enzyme, the phosphorylation of DGKδ1-ΔNTR was markedly reduced, but still persisted in PMA-stimulated COS-7 cells. On the other hand, DGKδ1 N-terminal region (DGKδ1-NTR) alone was also phosphorylated in a PMA-dependent manner (Figure 1C). Because DGKδ1-NTR contains the DGKδ1-specific sequences in addition to the PH domain, we next tested whether or not the PH domain alone was phosphorylated. As shown in Figure 1(D), the DGKδ1-PH domain alone was phosphorylated by PMA stimulation. These results indicate that DGKδ1 has PMA-dependent phosphorylation sites in both the PH domain (amino acid residues 10–107) and the remaining C-terminal region (amino acid residues 109–1170).
Figure 1. Phosphorylation of DGKδ1-PH domain in response to PMA stimulation.
(A) A schematic representation of the DGKδ1 constructs used for the phosphorylation assay. (B–D) COS-7 cells were transfected with a plasmid encoding 3×FLAG–DGKδ1 (B), 3×FLAG–DGKδ1-ΔNTR (B), 3×FLAG–DGKδ1-NTR (C) or 3×FLAG–DGKδ1-PH domain (D). After serum-starvation and 32P-labelling, cells were incubated for 30 min in the presence of 100 nM PMA (+) or 0.1% DMSO (−). FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibody and separated by SDS/PAGE. The radioactive signal was visualized by a BAS1800 Bio-Image Analyzer (upper panels), and the immunoprecipitated 3×FLAG-tagged proteins were analysed by Western blotting using anti-FLAG antibody (lower panels). Representatives of three repeated experiments are shown. PHD, PH domain.
Identification of phosphorylation sites within the DGKδ1-PH domain
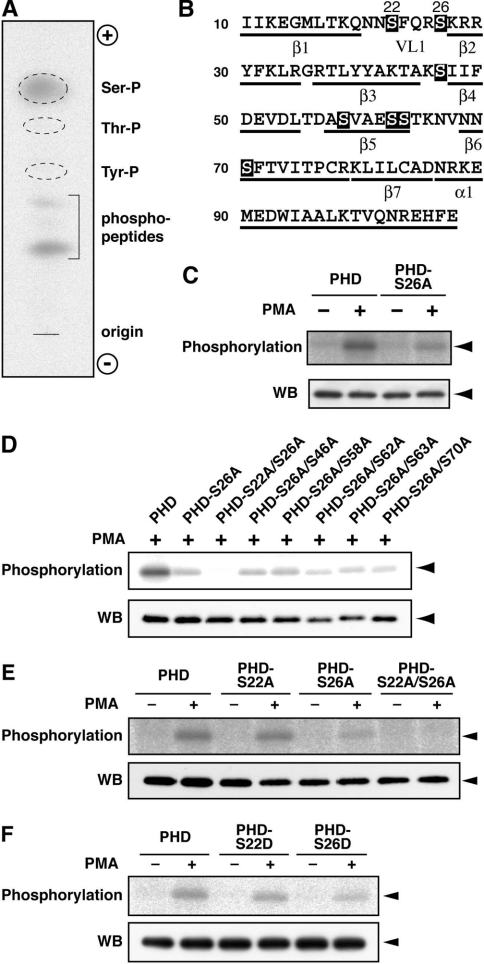
We next attempted to identify the phosphorylation site(s) within the DGKδ1-PH domain. The domain contains seven serine, seven threonine and three tyrosine residues (Figure 2B). To determine which amino acid residues are phosphorylated in PMA-stimulated cells, we first carried out a phosphoamino acid analysis of the DGKδ1-PH domain. As shown in Figure 2(A), we detected radio-active phosphoserine. However, no phosphorylation signal of threonine and tyrosine residues was detected.
Figure 2. Phosphorylation at Ser-22 and Ser-26 in the DGKδ1-PH domain.
(A) Phosphoamino acid analysis of the DGKδ1-PH domain. The analysis was performed as described in the Experimental section. Ser-P, phosphoserine; Thr-P, phosphothreonine; Tyr-P, phosphotyrosine; phosphopeptides, undigested phosphopeptides. (B) Primary and predicted secondary structure of the DGKδ1-PH domain. Serine residues are highlighted. α, α-helix; β, β-sheet; VL1, variable loop 1. (C–F) Effects of point mutations on phosphorylation of the DGKδ1-PH domain. COS-7 cells were transfected with expression plasmids as indicated. After starvation and 32P-labelling, cells were incubated for 30 min in the presence of 100 nM PMA (+) or 0.1% DMSO (−). FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibody and analysed by SDS/PAGE. The radioactive signal was visualized by a BAS1800 Bio-Image Analyzer (upper panels), and the immunoprecipitated 3×FLAG-tagged proteins were analysed by Western blotting using anti-FLAG antibody (lower panels). The position of the DGKδ1-PH domain and its mutants is indicated by arrowheads. Representatives of three repeated experiments are shown. PHD, PH domain.
