Abstract
In Drosophila oocytes and neuroblasts, the double-stranded RNA binding protein Staufen assembles into ribonucleoprotein particles, which mediate cytoplasmic mRNA trafficking and translation. Two different mammalian orthologues also appear to reside in distinct RNA-containing particles. To date, relatively little is known about the molecular composition of Staufen-containing ribonucleoprotein complexes. Here, we have used a novel one-step affinity purification protocol to identify components of Staufen 1-containing particles. Whereas the nucleocytoplasmic RNA-binding protein nucleolin is linked to Staufen in an RNA-dependent manner, the association of protein phosphatase 1, the microtubule-dependent motor protein kinesin and several components of the large and small ribosomal subunits with Staufen ribonucleoprotein complexes is RNA-independent. Notably, all these components do not co-purify with a second RNA-binding protein, hnRNPK (heterogeneous ribonucleoprotein K), demonstrating the high specificity of the purification protocol. Furthermore, pull-down and immunoprecipitation experiments suggest a direct interaction between Staufen 1 and the ribosomal protein P0 in vitro as well as in cells. In cell fractionation and sucrose gradient assays, Staufen co-fractionates with intact ribosomes and polysomes, but not with the isolated 40 S ribosomal subunit. Taken together, these findings imply that, in the cytoplasm of mammalian cells, an association with the ribosomal P-stalk protein P0 recruits Staufen 1 into ribosome-containing ribonucleoprotein particles, which also contain kinesin, protein phosphatase 1 and nucleolin.
Keywords: ribonucleoprotein particle, ribosome, RNA sorting, RNA-binding protein, translation
Abbreviations: dsRBD, double-stranded RNA-binding domain; EF1α, elongation factor 1α; (E)GFP, (enhanced) green fluorescent protein; FMRP, fragile X mental retardation protein 1; GKAP/SAPAP1, guanylate kinase-associated protein/SAP90/PSD-95-associated protein; GST, glutathione S-transferase; HEK, human embryonic kidney; hnRNP(K/U), heterogeneous ribonucleoprotein K or U respectively; IP buffer, immunoprecipitation buffer; PP1, protein phosphatase-1; (m)RNP, (messenger) ribonucleoprotein particle; NFAR, nuclear factor associated with double-stranded RNA; PABP, poly(A)+-binding protein; RHA, RNA helicase A; rStau, rat Staufen; SSTRIP, somatostatin receptor-interacting protein; Stau1, Staufen 1; Stau2, Staufen 2
INTRODUCTION
In many different cell types, a locally regulated protein synthesis in restricted cytoplasmic subregions of the cell appears to contribute to the establishment and maintenance of cell polarity [1,2]. Both cytoplasmic trafficking and site-specific translation of mRNAs in various cell types seems to be regulated by an ordered interaction of cis-acting RNA sequences with trans-acting proteins. In Drosophila, the double-stranded RNA-binding protein Staufen plays an essential role in the cytoplasmic localization of oskar and bicoid [1] mRNAs in oocytes, and of prospero transcripts in dividing neuroblasts [3]. Two mammalian Staufen orthologues, Stau1 and Stau2 (Staufen 1 and Staufen 2 respectively), from mouse, rat and human sources, are expressed in many tissues [4–10]. Stau1 co-localizes with the rough endoplasmic reticulum [5–7,11] and microtubules [6,9,11], and binds to RNA both in vitro and in vivo [6,7,9,10]. In neurons, Stau1 and Stau2 appear to form distinct RNPs (ribonucleoprotein particles), which move along dendritic microtubules [8,10,12–15]. For the present study, we have developed a novel one-step affinity purification protocol to unravel the molecular composition of mammalian Stau1-containing complexes. Stau1 RNPs are shown to contain the RNA-binding shuttling protein nucleolin, PP1 (protein phosphatase 1), the motor protein kinesin, and several components of the large and small ribosomal subunits, whereas none of these components co-purified with the hnRNPK (heterogeneous ribonucleoprotein K). Further experiments demonstrate an interaction between Stau1 and the ribosomal protein P0, as well as a co-fractionation of Stau1 with intact ribosomes and polysomes. Thus these findings suggest a P0-mediated recruitment of Stau1 to ribosome-containing RNPs, which may be involved in mRNA trafficking and translation.
EXPERIMENTAL
Construction of expression vectors
cDNA inserts were amplified via PCR with oligonucleotides containing restriction enzyme sites [9] [GenBank® accession numbers: rStau1 (rat Stau1), AF290989; rat nucleolin, AH002217; rat PP1α, S78215; rat hnRNPK, NM002140; rat Sharpin, AF203906; and human P0, NM_001002]. Digested PCR products were inserted into accordingly prepared prokaryotic expression vector pGEX2T (Amersham Biosciences, Uppsala, Sweden) or eukaryotic expression vectors pCMVtag2B (Stratagene GmbH, Heidelberg, Germany) and pEGFP-N1 (where ‘EGFP’ is enhanced green fluorescent protein; ClonTech Laboratories, Palo Alto, CA, U.S.A.). Vectors pGEX2T-rStau1, pGEX2T-PP1, pCMVtag2B-Stau1, pCMVtag2B-Nuc, pStau1-EGFP and pP0-EGFP contain the full-length coding regions of the respective Stau1, PP1α, nucleolin and P0 cDNAs. The P0 vector was kindly given by Dr Marek Tchórzewski (Department of Molecular Biology, Lubin, Poland). Number ranges in the names of fusion proteins encoded by different vectors refer to the N- and C-terminal amino acid positions of the corresponding endogenous proteins. The PDZ domain of SSTRIP (somatostatin receptor-interacting protein) [16] was PCR-amplified with two oligonucleotides (5′-TTTGGATCCTGGCTCCTACGACAGC-3′ and (5′-TTTGCGGCCGCTTACACTGCCTCATCC-3′), digested with BamHI/NotI and subcloned into pEGFP-N1 (ClonTech Laboratories) to create pPDZ-N1. Coding regions of rStau1 and hnRNPK cDNAs were PCR-amplified, digested with EcoRI/BamHI and XhoI/EcoRI respectively, and subcloned into pPDZ-N1 to generate pStau1-PDZ and phnRNPK-PDZ. An EcoRI/BamHI fragment from pGW1-Sharpin [17] was inserted into pPDZ-N1 to generate pSharp-PDZ.
