Abstract
The human HO-1 (haem oxygenase-1) gene encodes a microsomal enzyme responsible for the breakdown of haem, and is also cytoprotective in response to various cellular insults. HO-1 transcription is induced by a vast array of compounds including, but certainly not limited to, haem and heavy metals such as cadmium. In the present study, we show that upstream stimulatory factors, USF1 and USF2, ubiquitous proteins belonging to the basic helix–loop–helix-leucine zipper family of transcription factors, constitutively bind to the class B E-box located in the proximal promoter of the human HO-1 gene and are responsible for the enhancement of HO-1 gene transcription in human renal proximal tubular epithelial cells. Dimethylsulphate in vivo footprinting studies have identified three protected guanine residues in the E-box of the HO-1 proximal promoter. One of these guanine contact points is essential for USF binding, and when mutated mimics a deletion mutation of the entire E-box palindrome sequence encompassing all three guanine contact points. Binding of USF1 and USF2 to the HO-1 E-box was confirmed by chromatin immunoprecipitation and gel-shift assays. Furthermore, we show that overexpression of USF1 or USF2 enhances the basal expression of HO-1 and that expression of a USF dominant negative form reduces its expression. These results demonstrate for the first time that USF proteins bind to the human HO-1 promoter in vivo and are required for high-level expression of HO-1 by haem and cadmium in human renal epithelial cells.
Keywords: chromatin immunoprecipitation, E-box, haem oxygenase, in vivo footprinting, renal proximal tubular cells, upstream stimulatory factor
Abbreviations: AP-1, activator protein-1; bHLH-Zip, basic helix–loop–helix-leucine zipper; ChIP, chromatin immunoprecipitation; DMS, dimethylsulphate; EMSA, electrophoretic mobility-shift assay; FBS, foetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HA, haemagglutinin; hGH, human growth hormone; HO-1, haem oxygenase-1; HPTC, human renal proximal tubule cell; LMPCR, ligation-mediated PCR; RT, reverse transcriptase; USF, upstream stimulatory factor
INTRODUCTION
HO-1 (haem oxygenase-1) is a 32 kDa microsomal enzyme that catalyses the rate-limiting step in the degradation of haem. The enzymic reaction results in the opening of the haem ring, with the liberation of equimolar amounts of iron, carbon monoxide and biliverdin. Two isoforms of HO have been characterized: HO-1, an inducible form, and HO-2, a constitutive isoform [1]. A putative isoenzyme, HO-3, isolated from rat brain and sharing approx. 90% homology with HO-2, has also been described [2]. HO-1 is highly inducible by numerous stress stimuli including haem, heavy metals, growth factors, H2O2, UV light, oxidized lipids, shear stress, nitric oxide, glucose deprivation, angiotensin II, as well as others (reviewed in [3]). The induction of HO-1 occurs as an adaptive and beneficial response to these otherwise injurious stimuli through the action of one or more of the products of the HO-1 reaction [4]. Previous studies have demonstrated that the induction of HO-1 by chemical inducers or selective overexpression is protective in vitro and in vivo in several models of tissue injury [5–10]. The importance of HO-1 expression has been further confirmed by studies in HO-1 knockout mice and in a patient with HO-1 deficiency, both of which exhibit a proinflammatory state and increased susceptibility to oxidant injury [8,10–12]. Recent studies have demonstrated the biological relevance of HO-1 gene expression in a variety of diseases including atherosclerosis, transplant rejection, lung injury, renal failure, sepsis, vascular restenosis, as well as others [7].
The molecular regulation of HO-1 by most stimuli is controlled at the transcriptional level and is species- and cell-specific [3]. Previous studies have reported the presence of both positive and negative regulatory sequences in the human HO-1 promoter [13]. Consensus-binding sites for nuclear factor κB, AP-1 (activator protein-1), AP-2, Sp1 (stimulating protein-1), USF (upstream stimulatory factor), c-Myc/Max and interleukin-6 response elements, as well as other transcription factors, have been reported in the promoter region of the human HO-1 gene [14–18]. Most of these results were derived from promoter-deletion analyses in transient transfection studies, in vitro DNase I footprinting or computer-based consensus-sequence predictions. In our efforts to identify functionally relevant protein–DNA interactions in an intact cell at a single nucleotide resolution, we performed in vivo footprinting using DMS (dimethylsulphate) coupled with LMPCR (ligation-mediated PCR) and found protected guanine nucleotides located at −39 to −44 bp in the proximal promoter of the human HO-1 gene in a region that corresponds to an E-box (CACGTG). This core sequence is known to be bound by several proteins including USFs [19–24]. USF proteins belong to the class of bHLH-Zip (basic helix–loop–helix-leucine zipper) transcription factors, which include nuclear mammalian proteins such as c-Myc, Max, Mad, MxiI and others [25]. In mammalian cells, two ubiquitously expressed genes, USF1 and USF2, have been well characterized and have pleiotropic effects in cells and tissues [22,23]. USFs have been implicated in the regulation of several genes including cyclin B1 [26], cathepsin B [27], L-pyruvate kinase [28], inducible nitric oxide synthase [29], polymeric immunoglobulin receptor [30], insulin-like growth factor-2 receptor [31], breast-cancer susceptibility gene 2 [32], as well as others [33,34].
The purpose of the present study was to identify sites of protein–DNA interaction in the human HO-1 promoter in the context of an intact cell. Using in vivo footprinting, ChIP (chromatin immunoprecipitation) and gel-shift assays, we have identified that both USF1 and USF2 bind to an E-box sequence located in the HO-1 proximal promoter and are required for basal and maximal gene activation.
EXPERIMENTAL
Reagents
Tissue culture media, serum and supplements were obtained from Invitrogen (Carlsbad, CA, U.S.A.). Haemin (iron protoporphyrin chloride), CdCl2 (cadmium chloride) and DMS were obtained from Sigma (St. Louis, MO, U.S.A.). Rabbit polyclonal antibodies against USF1, USF2, c-Myc, Mad and Max were from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). Restriction endonucleases and reagents for PCR, including synthetic oligonucleotides, were obtained from New England Biolabs (Beverly, MA, U.S.A.) and Invitrogen respectively.
Cell culture, plasmids and transfection
HK-2 cells (A.T.C.C., Manassas, VA, U.S.A.), an immortalized human proximal tubule epithelial cell line from normal adult kidney [35], were grown in 10 cm collagen-coated tissue culture plates in keratinocyte serum-free medium supplemented with 5 ng/ml recombinant epidermal growth factor and 40 μg/ml bovine pituitary extract (Invitrogen). Primary cultures of human renal proximal tubule cells (HPTCs; Clonetics, Walkersville, MD, U.S.A.) were grown in renal epithelial basal medium supplemented with FBS (foetal bovine serum) (5%), insulin (5 μg/ml), transferrin (10 μg/ml), tri-iodothyronine (6.5 ng/ml), cortisol (0.5 μg/ml), adrenaline (epinephrine; 0.5 μg/ml) and human epidermal growth factor (10 ng/ml), at 37 °C under 5% CO2 [10]. For HPTCs, media was changed to 0.5% FBS before stimulation.
The plasmid constructs, pHOGH/4.5 and pHOGL3/4.5, containing a −4.5 kb fragment of the 5′-flanking region of the human HO-1 promoter including the transcription start site, have been described previously [36,37]. Plasmid constructs for USF1 (psvUSF1), USF2 (psvUSF2) and pRc/CMV566 were gifts from Dr M. Sawadogo (University of Texas at Houston, Houston, TX, U.S.A.). The dominant-negative expression vector for USF, HA (haemagglutinin)-tagged A-USF, was kindly provided by Dr C. Vinson (National Cancer Institute, National Institutes of Health, Bethesda, MD, U.S.A.).