We next used alanine-scanning mutagenesis to determine which serine residues within the DGKδ1-PH domain are targets of endogenous protein kinase. The PMA-dependency of phosphorylation implies that cPKC and/or nPKC may phosphorylate the serine residue(s). It is known that PKC preferentially phosphorylates serine and threonine flanked with arginine and lysine at −3 to −1 and +1 to +3 positions [25]. In this context, Ser-26 is flanked with four arginine/lysine residues at the appropriate positions (Figure 2B). This serine residue was thus replaced with alanine (S26A). As shown in Figure 2(C), the DGKδ1-PH domain-S26A mutant showed a considerable loss of phosphorylation signal compared with the wild-type domain. However, because some phosphorylation signal still persisted with this mutant, we next attempted to identify the other serine residues that were phosphorylated. We further substituted alanine for one of six serine residues (Ser-22, Ser-46, Ser-58, Ser-62, Ser-63 and Ser-70) of the S26A mutant (Figure 2B). Each of these double mutants was expressed in COS-7 cells, and the phosphorylation levels were assessed. Only the S22A/S26A mutant showed a complete loss of phosphorylation signal (Figure 2D).
Although the phosphorylation level of the S26A mutant was markedly reduced, an alanine mutation at Ser-22 alone reduced only slightly the PH domain phosphorylation (Figure 2E). We further constructed S22D and S26D mutants, in which a negative charge (aspartic acid) was introduced to mimic phosphoserine. Reduction patterns of phosphorylation of the aspartate mutants were essentially the same as those of the corresponding alanine mutants (Figures 2E and 2F), suggesting that the phosphorylation of one of the two serine residues occurs independently of each other. These results indicate that Ser-26 is the major phosphorylation site within the PH domain in response to PMA stimulation. Essentially the same results were obtained using HEK-293 cells (results not shown).
cPKC is involved in the phosphorylation of Ser-22 and Ser-26 within the DGKδ1-PH domain
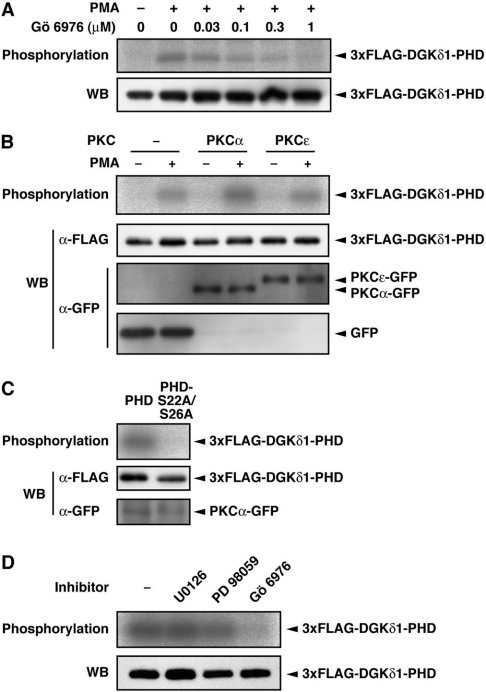
In the next set of experiments, we explored the endogenous protein kinase involved in the phosphorylation of the DGKδ1-PH domain. We showed previously that staurosporine, a non-selective PKC inhibitor, inhibits PMA-dependent phosphorylation of DGKδ1 [22]. In the present study, we tested the effects of a cPKC-selective inhibitor, Gö 6976, on phosphorylation. As shown in Figure 3(A), Gö 6976 clearly reduced the phosphorylation signal in a dose-dependent manner. Gö 6976 at a concentration of 1 μM achieved a complete loss of the signal. The result indicates that the DGKδ1-PH domain was phosphorylated through the cPKC-dependent pathway in PMA-stimulated cells.
Figure 3. Effects of a cPKC-selective inhibitor, Gö 6976, and PKCα expression on the DGKδ1-PH domain phosphorylation.
(A) COS-7 cells were transfected with a plasmid encoding the 3×FLAG–DGKδ1-PH domain. Cells were starved, 32P-labelled and incubated with Gö 6976 at the indicated concentrations for 30 min before PMA stimulation. Cells were stimulated with 100 nM PMA (+) or 0.1% DMSO (−) for 30 min in the presence of Gö 6976. (B) COS-7 cells were co-transfected with p3×FLAG–CMV–DGKδ1-PH domain and either pEGFP alone, pEGFP–PKCα or pEGFP–PKCε. After starvation and 32P-labelling, cells were incubated for 30 min in the presence of 100 nM PMA (+) or 0.1% DMSO (−). (C) COS-7 cells were co-transfected with pEGFP–PKCα and either p3×FLAG–CMV–DGKδ1-PH domain or -PH domain-S22A/S26A. After starvation and 32P-labelling, cells were incubated for 30 min in the presence of 100 nM PMA. (D) COS-7 cells were transfected with a plasmid encoding the 3×FLAG–DGKδ1-PH domain. Cells were starved, 32P-labelled and incubated with U0126 (50 μM), PD098059 (100 μM) or Gö 6976 (1 μM) for 30 min before PMA stimulation. Cells were treated with 100 nM PMA for 30 min in the presence of the inhibitors. FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibody and analysed by SDS/PAGE. The radioactive signal was visualized by a BAS1800 Bio-Image Analyzer (A–D, top panels), and the 3×FLAG-tagged proteins immunoprecipitated (A–D) and the GFP-fusion proteins in cell lysates (B and C) were analysed by Western blotting using anti-FLAG and anti-GFP antibodies (bottom panels, WB).