GST (glutathione S-transferase) pull-down assay
GST fusion proteins were expressed in Escherichia coli BL21 codon plus cells (Stratagene GmbH) and affinity-purified with glutathione–Sepharose 4B beads (Amersham Biosciences). Protein-coated beads were washed five times with STE buffer [10 mM Tris/HCl (pH 8.0)/150 mM NaCl/1 mM EDTA] containing 10 μM PMSF, and stored at 4 °C until further use in GST pulldown experiments. Adult rat brain was homogenized in RIPA buffer [150 mM NaCl/50 mM Tris/HCl (pH 8.0)/5 mM EDTA (pH 8.0)/0.5% sodium deoxycholate/1% Nonidet P-40/0.1% SDS] containing 0.1% (v/v) of a stock solution of the proteinase inhibitor cocktail Complete™ (Roche Diagnostics, Heidelberg, Germany) and 40 units/ml RNaseOUT (Invitrogen GmbH, Karlsruhe, Germany), followed by centrifugation at 15800 g and 4 °C for 15 min. Brain proteins from the supernatant fraction were allowed to bind to Sepharose-coupled GST fusion proteins for 2 h at 4 °C, followed by five washes with RIPA buffer. Bound proteins were eluted from the beads by boiling in sample buffer [18], before being subjected to Western blotting.
Cell culture and transfection
HEK (human embryonic kidney)-293 cells (A.T.C.C., Rockville, MD, U.S.A.) were grown and transfected with the eukaryotic expression vectors pStau1-PDZ, phnRNPK-PDZ, pCMVtag2B-Nuc, pEGFP-Nuc1–N310, pEGFP-Nuc309–Nuc713 and pP0-EGFP, as described previously [15].
Affinity purification of PDZ-tagged proteins
A synthetic peptide corresponding to the C-terminus of GKAP/SAPAP1 (guanylate kinase-associated protein/SAP90/PSD-95-associated protein; sequence IYIPEAQTRL) [19] was obtained from Genemed Synthesis Inc. (San Francisco, CA, U.S.A.). The peptide was coupled to NHS-activated Sepharose™ (Amersham Biosciences) at a concentration of 3 mg/ml Sepharose matrix. Six plates of transfected HEK-293 cells per construct were lysed in 1 ml RIPA buffer/culture dish (10 cm diameter) on ice for 15 min. In some experiments, lysates were incubated with 50 μg/ml RNase A (Type XII-A; Sigma–Aldrich, Taufkirchen, Germany) in lysis buffer for 15 min at 4 °C. Lysates were centrifuged (20000 g for 10 min) to remove insoluble matter. Supernatant fractions were incubated with 80 μl of GKAP–Sepharose slurry (60 μl of the precipitated bead-volume) for 2 h at 4 °C with slight agitation. Sepharose beads with bound PDZ-domain-containing proteins were precipitated by centrifugation (300 g for 1 min). Beads were washed five times (1 ml of RIPA buffer each wash). From the precipitated beads, bound proteins were eluted with 60 μl of sample buffer. Alternatively, cell lysis and protein purification were performed in IP buffer [immunoprecipitation buffer: 120 mM NaCl/50 mM Tris/HCl (pH 8.0)/1 mM EDTA/0.5% Nonidet P-40/0.1 mM PMSF/0.1% (v/v) of stock-solution proteinase inhibitor cocktail Complete™]. MS (mass spectrometry) was performed as described previously [20].
We have now extensively characterized the PDZ-tag affinity-purification technique. The GKAP C-terminus interacts selectively with the PDZ domain of SSTRIP/Shanks. As these proteins are highly expressed in neurons, but are only present at very low levels in cells derived from peripheral tissues, the GKAP matrix precipitates hardly any endogenous protein from cell lines, such as HEK-293 or HeLa cells. Owing to the high affinity of the peptide–PDZ domain interaction, rather stringent washing conditions, such as using 1 M NaCl, may be used, if required and tolerated by putative interaction partners of the PDZ-tagged bait. High selectivity of this procedure is demonstrated further by the fact that the Stau1-associated proteins identified in the present study are not detected in affinity purifications performed with several other PDZ-tagged proteins, including the neuronal scaffolding protein Sharpin (herein), Golgi-associated protein PIST, postsynaptic protein densin-180 and the tight junction protein MUPP1 (multi-PDZ protein 1; H.-J. Kreienkamp, C.-W. Liew, A. Quitsch and W. Wente, unpublished work). Since hnRNPK is an RNA-binding protein, similar to Stau1, we have deliberately included PDZ-tagged hnRNPK to evaluate further the specificity of individual proteins that co-purify with Stau1.
Cell fractionation and sucrose gradients
Preparation of different rat brain fractions was performed as described previously [21]. For HEK-293 cell fractionations, cells were scraped from culture plates in cold PBS, spun down and washed once in PBS [150 mM NaCl/20 mM sodium phosphate (pH 7.4)]. Cells were homogenized in cold HKM buffer [20 mM Hepes/HCl (pH 7.6)/100 mM KCl/20 mM MgCl2] supplemented with 0.5% Triton X-100, 1 mM PMSF, proteinase-inhibitor cocktail Complete™ [0.1% (v/v) of the stock solution] and 40 units/ml RNaseOUT (Invitrogen). After two subsequent centrifugation steps at 720 g and 15800 g for 10 min at 4 °C, the final supernatant was loaded on to a sucrose step gradient (5/10/15/20/25/30% sucrose) in HKM buffer. Gradients were centrifuged at 200000 g in a Sorvall TH614 rotor for 2 h at 4 °C. Fractions (0.5 ml) were collected from the top of the vial. Aliquots of each fraction were analysed for protein and RNA composition. Proteins were precipitated with 20% (w/v) trichloroacetic acid and cold acetone before Western blotting. RNA was extracted with peqGOLD TriFast (peqLab Biotechnology GmbH, Erlangen, Germany), and analysed on a non-denaturing 1.2% agarose gel.