HK-2 cells at a confluence of approx. 85% were transiently transfected using LIPOFECTAMINE™ 2000 (Invitrogen) and equimolar amounts of plasmid DNA using a batch transfection method [36,37]. Transfections were performed for 2 h in the above media, and after a 4–6 h recovery, cells were split into three individual 10 cm collagen-coated plates and allowed to recover for 24 h. Cells were induced with haemin (5 μM), CdCl2 (10 μM) or vehicle (0.5% DMSO), and RNA was isolated for Northern-blot analysis. In cells transfected with pHOGL3/4.5, reporter activity was measured by the luciferase assay as described previously [36]. In the luciferase experiments, equimolar amounts of A-USF or empty vector were co-transfected with pHOGL3/4.5 (1 μg) for 2 h, allowed to recover for 5 h and subsequently split into 12-well dishes. Cells were then induced with vehicle, CdCl2 or haemin for 18 h.
For transfections with USF1 and USF2 overexpression vectors, cells were grown on 10 cm dishes and co-transfected for 2 h with pHOGH/4.5 (4 μg) and equimolar amounts of empty vector, psvUSF1 or psvUSF2. Cells were allowed to recover for 5 h, and each plate of transfected cells was split into two 10 cm plates. The cells were then induced for 18 h with either vehicle (DMSO) or haemin (5 μM), and total RNA was extracted as described below. For the dominant-negative experiments, equimolar amounts of A-USF or empty vector were co-transfected with pHOGH/4.5 in 10 cm dishes for 6 h, allowed to recover overnight and induced with vehicle, CdCl2 or haemin for 12 h.
In vivo footprinting and LMPCR
Cells were grown to approx. 80% confluence on 15 cm plates and treated with haemin (5 μM), CdCl2 (10 μM) or DMSO for 2 h. Cells were washed once with room temperature (22 °C) PBS, followed by DMS (0.5% in PBS) treatment for 30 s with gentle agitation. In vivo footprinting and LMPCR were performed as described previously [38] with the exception that after the SDS/proteinase K treatment, one-third volume of saturated NaCl was added. The samples were centrifuged at 2250 g for 15 min and 2.5 vol. of cold 95% ethanol was added to the supernatant. DNA was collected by spooling and transferred into a tube containing 70% ethanol. Samples were centrifuged at 10000 g for 5 min, dried and finally resuspended in water. The primers used for in vivo footprinting and LMPCR are listed in Table 1.
Table 1. Sequences of oligonucleotide (oligo) primers.
| Primer type | Sequence (all listed as 5′→3′) |
|---|---|
| In vivo footprinting | |
| Linker adapter (equimolar amounts) | GCGGTGACCCGGGAGATCTGAATTC (also used in LMPCR) and GAATTCAGATC |
| For the non-transcribed strand | |
| Extension oligo | CAAGCAGTCAGCAGAGGATT |
| PCR and hybridization oligo | TGGCCAGACTTTGTTTCCCAAGGGTCA |
| For the transcribed strand | |
| Extension oligo | GTAAATTACCGTTCCTCCCT |
| PCR and hybridization oligo | TCGGGTTGCGGACGCTCCA |
| EMSAs | |
| Forward | CTGTTCCGCCTGGCCCACGTGACCCGCCGAGCATAA |
| Reverse | TTATGCTCGGCGGGTCACGTGGGCCAGGCGGAACAG |
| Site-directed mutagenesis | |
| pHOGH/4.5 E-box M1 | |
| M1 forward | CTGTTCCGCCTGGCCCACGTGAtCCGCCGAGCATAA |
| M1 reverse | TTATGCTCGGCGGaTCACGTGGGCCAGGCGGAACAG |
| pHOGH/4.5 E-box M2 | |
| M2 forward | CTGTTCCGCCTGGCCCAtGTGACCCGCCGAGCATAA |
| M2 reverse | TTATGCTCGGCGGGTCACaTGGGCCAGGCGGAACAG |
| pHOGH/4.5 E-box M3 | |
| M3 forward | CTGTTCCGCCTGGCCtACGTGACCCGCCGAGCATAA |
| M3 reverse | TTATGCTCGGCGGGTCACGTaGGCCAGGCGGAACAG |
| pHOGH/4.5 Δ E-box | |
| Del E-box F | CATCAGCTGTTCCGCCTGGCCGCCGAGCATAAATGTG |
| Del E-box R | CACATTTATGCTCGGCGGCCAGGCGGAACAGCTGATG |
| ChIP | |
| For the proximal E-box | |
| Forward | TTGCAACGCCCGGCCAGAAA |
| Reverse | TCGGGTTGCGGACGCTCCA |
| Nested hybridization oligo | ATAAATGTGACCGGCCGCGGCTC |
| For the internal E-box | |
| Forward | GATGACACTGAGGCTCAGA |
| Reverse | GCTGAAATCGGTCTTGGTTGATTTCAGCC |
| Nested hybridization oligo | GGTCACACAGTAAGTTCAGCCTGCTCT |
EMSA (electrophoretic mobility-shift assay)
Nuclear extracts were collected using the Pierce NE-PER kit (Rockford, IL, U.S.A.). Vehicle-treated and haemin-induced nuclear extracts (4 μg) were incubated in 15 μl of 1× binding buffer [4% (v/v) glycerol, 1 M MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 10 mM Tris (pH 7.5) and 50 μg/ml poly(dI-dC)·(dI-dC)] at room temperature for 20 min with 0.1 pmol of a 32P-labelled probe (approx. 35000 c.p.m.). The annealed probe consisted of equimolar amounts of the forward and reverse oligonucleotides (36 bp) that span the E-box sequence and are listed in Table 1. Competition was performed with 20-fold molar excess of unlabelled oligonucleotide added before the addition of labelled probe. Supershifts included 1 μg of anti-USF1 (Santa Cruz Biotechnology, sc-8983 or sc-229) or 1 μg of anti-USF2 (Santa Cruz Biotechnology, sc-862) incubated at room temperature for 15 min before the addition of labelled probe. Reactions were electrophoresed on a 5% (w/v) non-denaturing polyacrylamide gel in 0.5× TBE (Tris/borate/EDTA). Gels were fixed, dried and exposed to film.
Site-directed mutagenesis
The plasmid pHOGH/4.5 was used as the parental clone, and mutations in the E-box sequence were generated with the Quik Change XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) according to the manufacturer's instructions. The oligonucleotides used for the generation of mutated constructs are listed in Table 1. M1 refers to the first protected G residue as seen on the in vivo footprinting. Similarly, M2 and M3 refer to protected G residues labelled 2 and 3 based on the in vivo footprinting (Figure 1). These three protected G residues were changed to A residues for the mutagenesis experiments. Del E-box is a deletion from the first protected G up to and including the third protected G. All the mutations were verified by sequencing.
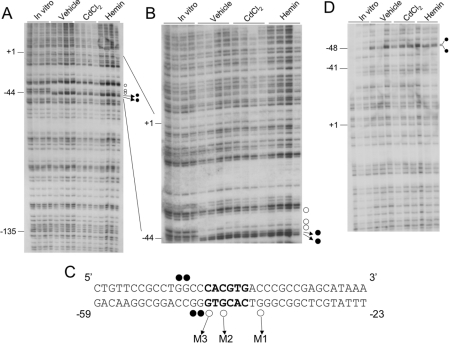
Figure 1. In vivo DMS footprinting of the human HO-1 proximal promoter in HK-2 cells.
(A) The transcribed strand of the human HO-1 promoter. Each lane is derived from an individual plate of HK-2 cells treated with vehicle (DMSO), CdCl2 (cadmium chloride, 10 μM) or haemin (5 μM) for 2 h. In vitro samples are derived from DNA purified from uninduced cells and then subsequently treated as described in the Experimental section. ○, Protected guanine residues; •, enhanced guanine residues. −135, −44 and +1 represent nucleotide positions relative to the transcription start site of the human HO-1 gene. (B) The same samples shown in (A) were electrophoresed for a longer run to visualize the E-box and distal sequences more closely. (C) The sequence of the human HO-1 proximal E-box (in bold) and flanking nucleotides. The guanine residues protected and enhanced in (A, B) are shown. The three protected guanine residues were assigned as M1, M2 and M3 and represent the sites for mutagenesis. (D) In vivo DMS footprinting of the non-transcribed strand of HO-1. •, Enhanced guanine residues at positions −47 and −48 bp as depicted in (C).