To substantiate the cPKC-dependent phosphorylation, we examined the effects of cPKC expression. We first investigated the expression patterns of endogenous PKC isoenzymes in COS-7 and HEK-293 cells by Western blotting. Among cPKC isoenzymes, only PKCα was highly expressed, whereas PKCs β and γ were not detected in the two cell lines (results not shown). Thus, in subsequent experiments, we focused on PKCα. The phosphorylation of the PH domain was appreciably enhanced by the overexpres-sion of PKCα (Figure 3B). In contrast, nPKC (ε-isoform), which was an nPKC isoform solely detectable in COS-7 and HEK-293 cells (results not shown), did not markedly affect the phosphorylation level. The double alanine mutant (DGKδ1-PH domain-S22A/S26A) was not phosphorylated in the PKCα-overexpressing cells (Figure 3C), indicating that at least one of Ser-22 and Ser-26 was indeed phosphorylated through the cPKC-dependent pathway.
PKC is known to activate the MEK [mitogen-activated protein kinase/ERK (extracellular-signal-regulated kinase) kinase]/ERK pathway [26,27]. However, MEK inhibitors, U0126 (50 μM) and PD098059 (100 μM) [28], failed to inhibit the phosphorylation (Figure 3D). In this experiment, we confirmed that U0126 (50 μM) and PD098059 (100 μM) inhibited phosphorylation of ERK by 95% and 70% respectively (results not shown). The result suggests that PKC-activated protein kinases including MEK, ERK and further downstream kinases of the ERK pathway are not involved in the phosphorylation of the DGKδ1-PH domain.
cPKC directly phosphorylates Ser-22 and Ser-26 of the DGKδ1-PH domain in vitro
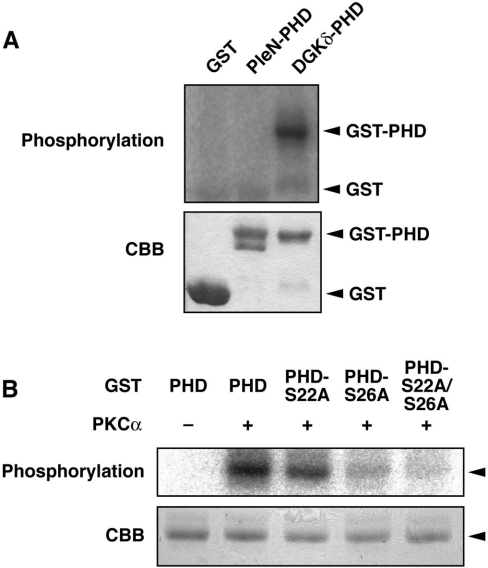
We next asked whether cPKC directly phosphorylates the DGKδ1-PH domain. The DGKδ1-PH domain fused with GST was purified and then incubated with purified rat PKCα in the presence of [γ-32P]ATP. As shown in Figure 4(A), the GST–DGKδ1-PH domain was phosphorylated by PKCα in vitro. On the other hand, the N-terminal PH domain of pleckstrin was not phosphorylated by PKCα, suggesting that the phosphorylation is specific for the DGKδ1-PH domain. We confirmed that the double alanine mutant (GST–DGKδ1-PH domain-S22A/S26A) failed to be phosphorylated by cPKC (Figure 4B). Moreover, the single alanine mutants (GST–DGKδ1-PH domain-S22A and -S26A) exhibited approx. 30% and 80% losses of phosphorylation signal respectively, compared with the wild-type PH domain. These results indicate that PKCα is capable of phosphorylating both Ser-22 and Ser-26 directly within the DGKδ1-PH domain, and that, consistent with the phosphorylation patterns obtained using intact cells (Figure 2E), the phosphorylation of Ser-26 in vitro by PKCα is dominant over that of Ser-22 in vitro.
Figure 4. In vitro phosphorylation of the DGKδ1-PH domain by PKCα.
(A) GST, GST–pleckstrin N-terminal PH domain (PleN-PHD) and GST–DGKδ1-PH domain (DGKδ-PHD) proteins were incubated with (+) or without (−) purified rat PKCα (>90% purity). (B) GST–DGKδ1-PH domain (PHD), GST–DGKδ1-PH domain-S22A (PHD-S22A), GST–DGKδ1-PH domain-S26A (PHD-S26A) or GST–DGKδ1-PH domain-S22A/S26A (PHD-S22A/S26A) proteins were incubated with (+) or without (−) PKCα. Subsequently, the phosphorylated products were separated by SDS/PAGE followed by Coomassie Brilliant Blue staining (CBB, lower panels). The radioactive signal was visualized by a BAS1800 Bio–Image Analyzer (upper panels). The position of the GST–DGKδ1-PH domain and its mutants is indicated by an arrowhead.
