Abstract
We have analysed the transcriptional regulation of the human histone H3 genes using promoter deletion series, scanning mutagenesis, specific mutagenesis and electrophoretic mobility-shift assay experiments. The promoters of five of the six examined histone H3 genes showed near-maximal activity at lengths of 133–227 bp: H3/d 198 bp, H3/h 147 bp, H3/k 133 bp, H3/m 227 bp, H3/n 140 bp (exception H3/i). To search for functional cis-elements within these regions, we performed scanning mutagenesis of the two histone H3 promoters H3/k and H3/m. Mutagenesis revealed that the functional framework of the histone H3 promoters consists of a TATA box and two tandemly arranged CCAAT boxes in relatively fixed positions. Alterations of the distance between the CCAAT boxes and of the distance between the CCAAT boxes and the TATA box resulted in significant loss of activity. In electrophoretic mobility-shift assay experiments, the factor CBF (CCAAT-binding factor)/NF-Y (nuclear factor-Y) bound to isolated CCAAT boxes of the H3/k promoter. This suggests that an initiation complex is formed on the histone H3 promoter that has a defined structure and limited flexibility, consisting of two molecules of CBF/NF-Y and further (general or specific) transcription factors.
Keywords: CCAAT-binding factor/nuclear factor-Y (CBF/NF-Y), CCAAT box, cell cycle, histone H3 gene, promoter structure, transcriptional regulation
Abbreviations: ATF, activating transcription factor; CBF, CCAAT-binding factor; Cdc, cell division cycle; EMSA, electrophoretic mobility-shift assay; HEK-293 cells, human embryonic kidney 293 cells; NF-Y, nuclear factor-Y; UTR, untranslated region
INTRODUCTION
The human genome contains 14 histone H3 genes, of which 11 belong to the replication-dependent group [1]. The replication-dependent group consists of histone H3 genes that are transcribed during the S phase of the cell cycle in co-ordination with DNA replication. In contrast, the replacement subtype H3.3 is encoded either by the H3.3A or the H3.3B gene [2]. Ten of the replication-dependent genes code for the H3 subtype histone H3.1 and one of them, H3/n, codes for the subtype histone H3.2, which contains a single amino acid variation (Ser-96 instead of Cys-96). In the present study, we exclusively refer to the 11 replication-dependent histone H3 genes.
The replication-dependent histone genes are devoid of introns and their mRNAs are not polyadenylated. Instead, they contain conserved 3′-end processing elements, including a 16 bp stem-loop motif which is involved in histone-specific 3′-end processing [3–5], nuclear export [6] and stability of the histone mRNA [7]. The replication-dependent histone genes are localized in four histone gene clusters on chromosomes 1 and 6, which show a non-regular arrangement of histone genes [1,8–11]. They are expressed in a cell-cycle-dependent manner, the regulation of which is both transcriptional and post-transcriptional (see [12–16] for reviews).
The transcriptional regulation of the replication-dependent histone genes is unique in each histone gene class. The promoters of the individual histone gene classes (H1, H2A, H2B, H3 and H4) are differently organized and, as far as we know, class-specifically regulated. In some cases, differences between the transcriptional regulation of individual genes of the same class were reported [17,18]. How the transcription of the various histone gene classes is quantitatively regulated with regard to the stoichiometry of the nucleosome and how a common cell-cycle regulation is accomplished is not yet understood.
Several studies have dealt with histone gene regulation and only few of them have concentrated on histone H3 genes. We systematically analysed the transcriptional regulation of the histone H3 genes. Since the histone H3 promoters show very few sequence homologies, we analysed whether they are jointly or individually regulated. In this analysis, we determined the cis-elements in the histone H3 promoters and the transcription factors that bind to them. We used promoter deletion series (with reporter gene assays), scanning mutagenesis (with reporter gene assays) and EMSAs (electrophoretic mobility-shift assays) to analyse the histone H3 promoters.