ChIP
ChIP was performed essentially as described in [39] with some modifications. HK-2 cells or HPTCs were grown to approx. 80% confluence on 15 cm plates and induced with haemin (5 μM), CdCl2 (10 μM) or vehicle (DMSO) for 2 h. Anti-USF1 (1 μg; sc-8983), anti-USF2 (sc-862), anti-c-Myc (sc-42), anti-Mad (sc-222) or anti-Max (sc-8011) was added and the samples rocked at 4 °C for 4 h. A no-antibody control was also included. PCR products were electrophoresed in a 1.8% agarose gel and transferred on to a Hybond XL membrane. The blots were then hybridized with a 32P-end-labelled nested oligonucleotide probe. The primers used for the ChIP assay are listed in Table 1.
RT (reverse transcriptase)–PCR
Total RNA (1 μg) derived from vehicle- and haemin-induced HK-2 cells were reverse-transcribed with oligo(dT) using a firststrand synthesis kit (Invitrogen). The cDNA was then diluted, and approx. 5 ng of this cDNA was used for PCR of USF2. PCR products were electrophoresed and visualized with ethidium bromide staining. The primers used for amplification of USF2 have been published previously [40].
RNA purification and Northern-blot hybridization
After transfection and induction, RNA was isolated as described previously [41]. A cDNA probe for hGH (human growth hormone) was radiolabelled with [α-32P]dATP using a random primer labelling kit, according to the manufacturer's instructions (Invitrogen) and used in the hybridization. Membranes were stripped and reprobed with cDNA probes for human HO-1 and human GAPDH (glyceraldehyde-3-phosphate dehydrogenase). To determine the expression levels quantitively, autoradiographs were scanned on an Hewlett-Packard ScanJet 4C using Adobe Photoshop software, and densitometry was performed using NIH Image 1.63 software. Expression of hGH/HO-1 was normalized to the GAPDH message and expressed as percentage of maximal expression. All experiments were repeated with at least 2–3 independent RNA preparations to show reproducibility.
Immunoblot and immunocytochemical analysis
For immunoblots, cells were split from a transfection described above and allowed to recover overnight. Cells were washed twice with PBS and lysed in Laemmli buffer. Samples were separated on a 10% (w/v) SDS/polyacrylamide gel and then transferred on to a Hybond C extra membrane. The membranes were incubated overnight at 4 °C with a 1:400 dilution of anti-USF1 or anti-USF2 antibodies in PBS containing 0.5% Tween 20 and 7% non-fat dry milk. After washing in PBS containing 0.5% Tween 20, filters were incubated in the same buffer containing a 1:10000 dilution of peroxidase-conjugated goat anti-rabbit IgG antibody (Jackson Immunoresearch Laboratories, West Grove, PA, U.S.A.) for 1 h at room temperature. Horseradish peroxidase activity was detected using ECL® according to the manufacturer's instructions (Amersham Biosciences, Piscataway, NJ, U.S.A.). Membranes were stripped and probed with an anti-actin antibody (1:1000 dilution) (Sigma) to confirm loading and transfer.
For immunofluorescence, HK-2 cells were plated on collagen-coated slides and transfected with empty vector or the HA-tagged A-USF construct. Cells were induced with haemin (5 μM) for 4 h, a time point at which HO-1 protein is induced (approx. 5–6-fold) in these cells by Western-blot analysis. Cells were fixed in cold methanol and washed in PBS. Dual-label immunocytochemistry was performed using Alexa 594-conjugated anti-HA (Molecular Probes) and anti-HO-1 (Stressgen Biotechnologies, Victoria, BC, Canada) antibodies for detection of A-USF and HO-1 respectively. HO-1 was detected using a goat anti-rabbit FITC conjugated secondary antibody (Jackson Immunoresearch Laboratories).
Statistical analysis
Results are expressed as means±S.E.M. For the luciferase data, analyses were performed using ANOVA and the Student–Newman–Keuls test. All results are considered significant at P<0.05.
RESULTS
In vivo DMS footprinting of the human HO-1 proximal promoter
The human HO-1 proximal promoter is GC-rich and contains computer-identified consensus DNA-binding sequences for nuclear factor κB, AP-1, Sp1 and AP-2. Using in vitro DNase I footprinting, it has been previously suggested that these consensus sequences are bound by their respective proteins [16]. Since limited information was available about the in vivo architecture of the human HO-1 gene and its transcriptional regulation, we performed in vivo footprinting on specific regions of the HO-1 promoter, particularly the proximal promoter. DMS has a preference for methylating guanine residues and to a lesser extent adenine residues [42]. One advantage of using DMS footprinting is that it is capable of detecting specific protein–DNA interaction sites in vivo in an intact cell.
As shown in Figures 1(A) and 1(B), we observed three protected guanine residues and two other enhanced guanine residues (G's) in the proximal promoter when compared with the band intensity apparent in the naked in vitro DMS-treated DNA samples. The DNA sequence surrounding this area on the transcribed (bottom) strand is 3′-GGGTGCACTG-5′ (Figure 1C). Within this sequence is a class B E-box motif (5′-CACGTG-3′) known to bind transcription factors belonging to the b-HLH-Zip family. Two of these contacts, denoted as M3 and M2, are located within the CACGTG palindrome sequence corresponding to −44 and −42 bp respectively from the transcription start site of the human HO-1 gene. Interestingly, the third contact, denoted as M1, is 2 bp outside of the 6 bp consensus E-box sequence and is located at −37 bp (Figure 1C). The findings of enhanced G residues immediately adjacent to a DMS footprint are consistent with similar observations in other genes such as manganese superoxide dismutase that have been reported previously [43]. Furthermore, the three footprints observed in Figures 1(A) and 1(B) are present in both haemin- and cadmium-treated cells as well as in the vehicle-treated control samples, suggesting that the factor(s) interacting with the HO-1 proximal E-box sequence are constitutively bound to the promoter DNA in vivo.
We also investigated the non-transcribed (top) strand of the human HO-1 proximal promoter corresponding to this region. As shown in Figure 1(D), we did not observe any footprints in this region either 5′ or 3′ to the E-box sequence. This suggests that most, if not all, of the DNA-binding proteins interact predominantly with the transcribed strand of the human HO-1 gene. However, at least two G residues were enhanced at positions −47 and −48 bp of the non-transcribed strand. Further examination of the transcribed strand by in vivo footprinting demonstrated no other enhanced or protected sites either proximal or distal to the E-box sequence extending beyond the transcription start site. These findings demonstrate that the proximal promoter of the human HO-1 gene is bound by transcription factors at the E-box motif and this is the only detectable sequence bound by proteins in vivo.
USF1 and USF2 bind to the human HO-1 E-box in vitro
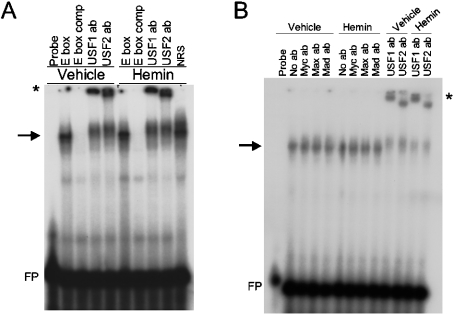
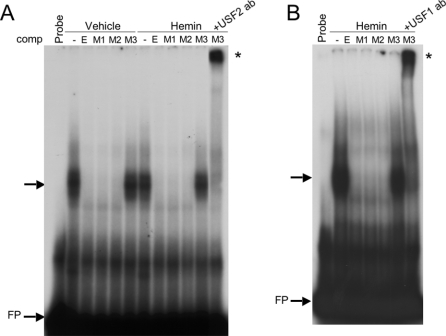
To identify the specific transcription factors that are bound to the canonical E-box in the human HO-1 proximal promoter, we performed EMSA using a 36 bp end-labelled double-stranded DNA probe containing the HO-1 E-box. As shown in Figure 2(A), a shifted complex was observed in vehicle-treated as well as in haemin-induced nuclear extracts. These two complexes appeared to be identical and could be competed out using 20-fold molar excess of unlabelled probe (Figure 2A, lanes 3 and 7 from the left). Antibodies against human USF1 and USF2 were both capable of supershifting this complex (Figure 2A, lanes 4, 5, 8 and 9 from the left). Similar results were also observed in nuclear extracts treated with cadmium (results not shown). To test the specificity of the protein–DNA interactions, we also performed EMSA using antibodies against the other members of the b-HLH-Zip family of transcription factors, including c-Myc, Mad and Max. As shown in Figure 2(B), these antibodies failed to supershift the protein–DNA complex, in contrast with the supershift observed for both USF1 and USF2.