EGF-dependent phosphorylation at Ser-22 and Ser-26 within the DGKδ1-PH domain
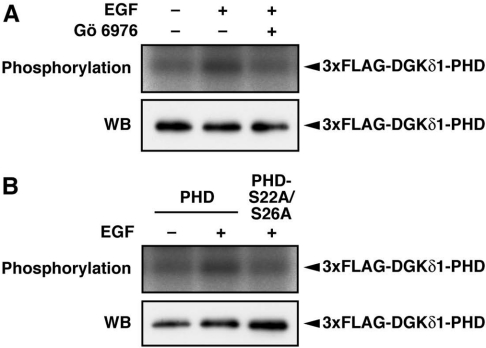
We examined whether, in addition to phorbol ester stimulation, Ser-22 and Ser-26 were phosphorylated by physiological stimulation such as growth factors. When COS-7 cells were stimulated with EGF, the PH domain expressed was phosphorylated (Figure 5A). This phosphorylation was markedly inhibited by Gö6976, indicating that cPKC plays a critical role in the signalling pathway from the EGF receptor stimulation to the DGKδ1-PH domain phosphorylation. The double alanine mutant (DGKδ1-PH domain-S22A/S26A) exhibited a markedly decreased phosphorylation level (Figure 5B). These results indicate that at least one of Ser-22 and Ser-26 was phosphorylated via cPKC in EGF-stimulated cells.
Figure 5. Phosphorylation of the DGKδ1-PH domain by EGF stimulation.
(A) COS-7 cells were transfected with a plasmid encoding 3×FLAG–DGKδ1-PH domain (PHD). Cells were starved, 32P-labelled, and incubated with Gö 6976 (1 μM) for 30 min before EGF stimulation. Cells were incubated with (+) or without (−) 100 ng/ml EGF for 10 min in the presence of Gö 6976. (B) COS-7 cells were transfected with either p3×FLAG–CMV–DGKδ1-PH domain (PHD) or -PH domain-S22A/S26A (PHD-S22A/S26A). After starvation and 32P-labelling, cells were incubated with (+) or without (−) 100 ng/ml EGF for 10 min. FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibody and aliquots were analysed by SDS/PAGE. The radioactive signal was visualized by a BAS1800 Bio-Image Analyzer (upper panels), and the immunoprecipitated 3×FLAG-tagged proteins were analysed by Western blotting using anti-FLAG antibody (lower panels).
Effects of PKC inhibitors on the translocation of the DGKδ1-PH domain
We next attempted to reveal the physiological relevance of the phosphorylation of DGKδ1 at Ser-22 and Ser-26. We reported previously that in response to PMA stimulation, DGKδ1 is translocated from the cytoplasm to the plasma membrane in HEK-293 cells [22], and that the DGKδ1-PH domain is necessary and sufficient for the DGKδ1 translocation [17]. Thus we examined the effects of PKC inhibitors on intracellular localization of the DGKδ1-PH domain in HEK-293 cells.
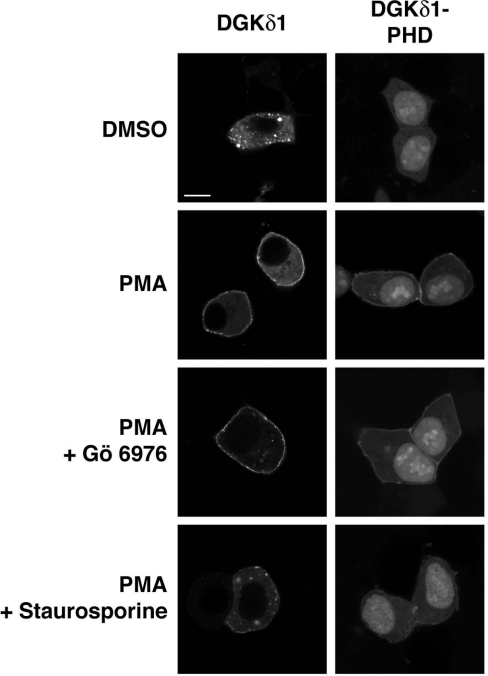
The YFP-tagged DGKδ1-PH domain exhibited cytoplasmic/nucleoplasmic localization in the resting cells (Figure 6). However, as reported previously [17], the DGKδ1-PH domain was partly translocated from the cytoplasm to the plasma membrane upon PMA stimulation. The subcellular localization of YFP alone was not affected by PMA stimulation (see Fig. 7). As reported previously for full-length DGKδ1 [22], the cell treatment with a non-selective PKC inhibitor, staurosporine, markedly inhibited the translocation of the PH domain (Figure 6). In contrast, Gö 6976 unexpectedly failed to block the PMA-dependent translocation. Essentially the same inhibitory pattern was obtained using full-length DGKδ1. These results strongly suggest that cPKC-dependent phosphorylation is not involved in the positive regulation of the translocation, and that unknown mechanisms, which are dependent on PMA and independent of cPKC, operate in this event.
Figure 6. Effects of PKC inhibitors on the subcellular distribution of DGKδ1 and its PH domain.
HEK-293 cells were transfected with a plasmid encoding YFP–DGKδ1 or YFP–DGKδ1-PH domain (PHD). After starvation, cells were pre-incubated with 1 μM Gö 6976 or 300 nM staurosporine for 30 min. Cells were stimulated with 100 nM PMA for 30 min in the presence of Gö 6976 or staurosporine. After stimulation, HEK-293 cells were fixed with 3.7% (w/v) formaldehyde and then mounted on to glass slides. Cells were examined using an inverted confocal laser scanning microscopy (Zeiss LSM 510). Scale bar, 10 μm. Representatives of three repeated experiments are shown.