Antibodies, immunoprecipitation and Western blotting
Generation of affinity-purified Stau1 antibodies has been described previously [15]. Antibodies directed against P0 (human polyclonal), kinesin (rabbit polyclonal) and PABP [poly(A)+-binding protein; rabbit polyclonal] were kindly given by Dr Morris Reichlin (Oklahoma Medical Research Foundation, Oklahoma City, OK, U.S.A.), Dr Mark McNiven (Center for Basic Research in Digestive Diseases, Rochester, MN, U.S.A.), and Dr Evita Mohr (Institute for Cell Biochemistry and Clinical Neurobiology, University-Hospital Hamburg-Eppendorf, Hamburg, Germany) respectively. Commercially available antibodies were used for the detection of nucleolin (mouse monoclonal C23 MS-3 antibody; Santa Cruz Biotechnology Inc., Heidelberg, Germany), L7a (mouse polyclonal L7a/SPA; Abcam Limited, Cambridge, U.K.), EF1α (elongation factor 1α; mouse monoclonal; Upstate, Lake Placid, NY, U.S.A.), PP1α (mouse monoclonal; Transduction Laboratories, Lexington, KY, U.S.A.), FMRP (fragile X mental retardation protein 1; mouse monoclonal; Euromedex, Mundelsheim, Germany), dynein intermediate chain (mouse monoclonal; Sigma) and α-tubulin (mouse monoclonal; Sigma). Recombinant proteins were detected with mouse monoclonal anti-FLAG antibody (Stratagene GmbH) and affinity-purified rabbit anti-GFP antibody (Abcam Limited, Cambridge, U.K.). As secondary antibodies, we used horseradish-peroxidase-conjugated donkey anti-human IgG and goat anti-rabbit IgG (Jackson Immuno-research Laboratories, West Grove, PA, U.S.A.) and sheep anti-mouse IgG (Amersham Biosciences). For immunoprecipitation, cells were lysed 36 h after transfection in IP buffer, kept on ice for 30 min and centrifuged at 15800 g for 15 min. Supernatant fractions were incubated at 4 °C for 4–14 h with 20 μl of EZview Red anti-FLAG M2 Affinity Gel (Sigma–Aldrich Chemie GmbH, Steinheim, Germany) or 5 μg of polyclonal antibody against the EGFP (anti-EGFP-peptide antibody; Clontech) and 25 μl of Protein A–agarose. Samples were centrifuged, washed and resuspended in sample buffer. For Western blots, primary antibodies directed against rStau1, P0 and kinesin were diluted 1:500 to 1:1000. Other antibodies were used at concentrations recommended by the manufacturer. Blots were analysed with the ECL®-System (Roche Diagnotics GmbH, Mannheim, Germany).
RESULTS
Purification of Stau1 ribonucleoprotein complexes
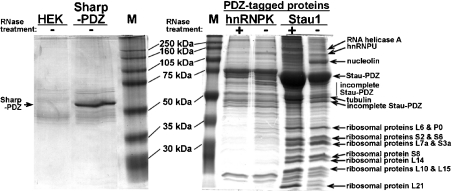
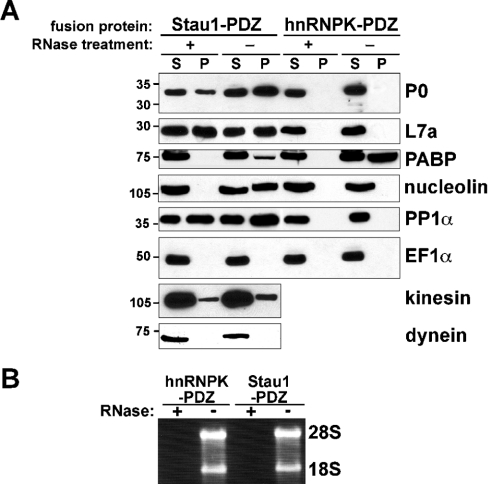
To identify components of Staufen RNPs assembled in vivo, we employed a novel one-step purification protocol. C-terminally PDZ-tagged Stau1, hnRNPK and Sharpin were transiently expressed in HEK-293 cells. After cell lysis, PDZ proteins were purified from both untreated and RNase-digested extracts using Sepharose beads coated with a 10-amino-acid residue peptide reflecting the C-terminus of GKAP/SAPAP1. This peptide binds with high affinity and specificity to the PDZ domain of the recombinant fusion proteins. After washing, affinity-purified proteins were separated by SDS/10%-PAGE, and prominent bands shown in Figure 1 were isolated and analysed via MS (for a summary of the identified proteins, see Table 1). RNase-insensitive interacting proteins of approx. 20–34 kDa were identified as ribosomal proteins L21, L10, L15, L14, S8, L7a, S3a, S6, S2, L6 and P0 (Figure 1). Larger bands contained β4-tubulin (50 kDa), PDZ-tagged rStau1 (70 kDa), the nucleocytoplasmic RNA-binding protein nucleolin (105 kDa) [22], the nuclear factor associated with double-stranded RNA (NFAR; 110 kDa) [23], hnRNPU (120 kDa) [24] and the RNA helicase A (150 kDa) [25]. RNase treatment strongly decreased the intensity of the nucleolin-containing band. Notably, none of these proteins was identified in affinity-purified material from untransfected HEK-293 cells or from cells expressing PDZ-tagged RNA-binding protein hnRNPK [24] or the PDZ-tagged neuronal protein Sharpin [17], with the exception of hnRNPU, which was also found in the hnRNPK complex (Figure 1). RNase treatment increased the efficiency of affinity purification of tagged Stau1 (Figure 1). This may be due to an increased accessibility of PDZ domains in the RNase-treated complex. Essentially identical data were obtained with both IP and RIPA buffer (results not shown). RIPA buffer contains 0.1% SDS and 0.5% deoxycholate, and disrupts relatively weak protein–protein interactions. Next, we verified the MS data and investigated the presence of other candidate components in Stau1 RNPs. Proteins that were co-affinity-purified with Stau1 and hnRNPK fusion proteins from both non- and RNase-treated cell extracts were separated by SDS/PAGE and analysed via Western blotting with different antibodies (Figure 2A). Agarose gel analysis of the RNA isolated from RNase-treated and untreated cell lysates shows complete RNA degradation in the treated fractions, as indicated by the disappearance of both 28 S and 18 S rRNA bands (Figure 2B). In Western blots, the ribosomal proteins P0 and L7a were detected in Stau1 precipitates from both untreated and RNase-digested cell lysates (Figure 2A, lanes P). In contrast, nucleolin and the PABP [24] were detected in the Stau1 precipitate before, but not after, RNase digestion, indicating that their association with this complex involves RNA–protein interactions. In a previous study [15], we have identified PP1 as a component of Stau1 complexes in the rat brain that directly interacts with Stau1. Western blot analysis of affinity-purified cell extracts performed in the present study shows that, in HEK-293 cells, PP1α also associates with Stau1 RNPs in an RNA-independent manner (Figure 2A). P0, L7a, nucleolin and PP1 were not found in fractions that were affinity-purified with the structurally unrelated RNA-binding protein hnRNPK (Figure 2A), indicating that these proteins specifically associate with Stau1 complexes in living cells. PABP, however, associates with both Stau1 and hnRNPK RNPs in an RNA-dependent manner. The plus-end-directed microtubule-associated motor protein kinesin 1 is present in both untreated and RNase-treated Stau1 RNPs (Figure 2A). This is in agreement with the finding that, in cultured neurons, Stau1 particles move bidirectionally along dendritic microtubules [9,11,13]. In contrast, the minus-end-directed motor dynein is absent from Stau1 RNPs. Moreover, the second most abundant protein of eukaryotic cells, EF1α [26], is not found in any of the four affinity-purified fractions, supporting the high specificity of the affinity-purification protocol.