Figure 2. USF1 and USF2 bind to the HO-1 E-box in vitro by EMSA.
(A) Nuclear extracts from uninduced (vehicle) and haemin (5 μM)-induced HK-2 cells were incubated with a labelled probe containing the HO-1 E-box and surrounding nucleotides (see Table 1 for the probe sequence). USF1 and USF2 polyclonal antibodies were added in samples shown for supershift analysis before the addition of a labelled probe. NRS (normal rabbit serum) is shown as an IgG control. (B) EMSA using the same HO-1 E-box probe as shown in (A) with uninduced and haemin-treated nuclear extracts and anti-c-Myc, -Mad and -Max antibodies. USF1 and USF2 supershifts in (A, B) are indicated by an *. FP, free probe.
USF1 and USF2 are associated with the human HO-1 proximal promoter E-box in vivo
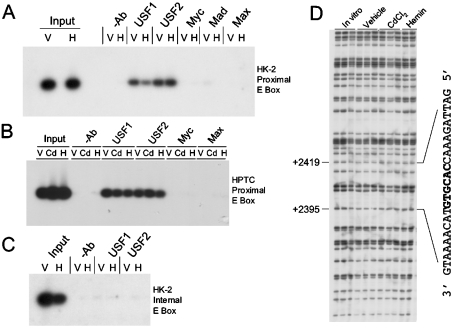
To confirm the presence of USF1 and USF2 binding to the human HO-1 proximal promoter in vivo, we evaluated uninduced and haemin-induced HK-2 cells by ChIP. Figure 3(A) shows that both USF1- and USF2-specific antibodies pull down protein–DNA complexes containing the human HO-1 E-box, indicating that USF1 and USF2 are constitutively associated with the E-box region. Consistent with the EMSA results in Figure 2, we observed no substantial signal in our ChIP studies when using antibodies directed against c-Myc, Max or Mad (Figure 3A). As a further confirmation of the results observed with the ChIP assays in the transformed HK-2 cell line, we also performed similar studies in primary HPTCs. As shown in Figure 3(B), both USF1 and USF2 also associate with DNA containing the HO-1 proximal E-box in the uninduced, cadmium- or haemin-induced cells. Consistent with the results obtained in HK-2 cells, c-Myc or Max antibodies failed to immunoprecipitate this region of the DNA in HPTCs.
Figure 3. USF1 and USF2 bind to the proximal HO-1 E-box in vivo, but not to an internal E-box.
(A) ChIP of the HO-1 proximal E-box of HK-2 cells induced with haemin or vehicle. PCR for all ChIP assays was performed on immunoprecipitated samples, electrophoresed, transferred on to a nylon membrane and subsequently hybridized with a nested oligonucleotide as described in the Experimental section. (B) ChIP of primary HPTCs treated with haemin (H, 5 μM), cadmium chloride (Cd, 10 μM) or vehicle (V). (C) ChIP performed on the human HO-1 internal E-box located in intron 2. (D) DMS in vivo footprinting of the HO-1 internal E-box-transcribed strand. The E-box and flanking sequences are illustrated.
Furthermore, to verify the specificity of the ChIP assay and to exclude any PCR-generated artifacts, we performed these studies on a canonical E-box sequence located in intron 2 of the human HO-1 gene 2.4 kb downstream of the transcription start site. Previous studies have suggested that USFs can bind to downstream internal E-box sequences and regulate gene transcription, e.g. the human β-globin gene [44]. As shown in Figure 3(C), no signal was obtained using the same USF1- or USF2-immunoprecipitated complexes when PCR was performed using primers bracketing this internal E-box sequence. These results demonstrate the specificity of both USF1 and USF2 to the E-box located in the human HO-1 promoter. These results were also confirmed by the absence of any protected footprints or enhancements detected by in vivo footprinting performed on the transcribed strand of this internal E-box (Figure 3D).
USF2b is the predominant form expressed in HK-2 cells
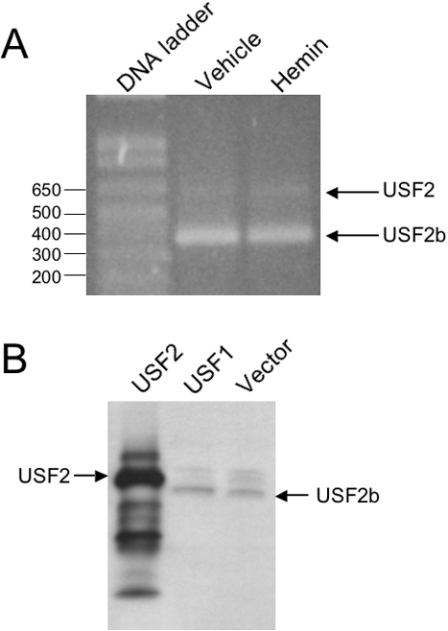
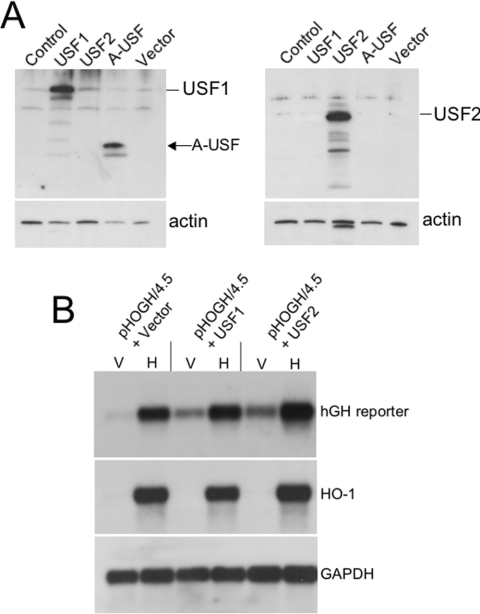
USF2 has been shown to exist in at least two forms, a 44 kDa protein originally identified in HeLa cell extracts [19,23] and a smaller form (approx. 38 kDa) derived from alternative splicing of the fourth exon of USF2 [40]. This latter isoform has been referred to as USF2b or USF2Δ4. Viollet et al. [40] have reported that various human cell lines express different amounts of each USF2 isoform. To determine the predominant isoform of USF2 in HK-2 cells, we performed RT–PCR on RNA derived from vehicle- and haemin-induced cells. Using primers previously described [40] that bracket the fourth exon of the human USF2 sequence, we confirmed that the smaller USF2b PCR product (382 bp) was obtained predominantly over the full-length (containing exon 4) USF2 form (583 bp) (Figure 4).
Figure 4. USF2b is the predominant form of USF2 expressed in HK-2 cells.
(A) Total RNA was purified from vehicle or haemin-treated HK-2 cells, then reverse-transcribed and used as template for PCR. Primers used (see the Experimental section) flank the fourth exon of human USF2. PCR products lacking the fourth exon (USF2b) and containing the fourth exon (USF2) are shown. Sizes of the DNA ladder are indicated in bp. (B) Western-blot analysis of HK-2 cells transiently transfected with USF2, USF1 or empty vector. Anti-USF2 antibody confirms USF2 overexpression (mass, approx. 44 kDa; lane 1). Endogenous USF2- (marked as USF2b) in the USF1-(lane 2) and empty vector- (lane 3) transfected cells is recognized as a lower-molecular-mass (approx. 38 kDa) protein.
Taken together, the DMS in vivo footprinting, ChIP and EMSA results shown so far strongly suggest that the human HO-1 E-box in proximal tubule cells is bound constitutively by USF1 and USF2b in vivo, and to a lesser extent by the USF1 and USF2 heterodimer.