Effects of mutations at Ser-22 and Ser-26 on the translocation of DGKδ1-PH domain
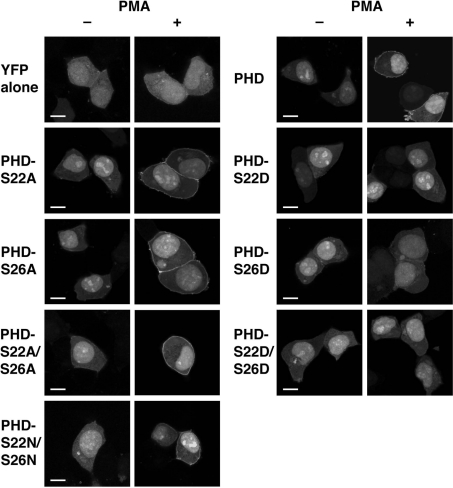
To analyse further the functional relationship between the phosphorylation of the PH domain and the PMA-dependent translocation, we examined subcellular distribution of the alanine mutants (YFP–DGKδ1-PHD-S22A, -S26A and -S22A/S26A), in which Ser-22 and/or Ser-26 were replaced with alanine to prevent phosphorylation. As observed for the experiment using Gö 6976, the alanine mutants were translocated markedly from the cytoplasm to the plasma membrane by PMA stimulation (Figure 7), suggesting further that the cPKC-dependent phosphorylation does not participate in the enhancement of translocation.
Figure 7. Effects of alanine/asparagine and aspartate mutations of Ser-22 and Ser-26 on subcellular distribution of the DGKδ1-PH domain.
HEK-293 cells were transfected with plasmids encoding YFP alone, YFP–DGKδ1-PH domain (PHD), YFP–DGKδ1-PH domain-S22A (PHD-S22A), YFP–DGKδ1-PH domain-S26A (PHD-S26A), YFP–DGKδ1-PH domain-S22A/S26A (PHD-S22A/S26A), YFP–DGKδ1-PH domain-S22D (PHD-S22D), YFP–DGKδ1-PH domain-S26D (PHD-S26D), YFP–DGKδ1-PH domain-S22D/S26D (PHD-S22D/S26D) or YFP–DGKδ1-PH domain-S22N/S26N (PHD-S22N/S26N) as indicated. After starvation, cells were incubated for 30 min in the presence of 100 nM PMA (+) or 0.1% DMSO (−). After stimulation, HEK-293 cells were fixed with 3.7% (w/v) formaldehyde and then mounted on to glass slides. Cells were examined using an inverted confocal laser scanning microscopy (Zeiss LSM 510). Scale bar, 10 μm. Representatives of three repeated experiments are shown.
We next examined subcellular distribution of the aspartate mutants (YFP–DGKδ1-PHD-S22D, -S26D and -S22D/S26D), in which one or two aspartate residues were introduced to mimic phosphoserine. In contrast with the alanine mutants, the aspartate mutants localized at the cytoplasm in resting cells and failed to translocate even in PMA-stimulated cells. In contrast, YFP–DGKδ1-PHD-S22N/S26N, in which asparagine was substituted for Ser-22 and Ser-26, was translocated markedly to the plasma membrane. These results suggest that the phosphorylation of one of Ser-22 and Ser-26 prevents the translocation and/or enhances dissociation from the plasma membrane following the enzyme translocation. Western blot analysis showed that the expression levels of the mutants were almost the same (Figure 2; and results not shown). Therefore we consider that the differences of localization are not caused by variable expression levels of the enzyme constructs.
Effects of mutations at Ser-22 and Ser-26 on translocation of full-length DGKδ1
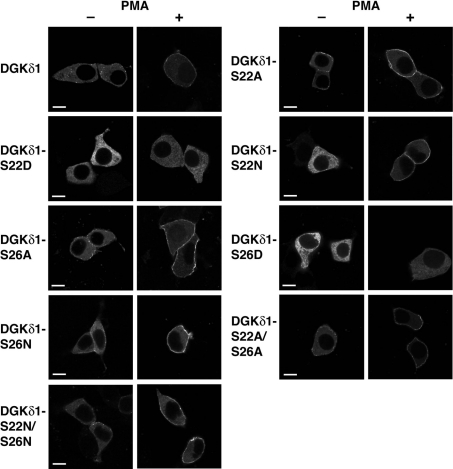
We next examined whether the phosphorylation of Ser-22 or Ser-26 indeed affected the subcellular localization of full-length DGKδ1. As reported previously [22], wild-type DGKδ1 was translocated from the cytoplasm to the plasma membrane by PMA stimulation (Figure 8). However, the aspartate mutants, YFP-DGKδ1-S22D and -S26D, remained at the cytoplasm even in PMA-stimulated cells. In contrast with the aspartate mutants, alanine mutants, YFP–DGKδ1-S22A, -S26A and -S22A/S26A, failed to block the PMA-dependent translocation. Moreover, all of the asparagine mutants, YFP–DGKδ1-S22N, -S26N and -S22N/S26N, clearly underwent the PMA-induced translocation, indicating that the negative charge on the aspartate residue is essential to prevent the translocation. These results suggest that the phosphorylation of either Ser-22 or Ser-26 negatively regulates the translocation of full-length DGKδ1 via its PH domain in agreement with the results of the expression of the PH domain alone (Figure 7).
Figure 8. Effects of alanine/asparagine and aspartate mutations of Ser-22 and Ser-26 on the subcellular distribution of full-length DGKδ1.