Figure 1. One-step affinity purification of Stau1 ribonucleoprotein complexes.
C-terminally tagged Sharpin–PDZ, hnRNPK–PDZ and Stau1–PDZ were affinity-purified with GKAP-coated Sepharose beads from untreated (−) and RNase-digested (+) extracts of transfected HEK-293 cells lysed in RIPA buffer. After washing, proteins were separated via gel electrophoresis. Prominent bands were excised (arrows) and proteins were identified by MS. Stau1 RNPs contain the RNA-binding proteins RNA helicase A, hnRNPU, NFAR and nucleolin, as well as several proteins from the large and small ribosomal subunit, all of which are not identified in precipitates obtained from untransfected (HEK), Sharpin–PDZ (Sharp-PDZ) or hnRNPK–PDZ-expressing cells. The increase in affinity-purification efficiency of tagged Stau1 after RNase treatment probably reflects an increased accessibility of PDZ domains in the complex. Incomplete Stau-PDZ: short variants of PDZ-tagged rStau1, which may result from partial degradation. M, marker lane.
Table 1. Proteins associated with Stauf1-containing complexes.
Note that EF1α was not detected as a component of Staufen RNPs, either via MS or Western blotting, and served as a negative control. The band sizes (kDa) shown in the final data column were as seen in the gel. Explanation of synbols: +, detected; −, not detected; n.d., not determined.
| Protein | GenBank® accession no. | MS | Western blot | Band size (kDa) |
|---|---|---|---|---|
| RNAH | NP_076950 | + | n.d. | 150 |
| hnRNPU (SAFA) | NP_114032 | + | + | 120 |
| NFAR | Q12906 | + | n.d. | 110 |
| Kinesin heavy chain | A41919 | − | + | 110 |
| Nucleolin | NP_005372 | + | + | 105 |
| PABP | P11940 | − | + | 75 |
| FMRP | Q06787 | − | + | 75 |
| Dynein intermediate chain | O14576 | − | + | 70 |
| β4-Tubulin | Q13509 | + | n.d. | 50 |
| α-Tubulin | P05209 | − | + | 50 |
| EF1α | CAA34756 | − | − | 50 |
| Ribosomal protein P0 | NP_000993 | + | + | 34 |
| Ribosomal protein L6 | AAH20679 | + | n.d. | 34 |
| Ribosomal protein S2 | NP_002943 | + | n.d. | 32 |
| Ribosomal protein S6 | P10660 | + | + | 32 |
| Ribosomal protein S3a | L13802 | + | n.d. | 30 |
| Ribosomal protein L7a | NP_000963 | + | + | 30 |
| Ribosomal protein S8 | S11415 | + | n.d. | 28 |
| Ribosomal protein L14 | P50914 | + | n.d. | 27 |
| Ribosomal protein L10 | S34425 | + | n.d. | 26 |
| Ribosomal protein L15 | P61313 | + | n.d. | 26 |
| Ribosomal protein L21 | S55913 | + | n.d. | 20 |
Figure 2. Components of Stau1 RNPs.
(A) Western blot analysis of Stau1–PDZ and hnRNPK–PDZ precipitates (P) and supernatant fractions (S) from untreated (−) and RNase-treated (+) HEK-293 cell lysates performed with the indicated antibodies. The ribosomal proteins P0 and L7a, as well as PP1 and kinesin, associate with Stau1 RNPs, but not hnRNPK complexes, in an RNA-independent manner. In contrast, the interaction of Stau1 with nucleolin and PABP is RNA-dependent. Whereas nucleolin is only found in the Stau1–PDZ pull-down, PABP is also detected in the hnRNPK–PDZ precipitate. In contrast with kinesin, the motor protein dynein is not found in the Stau1–PDZ pull-down. The prominent cytoplasmic protein EF1α is detected in none of the precipitates. (B) Both the 28 S and 18 S rRNAs are present in untreated (−), but not RNase-digested (+), lysates.
Stau1 associates with nucleolin in an RNA-dependent manner
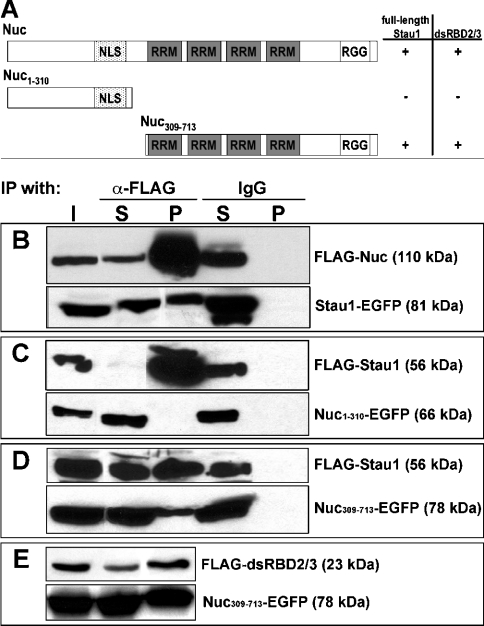
To verify the recruitment of nucleolin into Stau1 RNPs, HEK-293 cells were co-transfected with vectors encoding Stau1–EGFP and FLAG–nucleolin, followed by immunoprecipitation (Figure 3). Western blot analysis with anti-GFP and anti-FLAG antibodies revealed the presence of FLAG–nucleolin and Stau1–EGFP in anti-FLAG immunoprecipitates (Figure 3B), indicating an association of both proteins in transfected cells. To map further the domains involved in this interaction, we co-expressed two different EGFP-tagged subregions of nucleolin together with FLAG–Stau1. Whereas FLAG–Stau1 and Nuc309–Nuc713–EGFP were clearly detected in anti-FLAG immunoprecipitates (Figures 3C and 3D), Nuc1–N310–EGFP was not present (Figure 3C). In addition, immunoprecipitation was performed with anti-FLAG antibody and extracts from cells co-expressing Nuc309–Nuc713–EGFP and FLAG–dsRBD2/3, possessing only dsRBDs (double-stranded RNA-binding domains) 2 and 3 of Stau1. The precipitate contained both FLAG–dsRBD2/3 and Nuc309–Nuc713–EGFP (Figure 3E), showing that RNA-binding domains of both proteins are involved in their cytoplasmic association. None of the recombinant proteins were precipitated with a rabbit IgG control antiserum (Figures 3B–3E).