DMS-protected guanine residue at −44 of the human HO-1 promoter is essential for USF binding and is required for enhanced transcription
To investigate the significance of the protected guanine bases (M1, M2 and M3) in the HO-1 E-box (Figure 1C), we performed EMSA competition studies using specific mutations of each of these guanine residues changed to an adenine base. We chose to examine these bases (M1, M2 and M3) rather than the enhanced guanine residues because they represent direct protein–DNA contact points. Complementary oligonucleotides containing these individual mutations were synthesized, annealed and used as unlabelled competitors in EMSAs using the same end-labelled E-box probe utilized in Figure 2. As shown in Figure 5, a 20-fold molar excess of the unlabelled M1 or M2 mutant was capable of competing out the shift of the wild-type E-box probe in both vehicle- and haemin-treated nuclear extracts as efficiently as the unlabelled wild-type E-box oligonucleotide (Figure 5, lanes 3–5 and 8–10). The results indicate that these two guanine residues (M1 and M2) located at −42 and −37 of the HO-1 promoter are not required for efficient USF binding to the E-box in vitro. The M3 competitor, however, was not capable of competing out the USF-E-box shift, indicating that the guanine at −44 of the HO-1 promoter is essential for efficient USF binding to the HO-1 E-box in vitro. Furthermore, closer examination of these results showed that the protein–DNA complex shift was actually comprised of two bands, a more intense faster migrating band and a lower intensity, slower migrating band. Given our RT–PCR results (Figure 4) showing that the smaller USF2b is the more abundant USF2 isoform in HK-2 cells, it is possible that the faster migrating protein–DNA complex may contain the alternatively spliced variant, USF2b, whereas the slower migrating complex contains USF2.
Figure 5. Importance of the guanine at −44 bp for DNA–protein binding.
(A) EMSA using the HO-1 proximal E-box with either vehicle- or haemin (5 μM)-treated nuclear extracts. Shifted DNA–protein complexes are indicated by an arrow, supershifted complexes are indicated with *. Unlabelled competitors are: E, E-box; M1, M2 and M3 represent G→A mutations of the three DMS-protected G bases shown in Figures 1(A) and 1(B). USF2 antibody was added after the addition of the unlabelled M3 competitor. (B) EMSA as seen in (A) using anti-USF1 antibody for supershift analysis. FP, free probe.
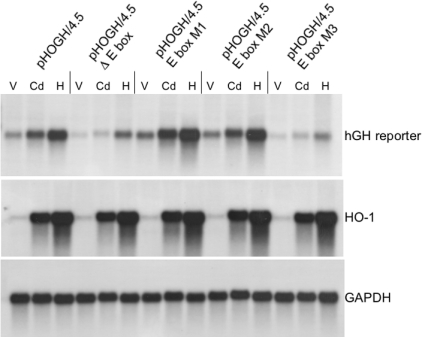
To investigate further the functional significance of the three protected guanine residues detected by in vivo footprinting, we performed site-directed mutagenesis of the human HO-1 promoter in a hGH reporter system. We have shown previously that a −4.5 kb fragment of the human HO-1 promoter is necessary, at least in part, for gene activation in response to haemin and cadmium in human renal proximal tubular cells in transient transfection studies [36]. Each of the three guanine residues was individually mutated to an adenine in pHOGH/4.5. Another mutant that eliminates the E-box starting from the M3 contact point up to and including the M1 contact point was also made. In these experiments, cells were transiently transfected using a batch transfection method as described in the Experimental section. As shown in Figure 6, deletion of the E-box in the 4.5 kb human HO-1 promoter construct significantly decreased expression of the hGH reporter when induced with either cadmium or haemin. In contrast, the M1 and M2 mutations did not decrease hGH expression. However, it is interesting that the M2 mutant appeared to increase expression by approx. 10–20% under all conditions tested. In contrast, the M3 mutation mimicked the results observed with deletion of the E-box. These results demonstrate that the guanine base at −44 relative to the transcription start site is essential for enhanced expression of the HO-1 promoter. Furthermore, the deletion of the E-box and the M3 mutations did not completely abolish cadmium- or haemin-mediated induction of the reporter gene, but significantly decreased the level of transcription. These results indicate that other factor(s) and hence other DNA sequences are required for the actual induction of the human HO-1 gene by cadmium or haemin, and that these factors act in concert with USFs bound to the E-box to drive transcription of HO-1 to a high level. This function for USFs to participate as an enhancer of transcription has been reported previously for other genes [30,45,46].
Figure 6. Functional significance of the guanine at −44 bp in a promoter-reporter transcription assay by Northern-blot analysis.
HK-2 cells were transfected with pHOGH/4.5, pHOGH/4.5 Δ E-box (entire E-box deleted), pHOGH E-box M1, pHOGH E-box M2 or pHOGH E-box M3, representing single bp mutations at M1, M2 and M3 respectively, then split into three plates as described in the Experimental section. Cells were treated with vehicle (V), cadmium (Cd, 10 μM) or haemin (H, 5 μM), and total RNA was extracted, blotted and sequentially hybridized with a 32P-labelled hGH cDNA (hGH), human HO-1 as a control for induction and GAPDH as a control for loading and transfer of RNA.
Effects of USF1/USF2 overexpression on HO-1 gene expression
We then tested the effects of overexpression of USF1 or USF2 with pHOGH4.5 in HK-2 cells induced with haemin (5 μM). Overexpression of USF1 and USF2 was confirmed by Western blotting (Figure 7A). As shown in Figure 7(B), both USF1 and USF2 increased basal expression of the reporter gene in vehicle-treated cells, without a significant effect on haemin-mediated induction. We did not observe an increase in endogenous HO-1 gene expression with USF1/USF2 overexpression. This is consistent with the findings that both USF1 and USF2 are constitutively bound to the E-box sequence in the proximal HO-1 promoter rendering the E-box saturated. Hence, any further overexpression does not affect endogenous HO-1 gene transcription in HK-2 cells.
Figure 7. Effect of USF1 and USF2 overexpression on HO-1 gene expression.
(A) Confirmation of USF1 and USF2 overexpression by Western-blot analysis. HK-2 cells were transiently transfected with the indicated plasmids and duplicate blots were made out of the total cellular protein. Anti-USF1 antibody confirms USF1 and A-USF overexpression (left panel) and anti-USF2 confirms USF2 overexpression (right panel). USF1 and USF2 are recognized as positive bands at approx. 43 and 44 kDa respectively. Blots were stripped and probed with an anti-actin antibody as a loading control. Actin was identified as approx. 46 kDa protein. The band in lane 3 in the lower right panel represents residual USF2 signal not completely stripped from the blot. (B) Northern-blot analysis of HK-2 cells co-transfected with pHOGH/4.5 and either USF1 or USF2 overexpression vectors. V, vehicle-treated cells; H, haemin (5 μM)-treated cells. The blots were stripped and reprobed with a HO-1 cDNA as a control for induction and GAPDH as a control for loading and transfer of RNA.
Effects of dominant-negative A-USF on HO-1 gene expression
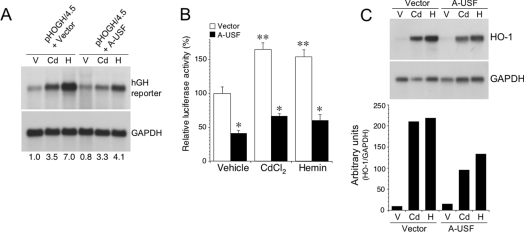
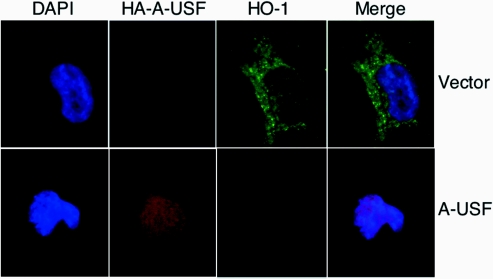
We next examined the effect of a USF dominant negative (A-USF), which inhibits dimerization of USF1 and USF2, thereby preventing DNA binding and gene activation [47,48] on HO-1 reporter constructs (pHOGH/4.5 or pHOGL/34.5). Specific overexpression of A-USF was confirmed by immunoblot analysis (Figure 7A). A-USF expression attenuated activation of the hGH reporter (Figure 8A) as well as a luciferase reporter construct containing the same −4.5 kb human HO-1 promoter fragment (Figure 8B). In addition, A-USF expression also reduced endogenous haemin (5 μM)- and cadmium (10 μM)-mediated HO-1 induction (Figure 8C). To investigate further the effects of A-USF on endogenous HO-1 protein expression, we used dual-label immunofluorescence in HK-2 cells transiently transfected with empty vector or the HA-tagged A-USF construct. As shown in Figure 9, significant HO-1 protein expression (green) was detected in cells transfected with empty vector after haemin (5 μM) stimulation. However, in cells showing nuclear staining for the HA-tagged A-USF (red), no HO-1 protein was detectable. These results demonstrate the important role of USF1 and USF2 in mediating transcription of the human HO-1 gene in HK-2 cells.