EXPERIMENTAL
Cell culture
HEK (human embryonic kidney)-293 cells were obtained from DSMZ (Braunschweig, Germany) and were grown in Dulbecco's modified Eagle's medium with 10% (v/v) fetal bovine serum. Culture conditions were 37 °C and 5% CO2.
Reporter gene assays
Reporter gene assays were performed with the Dual-Luciferase® Reporter Assay system (Promega, Madison, WI, U.S.A.). Promoter fragments were cloned into the pGL3-basic vector in such a way that the fusion site between the promoter and the firefly luciferase-coding strand (luc+) was the ATG of the start codon. Plasmid DNA was purified using the EndoFree® Plasmid Maxi kit (Qiagen, Hilden, Germany) and quantified spectrophotometrically. The endotoxin-free plasmid DNA was then transfected into the adherently growing HEK-293 cells in six-well plates with the Effectene™ Transfection system (Qiagen) according to the manufacturer's instructions. Cells were transfected at 40–80% confluence and incubated for 18 h. For standardization, 10 ng of the Renilla luciferase vector pRLCMV was co-transfected. Cell lysis and measurement of luciferase activities were performed according to the manufacturer's instructions. For each promoter construct at least four independent experiments were performed.
Mutagenesis
Mutagenesis of promoters was performed using the GeneEditor™ in vitro site-directed mutagenesis system (Promega) according to the manufacturer's instructions. The oligonucleotide sequences used in the mutagenesis reactions are listed as follows, and the underlined nucleotides represent mutated bases. Mutagenic primers used for the generation of K5mut1–K5mut8 (H3/k promoter mutants 1–8): K5mut1, 5′-GGAGACCATGAACTGCTAATGAGTGACTCGGAATAGGTGAACAACAAAAATTTGAG-3′; K5mut2, 5′-GAACTGCTAAGTCTGTCAGAGGAAGCTTGTCCACACAAAAATTTGAGTCCTTCGCC-3′; K5mut3, 5′-GTCAGAGGAATAGGTGAACAACAACCCGGGTCTGCCTTCGCCAATCCGGTTACTGTTGG-3′; K5mut4, 5′-GGTGAACAACAAAAATTTGAGTCCTTATAACCGAATGTTACTGTTGGGTAGGCCTTCAGC-3′; K5mut5, 5′-GAGTCCTTCGCCAATCCGGTTAAGTGGTTTGCGGCCTTCAGCATACTTTTGTCC-3′; K5mut6, 5′-CCGGTTACTGTTGGGTAGGCCGGACTACGCATTTTGTCCAATCAGCTTCAGACTCTC-3′; K5mut7, 5′-GGGTAGGCCTTCAGCATACTTTTTGAACCGACTAGTCAGACTCTCACTATAAATAAGCGGC-3′; K5mut8, 5′-GCATACTTTTGTCCAATCAGCTTCAGCAGAGACAGCGCAATAAGCGGCTAGCTTTCTCTTTCTCC-3′.
Mutagenic primers used for the generation of M4mut1–M4mut18 (H3/m promoter mutants 1–18): M4mut1, 5′-GCAGCTAAGGGGTTAACAAAAGTCATGACGAGTAGCTACGGTAATGGGCAGG-3′; M4mut2, 5′-GGGCAGGAGCCTCTCTTAATCTTACCAACTTACCAGAGATGGACCAATCCAAGAAGGGC-3′; M4mut3, 5′-CTCTTAATCTGCAACCAGGCACAGATCGTTCAACCGAAAAGAAGGGCGCGGGGATTTTTG-3′; M4mut4, 5′-GCACAGAGATGGACCAATCCAAGACTTTATATTTTCTTTTTGAATTTTCTTGGGTCCAATAG-3′; M4mut5, 5′-CCAAGAAGGGCGCGGGGATTTTGTCCGGGGAGTGGGTCCAATAGTTGGTGGTCTG-3′; M4mut6, 5′-GGATTTTTGAATTTTCTTGGGTAACCGAGTTGGTGGTCTGACTC-3′; M4mut8, 5′-CCAATAGTTGGTGGTCTGACTCGCGCCCCGAAGAGTAGCTCTTTCCTTTCC-3′; M4mut9, 5′-GGTGGTCTGACTCTATAAAAGAAGCTGCTAGAGGGAAGTTCCTCCACAGACGTCTCTGCAGGC-3′; M4mut10, 5′-GAAGAGTAGCTCTTTCCTTTCCGAACACTCATGAGAGGCAGGCAAGCTTTTCTGTGGTTTTGCC-3′; M4mut11, 5′-CCTTTCCTCCACAGACGTCTCTGCAGTACCTAGGGGAGTGTTGGGTGCCATGGAAGACGCCAAAAAC-3′; M4mut16, 5′-GAATTTTCTTGGGTCCAATAGTTGTGTTGAGGACTCTATAAAAGAAGAGTAGCTCTTTCC-3′; M4mut18, 5′-GGGGTTAACAAAATGACGTCAGAGACTTGTCGGTAATGGGCAGGAGCCTCTC-3′.