Figure 3. Mapping of the interaction domains in Stau1 and nucleolin.
(A) Schematic representation of nucleolin, including the nuclear localization signal (NLS) as well as two different types of RNA-binding domains (RRM and RGG). Table 1 shows whether full-length Stau1 or its dsRBDs 2 and 3 alone co-immunoprecipitate with full-length nucleolin or C- and N-terminal portions of this protein. (B–E) Western blots of anti-FLAG immunoprecipitates obtained from HEK-293 cells co-expressing two different recombinant proteins (shown on the right). Fusion proteins were detected with either anti-FLAG or anti-GFP antibodies. I, input; S, supernatant fraction; P, pellet fraction. The interaction between Stau1 and nucleolin (Nuc) appears to involve RNA-binding domains in both proteins.
Stau1 co-fractionates with intact ribosomes and polysomes
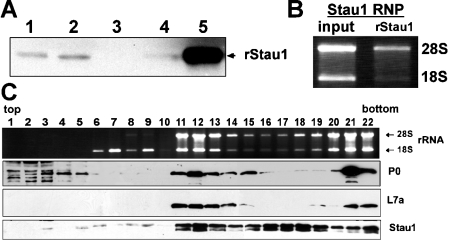
The presence of several ribosomal proteins in the Stau1-PDZ-purified material indicates that Stau1 associates with ribosomes. To evaluate further this observation, we investigated the abundance of Stau1 in different subcellular rat brain fractions (Figure 4A). In a Western blot, minor amounts of Stau1 are detected in the crude lysate and the nuclear fraction, whereas it is hardly visible in the cytosolic portion. In contrast, Stau1 is strongly enriched in the polysome fraction, indicating that the vast majority of cytoplasmic Stau1 is recruited into large RNP complexes. In addition, total RNA isolated from Stau1-PDZ affinity-purified material contains both 18 S and 28 S ribosomal rRNAs, suggesting that Stau1 complexes include intact ribosomes (Figure 4B). To characterize Stau1 RNPs in more detail, we fractionated cytoplasmic extracts from HEK-293 cells in a 5–30% sucrose gradient (Figure 4C). From recovered fractions, total RNA was isolated to determine the distribution of 40 S and 60 S ribosomal subunits, as well as intact ribosomes and polysomes. Aliquots of each fraction were analysed further on Western blots using antibodies directed against Stau1, P0 and L7a. Analysis of the distribution of 18 S and 28 S rRNAs revealed that the first five fractions do not contain ribosomal subunits, whereas fractions 6–10 preferentially include the 40 S, but not the 60 S, subunit. Fractions 1–10 contain only trace amounts of Stau1, and essentially no L7a from the large ribosomal subunit. Note that Stau1 does not co-fractionate with the isolated 40 S ribosomal subunit (fractions 6–10). The ribosomal protein P0 is detected in the first few fractions of the gradient harbouring soluble cytosolic components, and co-migrates with Stau1 only in those heavier fractions containing intact ribosomes and polysomes. This is consistent with a previous observation that, in higher eukaryotes, the P-stalk protein P0 exists in a soluble cytoplasmic and a ribosome-associated pool [27]. Interestingly, fractions 11–14 and 21 and 22, which have a high concentration of intact ribosomes/polysomes as indicated by the accumulation of both ribosomal RNAs as well as the 60 S components P0 and L7a, are highly enriched in Stau1. However, in fractions 15–19, which possess only a moderate amount of ribosomes, Stau1 is still clearly detectable. α-Tubulin is only found in fractions 1–6 (results not shown). Taken together, these findings imply that Stau1 is a component of at least two distinct tubulin-free complexes, only one of which appears to contain significant amounts of intact ribosomes or polysomes. Stau1 does not bind efficiently to the small ribosomal subunit alone.
Figure 4. Stau1 co-fractionates with ribosomes.
(A) Western blot of different rat brain fractions with anti-Stau1 serum. Lane 1, nuclear extract; lane 2, crude lysate; lane 3, cytosolic fraction; lane 4, interphase. Stau1 is highly enriched in the polysome fraction (lane 5). (B) Agarose gel analysis of total RNA isolated from Stau1–PDZ affinity-purified material reveals that it contains 18 S and 28 S rRNAs. (C) Analysis of 500 μl fractions from a 5–30% sucrose gradient performed with crude lysate from HEK-293 cells. Gel analysis of isolated RNA (uppermost row) and Western blot detection of P0, L7a, and Stau1 in individual fractions (lower three rows) is shown.