Figure 8. Effect of dominant-negative A-USF expression on HO-1 reporter and mRNA expression.
(A) HK-2 cells were co-transfected with pHOGH/4.5 and either empty vector (pCMV566) or A-USF as described in the Experimental section and subsequently treated with vehicle (V), cadmium (Cd, 10 μM) or haemin (H; 5 μM) for 12 h. Total RNA was isolated and subjected to Northern-blot analysis with 32P-labelled cDNA probes specific for hGH or GAPDH. The numbers at the bottom refer to the relative band intensities as determined by densitometry and controlled for GAPDH from three independent experiments. (B) Effect of A-USF expression on luciferase reporter activity of pHOGL3/4.5. Results are expressed as means±S.E.M. *P<0.05 versus vector; **P<0.05 versus vehicle. (C) Northern-blot analysis of endogenous HO-1 gene induction with and without A-USF expression. HK-2 cells were co-transfected with either empty vector or A-USF and treated with V, Cd or H as in (A). Total RNA was isolated and subjected to Northern-blot analysis with 32P-labelled cDNA probes specific for HO-1 or GAPDH. A densitometric analysis of the autoradiogram corrected for the internal control (GAPDH) is shown in the graph below.
Figure 9. Effect of dominant-negative A-USF expression on HO-1 protein expression.
HK-2 cells were transiently transfected with empty vector (upper panel) or the dominant negative, HA-tagged A-USF vector (lower panels). Cells were stimulated 24 h after transfection with haemin (5 μM) for 4 h. Dual-label immunofluorescence was performed using Alexa 594-conjugated anti-HA (Molecular Probes) and anti-HO-1 (Stressgen Biotechnologies) antibodies for detection of A-USF and HO-1 respectively. HO-1 was detected using a goat anti-rabbit FITC-conjugated secondary antibody (Jackson Immunoresearch Laboratories). Results are representative of two independent experiments. DAPI, 4,6-diamidino-2-phenylindole.
DISCUSSION
The results of our present studies provide insights into the in vivo architecture of the proximal promoter of the human HO-1 gene in human renal proximal tubular cells. We have demonstrated that a 6 bp sequence containing an E-box motif, CACGTG, located at nt −44 to −39 bp is essential for maximal haemin- or cadmium-mediated human HO-1 promoter activity. Three guanine bases protected in vivo from DMS methylation were discovered, two of which reside in the canonical E-box motif, and the third protected guanine residing 2 bases outside of the E-box. We have identified that the proteins bound to this sequence are USF1 and USF2 and that they associate with the E-box potentially as a heterodimer, since both USF1 and USF2 antibodies supershift the protein–DNA complex in vitro and bind to the proximal HO-1 promoter in vivo, as confirmed by ChIP analysis. Furthermore, we demonstrate that the guanine at −44 of human HO-1 gene is essential for USF binding and enhances USF-driven expression of HO-1 in HK-2 cells. Moreover, we have demonstrated that overexpression of USF1 or USF2 increased basal HO-1 promoter-driven reporter expression, and a dominant negative form of USF (A-USF) can attenuate endogenous HO-1 mRNA and protein expression.
USF was initially purified from HeLa cell nuclear extracts and was shown to increase the transcription of the adenovirus major late promoter in an in vitro system wherein methidiumpropyl-EDTA-Fe(II) footprint analyses showed that USF interacted with the palindromic sequence GGCCACGTGACC [19]. Subsequent studies determined that USF activity comprised two separate proteins, USF1 and USF2, which can exist as either homo- or heterodimers [22,23]. There are two classes of the E-box sequence, Class A corresponding to CAGCTG and Class B to CACGTG. The outer C:G bp in both half sites is indispensable for native USF binding [49]. Our findings are consistent with the indispensability of the outer guanine residue, since mutation of this single nucleotide (M3) significantly attenuated human HO-1 gene expression (Figure 6). Interestingly, the importance of the C residue on the complementary strand of this core sequence was also reported by Sato et al. [18], who observed that mutation of the C to a T abolished binding of USF to a synthetic oligonucleotide. Furthermore, the three protected G residues on the human HO-1 E-box that we detected are also protected from DMS methylation in the adenovirus major late promoter E-box, as reported by Chodosh et al. [50] using an in vitro methylation interference assay.
The human HO-1 promoter is characterized by the presence of a non-canonical TATA box (CATAAA) located 10 bp downstream of the E-box and 28 bp upstream of the transcription start site. Our findings showing involvement of USFs in human HO-1 gene expression are similar to studies pertaining to the regulation of the human β-globin gene, which also has a non-canonical TATA-like motif (CATAAA) and E-box motifs that contribute to the efficient transcription of the β-globin gene [44,51]. Interestingly, whereas the β-globin gene is involved in haemoglobin biosynthesis, HO-1 is involved in the degradation of haem moieties including those present in haemoglobin.
Studies from our laboratory as well as others demonstrate significant differences in the regulation of human HO-1 versus the mouse HO-1 gene [3,52]. However, the proximal E-box sequence is conserved in the human, rodent and avian HO-1 genes, as shown in the alignment in Figure 10. Previous studies have implicated the involvement of USF in HO-1 gene transcription [18,53–55]. Sato et al. [53] initially identified a protein called haem oxygenase transcription factor that was homologous with USF and reported its involvement in rat HO-1 gene transcription using in vitro transcription and DNase I footprinting in rat glioma cells. These authors subsequently extended their observations to the human HO-1 gene, again using cell-free systems, and reported that partially purified USF from HeLa cells bound to the core sequence CACGTGAC corresponding to −44 to −37 bp of the human HO-1 promoter [18]. Studies by Maeshima et al. [55] have suggested a role for USF in the induction of the rat HO-1 gene after cadmium stimulation, but not by haemin. Our studies suggest that USF1 and USF2 are important for both cadmium- and haemin-mediated increased HO-1 induction in human renal proximal tubule cells. Nascimento et al. [54] performed in vitro DNase I footprinting on a 147 bp fragment of the human HO-1 proximal promoter extending from +24 to −123 bp promoter, and observed a protected region between −41 and −50 bp in both control and UV-A-radiation-treated samples. However, the protected region by DNase I footprinting included only the first 3 bp upstream of the core E-box sequence and the functional significance of these observations in modulating HO-1 gene expression was not examined [54]. Our studies using in vivo footprinting, ChIP, mutational and functional assays conclusively demonstrate a prominent functional role for USFs in the regulation of the human HO-1 gene.
Figure 10. Sequence alignment of the human, rat, mouse and chicken HO-1 proximal promoter regions showing the E-box and the flanking nucleotides.
The positions relative to the transcription start sites are indicated. The sequences are derived from GenBank® accession no. Z82244 for human, J02722 for rat, X56824 for mouse and U95209 for chicken HO-1 genes.
As additional confirmation, we have used A-USF, a dominant-negative mutant of USF, to prevent binding of endogenous USF1 and USF2 to the human HO-1 promoter. A-USF was derived from USF1 (hence its detection by anti-USF1 antibody in Figure 7A) by deleting all N-terminal activation domains and substituting acidic amino acids for the basic amino acids involved in DNA binding in native USF dimers [47,48]. The dimers between A-USF and native USF are more stable than DNA-bound native dimers, resulting in the removal of USF from its target DNA with modest expression of A-USF [47,48]. We observed inhibition of the human HO-1 promoter-reporter construct as well as endogenous HO-1 gene expression with dominant-negative expression of A-USF. As we expected, however, we did not observe the level of inhibition attained in the reporter assay using a deletion of the E-box or M3 (Figure 6). This is due to the relatively low transfection efficiency of the cells tested in the present study, as well as the inability of the dominant negative to displace all of the USF proteins from binding to the endogenous promoter.