HEK-293 cells were transfected with plasmids encoding YFP–DGKδ1 (DGKδ1), YFP–DGKδ1-S22A (DGKδ1-S22A), YFP–DGKδ1-S22D (DGKδ1-S22D), YFP–DGKδ1-S22N (DGKδ1-S22N), YFP–DGKδ1-S26A (DGKδ1-S26A), YFP–DGKδ1-S26D (DGKδ1-S26D), YFP–DGKδ1-S26N (DGKδ1-S26N), YFP–DGKδ1-S22A/S26A (DGKδ1-S22A/S26A) or YFP–DGKδ1-S22N/S26N (DGKδ1-S22N/S26N) as indicated. After starvation, cells were incubated for 30 min in the presence of 100 nM PMA (+) or 0.1% DMSO (−). After stimulation, HEK-293 cells were fixed with 3.7% (w/v) formaldehyde and then mounted on to glass slides. Cells were examined using an inverted confocal laser scanning microscopy (Zeiss LSM 510). Scale bar, 10 μm. Representatives of three repeated experiments are shown.
DISCUSSION
In the present study, we report that Ser-22 and Ser-26 of DGKδ1 within the PH domain are phosphorylated in PMA- and EGF-stimulated cells. The results presented here indicate that cPKC is involved in the phosphorylation of DGKδ1. Although we cannot rule out the possibility that an unknown protein kinase that functions downstream of cPKC phosphorylates the DGKδ1 PH domain in cells, it is likely that cPKC is a direct upstream kinase for Ser-22 and Ser-26. This conclusion is based on the observation that: (i) purified PKCα phosphorylated the purified DGKδ1-PH domain directly in vitro (Figure 4), (ii) the inhibition of MEK, which is the major downstream effector of PKC [26,27], did not affect the phosphorylation of the DGKδ1-PH domain (Figure 3D), (iii) Ser-22 and Ser-26 appreciably fulfil the canonical phosphorylation site motif for PKCα as discussed in the next paragraph, and (iv) the preference of the phosphorylation site (Ser-26>Ser-22) detected for the endogenous protein kinase was essentially the same as that exhibited by PKCα in vitro (Figures 2E and 4B). Because PKCα was the solely detectable cPKC in HepG2, HEK-293 and COS-7 cells (results not shown) where cellular or transfected DGKδ1 was phosphorylated by endogenous protein kinase [22], it is likely that PKCα mainly participates in the DGKδ1-PH domain phosphorylation in these cells. Although co-immunoprecipitation experiments were performed, we failed to detect any appreciable association of PKCα with DGKδ1 (results not shown). This suggests that the phosphorylation of DGKδ1 at Ser-22 and Ser-26 by PKCα occurs transiently, and that the complex between the two proteins, if any, cannot be recovered under detergent lysis conditions.
The flanking sequence of Ser-22 (QNNS22FQR, where underlines indicate residues identical with the corresponding residues in the canonical phosphorylation site motif for PKCα) and that of Ser-26 (FQRS26KRR) partly fulfil the canonical phosphorylation site motif for PKCα {[RXXS*F(R/K)(R/K)], where the asterisk indicates the phosphorylation site} [29]. It is known that each PKC isoform has an isoform-specific recognition motif [29]. Because the flanking sequences of Ser-22 and Ser-26 show no agreement with the canonical phosphorylation site motif for PKCε [R(K/E/R)XS*XXX], these Ser residues are predicted to be poor substrates of PKCε. Indeed, in contrast with PKCα, overexpressed PKCε did not significantly affect the phosphorylation level of the DGKδ1-PH domain (Figure 3B).
DGKδ1 lacking the PH domain was considerably phosphorylated (Figure 1B). Moreover, full-length DGKδ1 having double alanine or aspartate mutations of Ser-22 and Ser-26 was also phosphorylated (50–80% of control) (results not shown). The results indicate that DGKδ1 has at least one additional phosphorylation site outside of the PH domain. Staurosporine, a non-selective PKC inhibitor, completely inhibited PMA-dependent phosphorylation of full-length DGKδ1 [22], whereas Gö 6976, the cPKC-selective inhibitor, only partly inhibited the phosphorylation (approx. 30% inhibition) (results not shown). Thus PMA-dependent, staurosporine-sensitive and Gö 6976-insensitive protein kinase, for example nPKC, may participate in the phosphorylation of the additional site(s) present in the C-terminal portion of DGKδ1. In this context, the action of this DGK isoform would be controlled by multiple protein kinases in a complex manner. The physiological importance of the phosphorylation outside of the PH domain is unclear. Since any mutations at Ser-22 and Ser-26 did not affect the oligomer formation of DGKδ1 (results not shown), the additional phosphorylation site(s) may be involved in the dissociation of oligomer structure [22].
NMR and crystal structures have now been reported for eight different PH domains [23,24]. By alignment of the amino acid sequence of the DGKδ1-PH domain with those of the eight PH domains, Ser-22 and Ser-26 are predicted to locate in the first variable loop between the first and second β-sheets (see Figure 2B). Binding targets of most of the PH domains are negatively charged molecules such as PtdIns(4,5)P2 and PtdIns-(3,4,5)P3. NMR studies showed that the interaction between the phosphoinositide and the PH domain is mediated by the positively charged surface of the domain that contains variable loops 1, 2 and 3 [23,24]. Although target molecule(s) of the DGKδ1-PH domain have not yet been identified, it is easily imagined that phosphorylation, introduction of negative charges, within the variable loop 1 would directly affect affinity for putative target molecule(s) of the enzyme. Alternatively, conformational changes caused by the phosphorylation may be more important for target molecule recognition by the PH domain.