Stau1 interacts with the ribosomal protein P0
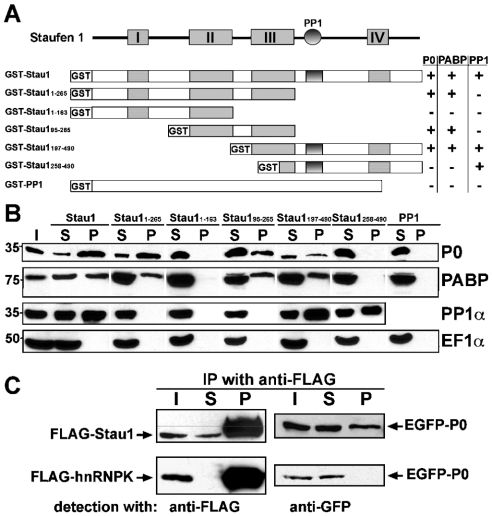
To verify the interaction between Stau1 and P0, one of the prominent ribosomal proteins isolated in the PDZ-affinity purification assay, we performed GST pull-down experiments (Figures 5A and 5B). Several distinct GST fusion proteins coupled with Sepharose beads were used in a pull-down assay with proteins obtained from an adult rat brain lysate. Fusion proteins containing full-length Stau1 (GST–Stau1) or Stau1 amino acid residues 1–265 (GST–Stau11–265), 95–265 (GST–Stau195–265) and 197–490 (GST–Stau1197–490) pulled-down P0 and PABP. In contrast, GST–Stau11–163 and GST–Stau1258–490 did not bind to P0 and PABP. Furthermore, PP1 was pulled-down with GST–Stau1, GST–Stau1197–490 and GST–Stau1258–490, all of which contain the previously characterized PP1 binding site [15]. However, the phosphatase was not isolated with GST–Stau11–265, GST–Stau11–163 and GST–Stau195–265, which lack this site, thus confirming the specificity of the pull-down assay. Moreover, GST alone or in fusion with full-length PP1α (GST–PP1α) pulled down neither P0 nor PP1 from rat brain extracts. None of the tested GST proteins bound to elongation factor EF1α.
Figure 5. Interaction of Stau1 with ribosomal protein P0.
(A) Schematic representation of Stau1, showing its dRBDs I–IV (boxes) and PP1 interaction domain (circle), as well as different GST fusion proteins. Table 1 summarizes whether the corresponding GST fusion proteins are able to pull-down rat brain P0, PP1 and PABP. (B) Western blots of GST pull-down experiments probed with specific antibodies against P0, PABP, PP1α and EF1α. Pull-downs were performed with the indicated GST fusion proteins and an adult rat brain lysate. P0 and PABP are only pulled-down by fusion proteins containing dsRBD III. PP1 precipitation requires the previously characterized PP1 interaction site. (C) Western blots of anti-FLAG immunoprecipitates obtained from HEK-293 cells co-expressing full-length or truncated versions of EGFP-tagged P0, together with either FLAG–Stau1 (upper row) or FLAG–hnRNPK (lower row). Recombinant proteins were detected with either anti-FLAG or anti-GFP antibodies. P0 co-immunoprecipitates with recombinant Stau1, but not with hnRNPK. I, input; S, supernatant fraction; P, pellet fraction.
Association of Stau1 and P0 was verified further by immunoprecipitation. From HEK-293 cell lysates, which contained P0–EGFP together with either FLAG–Stau1 or FLAG–hnRNPK, recombinant FLAG-tagged proteins were immunoprecipitated. Western blotting revealed that P0 co-precipitated with FLAG–Stau1, but not with FLAG–hnRNPK (Figure 5C). Taken together, these data suggest that, via an interaction that involves its third dsRBD and the ribosomal P-stalk protein P0, Stau1 can associate with ribosomes.
DISCUSSION
In neurons, Stau1 resides, at least in part, in large RNP complexes [12,14], and RNA-containing Stau1 particles move bidirectionally along dendritic microtubules [13]. To identify individual components of Stau1 RNA complexes, and thereby start to reveal cellular functions of these particles, we have developed a simple, yet highly specific one-step affinity purification protocol to isolate Stau1 RNPs. Parallel isolation and analysis of RNP complexes containing the RNA-binding protein hnRNPK or the neuronal protein Sharpin strongly underscores the high specificity of the purification assay. This is supported further by the absence of EF1α, the second most abundant protein in eukaryotic cells [26], in all affinity-purified fractions.
We identified the RNA-binding protein nucleolin as a prominent component of Stau1 RNP complexes. Stau1–nucleolin association is RNA-dependent, and involves RNA-binding domains in both proteins. Nucleolin is known to accumulate in nucleoli, and appears to play a role at multiple steps of ribosome biogenesis, including nucleocytoplasmic export [22]. Despite its nucleolar accumulation, it shuttles between nucleus and cytoplasm and is a component of distinct mRNPs (messenger RNPs) [22,28], such as those containing FMRP [29]. FMRP is another nucleocytoplasmic RNA-binding protein that associates with polysomes and appears to regulate translation [30]. It has been found in a complex together with Purα, Stau1 and the motor myosin Va [31]. Consistently, in the present study we detected FMRP and PABP in Stau1 RNPs by Western blotting (Figure 2 and Table 1). PABP plays a role in RNA stability, transport and translation [24]. Using MS, we identified NFAR as an additional component of Stau1 RNPs. Similar to Stau1, NFAR is a double-stranded RNA-binding protein that is present in many cell types and tissues [23]. Although it is mostly concentrated in the nucleus, it also associates with translationally quiescent mRNP complexes in the cytoplasm [32]. Thus Stau1, NFAR, nucleolin, FMRP and PABP may assemble into one complex regulating mRNA trafficking and translation. RNA helicase A (RHA), another RNA-binding protein identified in Stau1 RNPs herein, may be involved in RNP remodelling [14]. RHA is a nucleocytoplasmic shuttling protein that co-operates with Sam68 and Tap in the export of retroviral RNA from the nucleus [33]. Both RHA and Tap bind to the retroviral cis-element that mediates constitutive nuclear RNA export. It is tempting to speculate that the presence of RHA in cytoplasmic Stau1 RNPs reflects a function in cytoplasmic mRNA trafficking. Consistently, Stau1 is involved in the cytoplasmic selection, as well as subsequent recruitment and encapsidation, of genomic RNA from HIV type 1 into virus particles [34,35]. Further support for a role of RNA-binding nucleocytoplasmic shuttling proteins, such as RHA, nucleolin, NFAR and FMRP in cytoplasmic mRNA trafficking, comes from Drosophila. In fly oocytes, the nuclear processing history of oskar transcripts plays a significant role in regulating its cytoplasmic fate [36]. Thus some trans-acting factors appear to bind to a given RNA in the nucleus, remain RNA-associated after nuclear export, and thereby play a role in cytoplasmic mRNA translocation.
In mammalian neurons, endogenous Stau1 accumulates at microtubules [9,11], and recombinant Stau1 assembles into RNA-containing granules, which travel bidirectionally along dendritic microtubules [13]. The identification of tubulin and kinesin in Stau1–PDZ precipitates described in the present study suggests that this plus-end-directed microtubule-based motor protein mediates Stau1 RNP movement in different cell types. This is also consistent with the finding that 670 kDa Stau1-containing particles from rat brain co-migrate with kinesin heavy chain during gel filtration [12]. In contrast, in the present study the minus-end-directed microtubule motor dynein was not found in Stau1 RNPs. Consistently, Staufen RNPs from Xenopus laevis oocytes contain kinesin, but not dynein [37].