In summary, our studies demonstrate a prominent role for USF1 and USF2 in the regulation of the human HO-1 gene in human renal proximal tubular cells. Recent evidence has demonstrated the critical importance of HO-1 expression in mediating antioxidant, anti-inflammatory and anti-apoptotic effects (reviewed in [7]). We and others have demonstrated that induction of HO-1, by chemical inducers or selective overexpression, is cytoprotective both in vitro and in vivo in renal injury, which has been further substantiated by studies in HO-1 knockout mice (reviewed in [7]). Given the biological relevance of HO-1 induction in renal injury, further understanding of the molecular regulation of the HO-1 gene is paramount. We hypothesize that both USF1 and USF2 (its splice variant USF2b in particular) co-ordinate with other transcription factors and are critical in regulating the expression of the human HO-1 gene.
Acknowledgments
This work was supported by grants from the National Institutes of Health, R01 DK59600 and R01 HL68157 (to A.A.). We are grateful to Dr Michelle Sawadogo for the gift of expression vectors for USF1 and USF2 and Dr Charles Vinson for the HA-tagged A-USF expression vector. We thank all members of the Nick laboratory, Cynthia Dawson, Kimberly Durgan, Nathalie Hill-Kapturczak, Amie Mark, Pam Powell and Jorg Bungert, for helpful discussions.
References
- 1.Maines M. D. The heme oxygenase system: a regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 1997;37:517–554. doi: 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- 2.McCoubrey W. K., Jr, Huang T. J., Maines M. D. Isolation and characterization of a cDNA from the rat brain that encodes hemoprotein heme oxygenase-3. Eur. J. Biochem. 1997;247:725–732. doi: 10.1111/j.1432-1033.1997.00725.x. [DOI] [PubMed] [Google Scholar]
- 3.Sikorski E. M., Hock T., Hill-Kapturczak N., Agarwal A. The story so far: molecular regulation of the heme oxygenase-1 gene in renal injury. Am. J. Physiol. Renal Physiol. 2004;286:F425–F441. doi: 10.1152/ajprenal.00297.2003. [DOI] [PubMed] [Google Scholar]
- 4.Platt J. L., Nath K. A. Heme oxygenase: protective gene or Trojan horse. Nat. Med. 1998;4:1364–1365. doi: 10.1038/3947. [DOI] [PubMed] [Google Scholar]
- 5.Abraham N. G., Lavrovsky Y., Schwartzman M. L., Stoltz R. A., Levere R. D., Gerritsen M. E., Shibahara S., Kappas A. Transfection of the human heme oxygenase gene into rabbit coronary microvessel endothelial cells: protective effect against heme and hemoglobin toxicity. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6798–6802. doi: 10.1073/pnas.92.15.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwal A., Balla J., Alam J., Croatt A. J., Nath K. A. Induction of heme oxygenase in toxic renal injury: a protective role in cisplatin nephrotoxicity in the rat. Kidney Int. 1995;48:1298–1307. doi: 10.1038/ki.1995.414. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal A., Nick H. S. Renal response to tissue injury: lessons from heme oxygenase-1 gene ablation and expression. J. Am. Soc. Nephrol. 2000;11:965–973. doi: 10.1681/ASN.V115965. [DOI] [PubMed] [Google Scholar]
- 8.Nath K. A., Haggard J. J., Croatt A. J., Grande J. P., Poss K. D., Alam J. The indispensability of heme oxygenase-1 in protecting against acute heme protein-induced toxicity in vivo. Am. J. Pathol. 2000;156:1527–1535. doi: 10.1016/S0002-9440(10)65024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nath K. A., Balla G., Vercellotti G. M., Balla J., Jacob H. S., Levitt M. D., Rosenberg M. E. Induction of heme oxygenase is a rapid, protective response in rhabdomyolysis in the rat. J. Clin. Invest. 1992;90:267–270. doi: 10.1172/JCI115847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiraishi F., Curtis L. M., Truong L., Poss K., Visner G. A., Madsen K., Nick H. S., Agarwal A. Heme oxygenase-1 gene ablation or expression modulates cisplatin-induced renal tubular apoptosis. Am. J. Physiol. Renal Physiol. 2000;278:F726–F736. doi: 10.1152/ajprenal.2000.278.5.F726. [DOI] [PubMed] [Google Scholar]
- 11.Poss K. D., Tonegawa S. Reduced stress defense in heme oxygenase 1-deficient cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10925–10930. doi: 10.1073/pnas.94.20.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yachie A., Niida Y., Wada T., Igarashi N., Kaneda H., Toma T., Ohta K., Kasahara Y., Koizumi S. Oxidative stress causes enhanced endothelial cell injury in human heme oxygenase-1 deficiency. J. Clin. Invest. 1999;103:129–135. doi: 10.1172/JCI4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi S., Takahashi Y., Ito K., Nagano T., Shibahara S., Miura T. Positive and negative regulation of the human heme oxygenase-1 gene expression in cultured cells. Biochim. Biophys. Acta. 1999;1447:231–235. doi: 10.1016/s0167-4781(99)00156-6. [DOI] [PubMed] [Google Scholar]
- 14.Takeda K., Ishizawa S., Sato M., Yoshida T., Shibahara S. Identification of a cis-acting element that is responsible for cadmium-mediated induction of the human heme oxygenase gene. J. Biol. Chem. 1994;269:22858–22867. [PubMed] [Google Scholar]
- 15.Shibahara S., Sato M., Muller R. M., Yoshida T. Structural organization of the human heme oxygenase gene and the function of its promoter. Eur. J. Biochem. 1989;179:557–563. doi: 10.1111/j.1432-1033.1989.tb14583.x. [DOI] [PubMed] [Google Scholar]
- 16.Lavrovsky Y., Schwartzman M. L., Levere R. D., Kappas A., Abraham N. G. Identification of binding sites for transcription factors NF-κB and AP-2 in the promoter region of the human heme oxygenase 1 gene. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5987–5991. doi: 10.1073/pnas.91.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deramaudt B. M., Remy P., Abraham N. G. Upregulation of human heme oxygenase gene expression by Ets-family proteins. J. Cell. Biochem. 1999;72:311–321. [PubMed] [Google Scholar]
- 18.Sato M., Ishizawa S., Yoshida T., Shibahara S. Interaction of upstream stimulatory factor with the human heme oxygenase gene promoter. Eur. J. Biochem. 1990;188:231–237. doi: 10.1111/j.1432-1033.1990.tb15394.x. [DOI] [PubMed] [Google Scholar]
- 19.Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell (Cambridge, Mass) 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 20.Howcroft T. K., Murphy C., Weissman J. D., Huber S. J., Sawadogo M., Singer D. S. Upstream stimulatory factor regulates major histocompatibility complex class I gene expression: the U2DeltaE4 splice variant abrogates E-box activity. Mol. Cell. Biol. 1999;19:4788–4797. doi: 10.1128/mcb.19.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X., Sawadogo M. Antiproliferative properties of the USF family of helix-loop-helix transcription factors. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1308–1313. doi: 10.1073/pnas.93.3.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirito M., Walker S., Lin Q., Kozlowski M. T., Klein W. H., Sawadogo M. Members of the USF family of helix-loop-helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 1992;2:231–240. [PMC free article] [PubMed] [Google Scholar]
- 23.Sirito M., Lin Q., Maity T., Sawadogo M. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 1994;22:427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirito M., Lin Q., Deng J. M., Behringer R. R., Sawadogo M. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3758–3763. doi: 10.1073/pnas.95.7.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxevanis A. D., Vinson C. R. Interactions of coiled coils in transcription factors: where is the specificity? Curr. Opin. Genet. Dev. 1993;3:278–285. doi: 10.1016/0959-437x(93)90035-n. [DOI] [PubMed] [Google Scholar]