We have provided results suggesting that the phosphorylation of one of Ser-22 and Ser-26 negatively regulates the translocation of DGKδ1 to the plasma membrane (Figures 6–8). It is puzzling to see that PMA, while causing the membrane translocation of DGKδ1, also suppresses the translocation through the enzyme phosphorylation by cPKC. In this context, it can be assumed that these discrepant actions of PMA operate in a segregated manner. The existence of negative regulation of the translocation implies that the intracellular localization of DGKδ1 is timely and strictly regulated by the balance between enhancement and suppression of the translocation. Because the PMA-dependent translocation of DGKδ1 was clearly detected (Figures 6–8), it is likely that only a part of the enzyme is phosphorylated by cPKC at the PH domain.
Unfortunately, the present study did not reveal positive regulatory mechanisms of the DGKδ1-translocation. However, we can conclude that the PH domain alone is sufficient for the PMA-induced translocation, and that the positive regulation of translocation is independent of phosphorylation in the DGKδ PH domain because the double alanine mutations at Ser-22 and Ser-26 and a Gö 6976 treatment, which gave a complete loss of phosphorylation signal (Figures 2 and 3), failed to inhibit the translocation of the PH domain (Figures 6 and 7). Moreover, the phosphorylation analysis using PKC inhibitors (Figure 6) allows us to speculate that a PMA-dependent, staurosporine-sensitive and Gö 6976-insensitive factor modifies a certain targeting molecule other than DGKδ1 itself that recruits the enzyme to the plasma membrane. Although nPKC fulfils these three criteria, it is unclear whether this enzyme is indeed involved in the positive regulation of DGKδ1-translocation. Further studies searching for the molecule(s) responsible for recruiting the DGKδ1-PH domain by using, for example, the yeast two-hybrid system will address the positive-regulatory mechanisms of the translocation.
Recently, Luo et al. [30,31] reported that PKCα interacted with and phosphorylated DGKζ (type IV isoform). These proteins co-localized in the cytoplasm in resting cells. However, after addition of PMA, PKCα translocated from the cytoplasm to the plasma membrane, whereas DGKζ remained in the cytoplasm. In contrast, our results demonstrated that DGKδ1 is translocated to the plasma membrane in response to PMA stimulation. These results suggest that although both DGKδ1 and DGKζ are commonly phosphorylated by PKCα, they are spatiotemporally segregated from each other in PMA-stimulated cells.
In conclusion, our results provide evidence of the phosphorylation of the DGKδ1 PH domain by cPKC in PMA- and EGF-stimulated cells and suggest that the translocation of DGKδ1 to the plasma membrane is negatively and critically regulated by the phosphorylation. These results imply that the action of DGKδ1 is adeptly regulated in a complex manner.
Acknowledgments
This work was performed through Special Coordination Funds (to H. K.) and was also supported in part by grants (to F. S. and H. K.) from the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government, the Japan Foundation of Applied Enzymology (H. K.) and the Sapporo Medical University Foundation for Promotion of Medical Science (S.-i. I.).
References
- 1.Hodgkin M. N., Pettitt T. R., Martin A., Michell R. H., Pemberton A. J., Wakelam M. J. O. Diacylglycerols and phosphatidates: which molecular species are intracellular messengers? Trends Biochem. Sci. 1998;23:200–204. doi: 10.1016/s0968-0004(98)01200-6. [DOI] [PubMed] [Google Scholar]
- 2.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 3.Kanoh H., Yamada K., Sakane F. Diacylglycerol kinase: a key modulator of signal transduction? Trends Biochem. Sci. 1990;15:47–50. doi: 10.1016/0968-0004(90)90172-8. [DOI] [PubMed] [Google Scholar]
- 4.Hurley J. H., Newton A. C., Parker P. J., Blumberg P. M., Nishizuka Y. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ron D., Kazanietz M. G. New insights into the regulation of protein kinase C and novel phorbol ester receptors. FASEB J. 1999;13:1658–1676. [PubMed] [Google Scholar]
- 6.Kazanietz M. G. Novel ‘nonkinase’ phorbol ester receptors: the C1 domain connection. Mol. Pharmacol. 2002;61:759–767. doi: 10.1124/mol.61.4.759. [DOI] [PubMed] [Google Scholar]
- 7.Ohno S., Nishizuka Y. Protein kinase C isotypes and their specific functions: prologue. J. Biochem. (Tokyo) 2002;132:509–511. doi: 10.1093/oxfordjournals.jbchem.a003249. [DOI] [PubMed] [Google Scholar]
- 8.Exton J. H. Phosphatidylcholine breakdown and signal transduction. Biochim. Biophys. Acta. 1994;1212:26–42. doi: 10.1016/0005-2760(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 9.English D. Phosphatidic acid: a lipid messenger involved in intracellular and extracellular signalling. Cell. Signalling. 1996;8:341–347. doi: 10.1016/0898-6568(95)00076-3. [DOI] [PubMed] [Google Scholar]