Our data from the sucrose gradient experiment imply that, in HEK-293 cells, Stau1 is a cytoplasmic component of at least two distinct complexes, only one of which appears to contain significant amounts of ribosomes. Neither of the groups of Stau1 particles co-migrates with tubulin in sucrose gradients (results not shown). Interestingly, large RNA granules, which were biochemically isolated from cultured neurons, contain both Stau1 and densely packed clusters of ribosomes [14], and yet rat brain Stau1 is also present in ribosome-free fractions obtained by gel filtration [12]. Thus mammalian Stau1 appears to reside in at least two distinct RNP particles. Our sucrose gradients show further that endogenous Stau1 associates with intact ribosomes, and possibly also with the isolated large 60 S subunit, but Stau1 does not co-fractionate with the small ribosomal subunit alone. Consistently, P0, a component of the 60 S ribosomal subunit, was found to interact with Stau1 during PDZ-affinity purification, GST pull-down and co-immunoprecipitation experiments. However, a previous report shows co-migration of Stau1 from COS cell extracts with both isolated 40 S and 60 S subunits in sucrose gradients [38]. The same study also demonstrated that a third dsRBD in Stau1 is necessary for an RNA-independent binding to ribosomes. This is consistent with our GST pull-down data, identifying the same dsRBD as an essential region involved in the association with ribosomal stalk protein P0, as well as PABP. The ribosomal stalk is a distinct lateral protuberance positioned on the large ribosomal subunit [39]. In eukaryotes, it consists of the acidic P proteins P0, P1 and P2. The latter two form heterodimers that bind to the ribosome through P0. The stalk interacts with elongation factors, and is important for efficient translational activity. Distinct from other ribosomal proteins, P proteins predominantly reside in the cytoplasm, where they exist in both a soluble cytoplasmic and a ribosome-associated pool [27,39]. Thus the stalk constituents are thought to assemble on to ribosomal particles during the very last step of ribosome maturation, a process that takes place in the cytoplasm. Whether a recruitment of Stau1 to the ribosomal stalk, which is suggested by our work, may influence translation in mammalian cells awaits further analysis. It is noteworthy that the Drosophila Staufen orthologue plays a critical role in translational regulation of oskar mRNA in distinct cytoplasmic subregions of fly oocytes [40].
Interestingly, several Stau1-associated components described herein were also identified in a recently published paper by Villace et al. [41], in which a different tag was used to affinity-purify Stau1-containing particles from transfected human cells. Several proteins of both large and small ribosomal subunits, including P0, distinct nucleocytoplasmic RNA-binding proteins, such as nucleolin, RHA, hnRNPU and FMRP, and PABP, as well as tubulin and kinesin, were identified in both studies. These overlapping findings strongly underscore the high specificity of both purification methods, and support further a function of Stau1 during RNA localization and translation [24].
In a previous study we have shown that, via a direct protein–protein interaction, Stau1 binds PP1 in vivo in the rat brain [15]. Here, we show that PP1 is also a component of the Stau1 RNPs, which are assembled in HEK cells. In mammals, both proteins are expressed ubiquitously [5,6,9,42], and may thus interact with each other in numerous tissues and cell types. Since Stau1 does not regulate PP1 activity [15], it may instead function as a cytoplasmic targeting factor that recruits PP1 to particular RNPs, thereby aiming the fairly unselective phosphatase activity towards specific target substrates [42]. In this context, it is interesting to note that a number of ribosomal proteins, including the P-stalk proteins, are phosphorylated in vivo [43,44]. P protein phosphorylation regulates stalk formation [45]. Thus recruitment of Stau1, PP1, and P0 into a mutual complex may enable PP1 to regulate P protein phosphorylation, stalk assembly, and thus the translational activity of the ribosome. In the future, it will be interesting to identify those components of Stau1 RNPs whose functional regulation in vivo is governed by PP1 activity.
Acknowledgments
Financial support from the Deutsche Forschungsgemeinschaft (Ki488/2–6 to S.K. and Bo1718/1–1 to H.-J.K.) and Human Frontier Science Program Organization (RG0120/1999-B to S.K.) is acknowledged. The PDZ domain purification strategy is covered by German patent application 10 2004 017 273.0. This article is in part based on a doctoral study by C.B. in the Faculty of Biology, University of Hamburg. We thank Dr Morris Reichlin, Dr Mark McNiven and Dr Evita Mohr for generously providing antibodies, and Dr Marek Tchórzewski, Dr Eun Joon Kim (KAIST, South Korea) and John Jia En Chua (Institute for Cell Biochemistry and Clinical Neurobiology, University-Hospital Hamburg-Eppendorf, Hamburg, Germany) for providing expression vectors for P0, Sharpin-PDZ and hnRNPK-PDZ respectively.