- 26.Cogswell J. P., Godlevski M. M., Bonham M., Bisi J., Babiss L. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol. Cell. Biol. 1995;15:2782–2790. doi: 10.1128/mcb.15.5.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan S., Jane D. T., Dufresne M. J., Sloane B. F. Transcription of cathepsin B in glioma cells: regulation by an E-box adjacent to the transcription initiation site. Biol. Chem. 2003;384:1421–1427. doi: 10.1515/BC.2003.157. [DOI] [PubMed] [Google Scholar]
- 28.Wang H., Wollheim C. B. ChREBP rather than USF2 regulates glucose stimulation of endogenous L-pyruvate kinase expression in insulin-secreting cells. J. Biol. Chem. 2002;277:32746–32752. doi: 10.1074/jbc.M201635200. [DOI] [PubMed] [Google Scholar]
- 29.Gupta A. K., Kone B. C. USF-1 and USF-2 trans-repress IL-1β-induced iNOS transcription in mesangial cells. Am. J. Physiol. Cell Physiol. 2002;283:C1065–C1072. doi: 10.1152/ajpcell.00100.2002. [DOI] [PubMed] [Google Scholar]
- 30.Bruno M. E., West R. B., Schneeman T. A., Bresnick E. H., Kaetzel C. S. Upstream stimulatory factor but not c-Myc enhances transcription of the human polymeric immunoglobulin receptor gene. Mol. Immunol. 2004;40:695–708. doi: 10.1016/j.molimm.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Szentirmay M. N., Yang H. X., Pawar S. A., Vinson C., Sawadogo M. The IGF2 receptor is a USF2-specific target in nontumorigenic mammary epithelial cells but not in breast cancer cells. J. Biol. Chem. 2003;278:37231–37240. doi: 10.1074/jbc.M305791200. [DOI] [PubMed] [Google Scholar]
- 32.Wu K., Jiang S. W., Thangaraju M., Wu G., Couch F. J. Induction of the BRCA2 promoter by nuclear factor-κB. J. Biol. Chem. 2000;275:35548–35556. doi: 10.1074/jbc.M004390200. [DOI] [PubMed] [Google Scholar]
- 33.Chen W. G., West A. E., Tao X., Corfas G., Szentirmay M. N., Sawadogo M., Vinson C., Greenberg M. E. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J. Neurosci. 2003;23:2572–2581. doi: 10.1523/JNEUROSCI.23-07-02572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S. Y., Choi S. Y., Chao W., Volsky D. J. Transcriptional regulation of human excitatory amino acid transporter 1 (EAAT1): cloning of the EAAT1 promoter and characterization of its basal and inducible activity in human astrocytes. J. Neurochem. 2003;87:1485–1498. doi: 10.1046/j.1471-4159.2003.02128.x. [DOI] [PubMed] [Google Scholar]
- 35.Ryan M. J., Johnson G., Kirk J., Fuerstenberg S. M., Zager R. A., Torok-Storb B. HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994;45:48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- 36.Hill-Kapturczak N., Sikorski E., Voakes C., Garcia J., Nick H. S., Agarwal A. An internal enhancer regulates heme- and cadmium-mediated induction of human heme oxygenase-1. Am. J. Physiol. Renal Physiol. 2003;285:F515–F523. doi: 10.1152/ajprenal.00137.2003. [DOI] [PubMed] [Google Scholar]
- 37.Agarwal A., Shiraishi F., Visner G. A., Nick H. S. Linoleyl hydroperoxide transcriptionally upregulates heme oxygenase-1 gene expression in human renal epithelial and aortic endothelial cells. J. Am. Soc. Nephrol. 1998;9:1990–1997. doi: 10.1681/ASN.V9111990. [DOI] [PubMed] [Google Scholar]
- 38.Barbosa-Tessmann I. P., Chen C., Zhong C., Siu F., Schuster S. M., Nick H. S., Kilberg M. S. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 2000;275:26976–26985. doi: 10.1074/jbc.M000004200. [DOI] [PubMed] [Google Scholar]
- 39.Kang S. H., Vieira K., Bungert J. Combining chromatin immunoprecipitation and DNA footprinting: a novel method to analyze protein-DNA interactions in vivo. Nucleic Acids Res. 2002;30:e44. doi: 10.1093/nar/30.10.e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viollet B., Lefrancois-Martinez A. M., Henrion A., Kahn A., Raymondjean M., Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J. Biol. Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- 41.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 42.Hornstra I. K., Yang T. P. In vivo footprinting and genomic sequencing by ligation-mediated PCR. Anal. Biochem. 1993;213:179–193. doi: 10.1006/abio.1993.1407. [DOI] [PubMed] [Google Scholar]
- 43.Kuo S., Chesrown S. E., Mellott J. K., Rogers R. J., Hsu J. L., Nick H. S. In vivo architecture of the manganese superoxide dismutase promoter. J. Biol. Chem. 1999;274:3345–3354. doi: 10.1074/jbc.274.6.3345. [DOI] [PubMed] [Google Scholar]
- 44.Leach K. M., Vieira K. F., Kang S. H., Aslanian A., Teichmann M., Roeder R. G., Bungert J. Characterization of the human β-globin downstream promoter region. Nucleic Acids Res. 2003;31:1292–1301. doi: 10.1093/nar/gkg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moriuchi M., Moriuchi H., Margolis D. M., Fauci A. S. USF/c-Myc enhances, while Yin-Yang 1 suppresses, the promoter activity of CXCR4, a coreceptor for HIV-1 entry. J. Immunol. 1999;162:5986–5992. [PubMed] [Google Scholar]
- 46.Gobin S. J., Biesta P., Van den Elsen P. J. Regulation of human β2-microglobulin transactivation in hematopoietic cells. Blood. 2003;101:3058–3064. doi: 10.1182/blood-2002-09-2924. [DOI] [PubMed] [Google Scholar]
- 47.Krylov D., Kasai K., Echlin D. R., Taparowsky E. J., Arnheiter H., Vinson C. A general method to design dominant negatives to B-HLHZip proteins that abolish DNA binding. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12274–12279. doi: 10.1073/pnas.94.23.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qyang Y., Luo X., Lu T., Ismail P. M., Krylov D., Vinson C., Sawadogo M. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol. Cell. Biol. 1999;19:1508–1517. doi: 10.1128/mcb.19.2.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lennard A. C., Egly J. M. The bidirectional upstream element of the adenovirus-2 major late promoter binds a single monomeric molecule of the upstream factor. EMBO J. 1987;6:3027–3034. doi: 10.1002/j.1460-2075.1987.tb02608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chodosh L. A., Buratowski S., Sharp P. A. A yeast protein possesses the DNA-binding properties of the adenovirus major late transcription factor. Mol. Cell. Biol. 1989;9:820–822. doi: 10.1128/mcb.9.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wobbe C. R., Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol. Cell. Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shibahara S., Nakayama M., Kitamuro T., Udono-Fujimori R., Takahashi K. Repression of heme oxygenase-1 expression as a defense strategy in humans. Exp. Biol. Med. 2003;228:472–473. doi: 10.1177/15353702-0322805-08. [DOI] [PubMed] [Google Scholar]
- 53.Sato M., Fukushi Y., Ishizawa S., Okinaga S., Muller R. M., Shibahara S. Transcriptional control of the rat heme oxygenase gene by a nuclear protein that interacts with adenovirus 2 major late promoter. J. Biol. Chem. 1989;264:10251–10260. [PubMed] [Google Scholar]
- 54.Nascimento A. L., Luscher P., Tyrrell R. M. Ultraviolet A (320–380 nm) radiation causes an alteration in the binding of a specific protein/protein complex to a short region of the promoter of the human heme oxygenase 1 gene. Nucleic Acids Res. 1993;21:1103–1109. doi: 10.1093/nar/21.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maeshima H., Sato M., Ishikawa K., Katagata Y., Yoshida T. Participation of altered upstream stimulatory factor in the induction of rat heme oxygenase-1 by cadmium. Nucleic Acids Res. 1996;24:2959–2965. doi: 10.1093/nar/24.15.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]