- 10.Sakane F., Kanoh H. Molecules in focus: diacylglycerol kinase. Int. J. Biochem. Cell Biol. 1997;29:1139–1143. doi: 10.1016/s1357-2725(97)00037-x. [DOI] [PubMed] [Google Scholar]
- 11.Topham M. K., Prescott S. M. Mammalian diacylglycerol kinases, a family of lipid kinases with signaling functions. J. Biol. Chem. 1999;274:11447–11450. doi: 10.1074/jbc.274.17.11447. [DOI] [PubMed] [Google Scholar]
- 12.van Blitterswijk W. J., Houssa B. Properties and functions of diacylglycerol kinases. Cell. Signalling. 2000;12:595–605. doi: 10.1016/s0898-6568(00)00113-3. [DOI] [PubMed] [Google Scholar]
- 13.Kanoh H., Yamada K., Sakane F. Diacylglycerol kinases: emerging downstream regulators in cell signaling systems. J. Biochem. (Tokyo) 2002;131:629–633. doi: 10.1093/oxfordjournals.jbchem.a003144. [DOI] [PubMed] [Google Scholar]
- 14.Kai M., Sakane F., Imai S., Wada I., Kanoh H. Molecular cloning of a diacylglycerol kinase isozyme predominantly expressed in human retina with a truncated and inactive enzyme expression in most other human cells. J. Biol. Chem. 1994;269:18492–18498. [PubMed] [Google Scholar]
- 15.Ding L., Bunting M., Topham M. K., McIntyre T. M., Zimmerman G. A., Prescott S. M. Alternative splicing of the human diacylglycerol kinase ζ gene in muscle. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5519–5524. doi: 10.1073/pnas.94.11.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caricasole A., Bettini E., Sala C., Roncarati R., Kobayashi N., Caldara F., Goto K., Terstappen G. C. Molecular cloning and characterization of the human diacylglycerol kinase β (DGKβ) gene: alternative splicing generates DGKβ isotypes with different properties. J. Biol. Chem. 2002;277:4790–4796. doi: 10.1074/jbc.M110249200. [DOI] [PubMed] [Google Scholar]
- 17.Sakane F., Imai S., Yamada K., Murakami T., Tsushima S., Kanoh H. Alternative splicing of the human diacylglycerol kinase δ gene generates two isoforms differing in their expression patterns and in regulatory functions. J. Biol. Chem. 2002;277:43519–43526. doi: 10.1074/jbc.M206895200. [DOI] [PubMed] [Google Scholar]
- 18.Murakami T., Sakane F., Imai S. I., Houkin K., Kanoh H. Identification and characterization of two splice variants of human diacylglycerol kinase η. J. Biol. Chem. 2003;278:34364–34372. doi: 10.1074/jbc.M301542200. [DOI] [PubMed] [Google Scholar]
- 19.Sakane F., Imai S., Kai M., Wada I., Kanoh H. Molecular cloning of a novel diacylglycerol kinase isozyme with a pleckstrin homology domain and a C-terminal tail similar to those of the EPH family of protein tyrosine kinase. J. Biol. Chem. 1996;271:8394–8401. doi: 10.1074/jbc.271.14.8394. [DOI] [PubMed] [Google Scholar]
- 20.Ponting C. P. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 1995;4:1928–1930. doi: 10.1002/pro.5560040927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultz J., Ponting C. P., Hofmann K., Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997;6:249–253. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai S., Sakane F., Kanoh H. Phorbol ester-regulated oligomerization of diacylglycerol kinase δ linked to its phosphorylation and translocation. J. Biol. Chem. 2002;277:35323–35332. doi: 10.1074/jbc.M202035200. [DOI] [PubMed] [Google Scholar]
- 23.Lemmon M. A., Ferguson K. M. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 24.Maffucci T., Falasca M. Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism. FEBS Lett. 2001;506:173–179. doi: 10.1016/s0014-5793(01)02909-x. [DOI] [PubMed] [Google Scholar]
- 25.Newton A. C. Protein kinase C: structure, function, and regulation. J. Biol. Chem. 1995;270:28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 26.Morrison P., Saltiel A. R., Rosner M. R. Role of mitogen-activated protein kinase kinase in regulation of the epidermal growth factor receptor by protein kinase C. J. Biol. Chem. 1996;271:12891–12896. doi: 10.1074/jbc.271.22.12891. [DOI] [PubMed] [Google Scholar]
- 27.Marais R., Light Y., Mason C., Paterson H., Olson M. F., Marshall C. J. Requirement of Ras–GTP–Raf complexes for activation of Raf-1 by protein kinase C. Science. 1998;280:109–112. doi: 10.1126/science.280.5360.109. [DOI] [PubMed] [Google Scholar]
- 28.Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 29.Nishikawa K., Toker A., Johannes F. J., Songyang Z., Cantley L. C. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 30.Luo B., Prescott S. M., Topham M. K. Protein kinase Cα phosphorylates and negatively regulates diacylglycerol kinase ζ. J. Biol. Chem. 2003;278:39542–39547. doi: 10.1074/jbc.M307153200. [DOI] [PubMed] [Google Scholar]
- 31.Luo B., Prescott S. M., Topham M. K. Association of diacylglycerol kinase ζ with protein kinase Cα: spatial regulation of diacylglycerol signaling. J. Cell Biol. 2003;160:929–937. doi: 10.1083/jcb.200208120. [DOI] [PMC free article] [PubMed] [Google Scholar]