References
- 1.Zhou Y., King M. L. Sending RNAs into the future: RNA localization and germ cell fate. IUBMB Life. 2004;56:19–27. doi: 10.1080/15216540310001658886. [DOI] [PubMed] [Google Scholar]
- 2.Jansen R. P. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 3.Li P., Yang X., Wasser M., Cai Y., Chia W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. doi: 10.1016/s0092-8674(00)80504-8. [DOI] [PubMed] [Google Scholar]
- 4.Buchner G., Bassi M. T., Andolfi G., Ballabio A., Franco B. Identification of a novel homolog of the drosophila staufen protein in the chromosome 8q13–q21.1 region. Genomics. 1999;62:113–118. doi: 10.1006/geno.1999.6015. [DOI] [PubMed] [Google Scholar]
- 5.Marion R. M., Fortes P., Beloso A., Dotti C., Ortin J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickham L., Duchaine T., Luo M., Nabi I. R., DesGroseillers L. Mammalian Staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duchaine T., Wang H. J., Luo M., Steinberg S. V., Nabi I. R., DesGroseillers L. A novel murine staufen isoform modulates the RNA content of Staufen complexes. Mol. Cell. Biol. 2000;20:5592–5601. doi: 10.1128/mcb.20.15.5592-5601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duchaine T. F., Hemraj I., Furic L., Deitinghoff A., Kiebler M. A., DesGroseillers L. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J. Cell Sci. 2002;115:3285–3295. doi: 10.1242/jcs.115.16.3285. [DOI] [PubMed] [Google Scholar]
- 9.Monshausen M., Putz U., Rehbein M., Schweizer M., DesGroseillers L., Kuhl D., Richter D., Kindler S. Two rat brain Staufen isoforms differentially bind RNA. J. Neurochem. 2001;76:155–165. doi: 10.1046/j.1471-4159.2001.00061.x. [DOI] [PubMed] [Google Scholar]
- 10.Tang S. J., Meulemans D., Vazquez L., Colaco N., Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 11.Kiebler M. A., Hemraj I., Verkade P., Köhrmann M., Fortes P., Marion R. M., Ortin J., Dotti C. G. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J. Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallardo M., Deitinghoff A., Muller J., Goetze B., Macchi P., Peters C., Kiebler M. A. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köhrmann M., Luo M., Kaether C., DesGroseillers L., Dotti C. G., Kiebler M. A. Microtubule-dependent recruitment of staufen–green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krichevsky A. M., Kosik K. S. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 15.Monshausen M., Rehbein M., Richter D., Kindler S. The RNA-binding protein Staufen from rat brain interacts with protein phosphatase-1. J. Neurochem. 2002;81:557–564. doi: 10.1046/j.1471-4159.2002.00887.x. [DOI] [PubMed] [Google Scholar]
- 16.Zitzer H., Hönck H. H., Bächner D., Richter D., Kreienkamp H. J. Somatostatin receptor interacting protein defines a novel family of multidomain proteins present in human and rodent brain. J. Biol. Chem. 1999;274:32997–33001. doi: 10.1074/jbc.274.46.32997. [DOI] [PubMed] [Google Scholar]
- 17.Lim S., Sala C., Yoon J., Park S., Kuroda S., Sheng M., Kim E. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol. Cell. Neurosci. 2001;17:385–397. doi: 10.1006/mcne.2000.0940. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Kim E., Naisbitt S., Hsueh Y. P., Rao A., Rothschild A., Craig A. M., Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J. Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soltau M., Berhorster K., Kindler S., Buck F., Richter D., Kreienkamp H. J. Insulin receptor substrate of 53 kDa links postsynaptic shank to PSD-95. J. Neurochem. 2004;90:659–665. doi: 10.1111/j.1471-4159.2004.02523.x. [DOI] [PubMed] [Google Scholar]
- 21.Rehbein M., Kindler S., Horke S., Richter D. Two trans-acting rat-brain proteins, MARTA1 and MARTA2, interact specifically with the dendritic targeting element in MAP2 mRNAs. Brain Res. Mol. Brain Res. 2000;79:192–201. doi: 10.1016/s0169-328x(00)00114-5. [DOI] [PubMed] [Google Scholar]
- 22.Ginisty H., Sicard H., Roger B., Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 23.Saunders L. R., Barber G. N. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 24.Dreyfuss G., Kim V. N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 25.Lee C. G., Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J. Biol. Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 26.Condeelis J. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem. Sci. 1995;20:169–170. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- 27.Tsurugi K., Ogata K. Evidence for the exchangeability of acidic ribosomal proteins on cytoplasmic ribosomes in regenerating rat liver. J. Biochem. (Tokyo) 1985;98:1427–1431. doi: 10.1093/oxfordjournals.jbchem.a135410. [DOI] [PubMed] [Google Scholar]
- 28.Borer R. A., Lehner C. F., Eppenberger H. M., Nigg E. A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 29.Ceman S., Brown V., Warren S. T. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol. Cell. Biol. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaeffer C., Beaulande M., Ehresmann C., Ehresmann B., Moine H. The RNA binding protein FMRP: new connections and missing links. Biol. Cell. 2003;95:221–228. doi: 10.1016/s0248-4900(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 31.Ohashi S., Koike K., Omori A., Ichinose S., Ohara S., Kobayashi S., Sato T. A., Anzai K. Identification of mRNA/protein (mRNP) complexes containing Purα, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- 32.Brzostowski J., Robinson C., Orford R., Elgar S., Scarlett G., Peterkin T., Malartre M., Kneale G., Wormington M., Guille M. RNA-dependent cytoplasmic anchoring of a transcription factor subunit during Xenopus development. EMBO J. 2000;19:3683–3693. doi: 10.1093/emboj/19.14.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy T. R., Tang H., Xu W., Wong-Staal F. Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA. Oncogene. 2000;19:3570–3575. doi: 10.1038/sj.onc.1203676. [DOI] [PubMed] [Google Scholar]
- 34.Mouland A. J., Mercier J., Luo M., Bernier L., DesGroseillers L., Cohen E. A. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatel-Chaix L., Clement J. F., Martel C., Beriault V., Gatignol A., DesGroseillers L., Mouland A. J. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hachet O., Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature (London) 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- 37.Yoon Y. J., Mowry K. L. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131:3035–3045. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- 38.Luo M., Duchaine T. F., DesGroseillers L. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem. J. 2002;365:817–824. doi: 10.1042/bj20020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalo P., Reboud J. P. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 40.Micklem D. R., Adams J., Grunert S., St Johnston D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19:1366–1377. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villace P., Marion R. M., Ortin J. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 2004;32:2411–2420. doi: 10.1093/nar/gkh552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceulemans H., Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 43.Nusspaumer G., Remacha M., Ballesta J. P. Phosphorylation and N-terminal region of yeast ribosomal protein P1 mediate its degradation, which is prevented by protein P2. EMBO J. 2000;19:6075–6084. doi: 10.1093/emboj/19.22.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsurugi K., Collatz E., Todokoro K., Ulbrich N., Lightfoot H. N., Wool I. G. Isolation of eukaryotic ribosomal proteins. Purification and characterization of the 60 S ribosomal subunit proteins La, Lb, Lf, P1, P2, L13′, L14, L18′, L20, and L38. J. Biol. Chem. 1978;253:946–955. [PubMed] [Google Scholar]
- 45.Tchorzewski M., Boguszewska A., Dukowski P., Grankowski N. Oligomerization properties of the acidic ribosomal P-proteins from Saccharomyces cerevisiae: effect of P1A protein phosphorylation on the formation of the P1A–P2B hetero-complex. Biochim. Biophys. Acta. 2000;1499:63–73. doi: 10.1016/s0167-4889(00)00108-7. [DOI] [PubMed] [Google Scholar]