EMSAs
EMSAs were performed with double-stranded oligonucleotides with a length of 27–30 bp. The double-stranded oligonucleotides were generated by annealing of 500 pmol of each single-stranded complementary oligonucleotide in 20 μl of 0.1 M NaCl by heating the solution to 75 °C and slowly cooling it to room temperature (∼20 °C) within 2 h. The double-stranded oligonucleotides were radioactively labelled with [32P] in the following reaction mixture: 2 μl of double-stranded oligonucleotide solution (1.75 pmol/μl), 1 μl of 10× T4-kinase buffer, 5 μl of double-distilled water, 1 μl of T4-polynucleotide kinase and 1 μl of 5′-[γ-32P]ATP (aqueous solution, specific activity 3000 Ci/mmol and 10 mCi/ml). The mixture was incubated for 30 min at 37 °C and the labelled double-stranded oligonucleotide was purified with the Nucleotide Removal kit (Qiagen). The resulting solutions had activities of 75000–150000 c.p.m./μl. Band-shift experiments were performed using the Gel Shift Assay System kit (Promega). Binding reactions were set up as follows: 2 μl of 5× binding buffer [20% (v/v) glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM dithiothreitol, 250 mM NaCl, 50 mM Tris/HCl (pH 7.5), 0.25 mg/ml poly(dI-dC)·poly(dI-dC)], 2 μl of HeLa nuclear extract (2.4 mg/ml) or 2 μl of isolated NF-Y (nuclear factor-Y) complex (100 ng), competitor oligonucleotide (optional) and double-distilled water to a volume of 9 μl. The reaction mixture was preincubated for 10 min at room temperature, and then 1 μl of the labelled double-stranded oligonucleotide was added and mixed. The mixture was further incubated for 20 min and analysed on an EMSA gel. For the supershift experiments, the preincubated reaction mixture was further incubated for 10 min with 1 μl of CBF (CCAAT-binding factor)-A antiserum (1 μg/μl) (FL-207; Santa Cruz Biotechnology) before the addition of the labelled oligonucleotide. The EMSA gel had the following composition: 3.75 ml of 10× TBE (90 mM Tris, 90 mM boric acid, 2.5 mM EDTA, pH 8.3), 10 ml of acrylamide solution [30% (w/v), 0.8% (w/v) bisacrylamide], 3.13 ml of 60% glycerol, 57.5 ml of double-distilled water, 38 μl of N,N,N′,N′-tetramethylethylenediamine and 563 μl of 10% (w/v) ammonium persulphate for polymerization. The gel was run for 2 h at 400 V, dried and analysed quantitatively with the PhosphoImager system FLA-3000 (Fujifilm, Tokyo, Japan). The analysis software was ‘Aida 2.2’ (Raytest, Straubenhardt, Germany).
The sequences of the oligonucleotides are listed as follows (the sequences of the complementary oligonucleotides are not shown): KProx, 5′-TCAGCATACTTTTGTCCAATCAGCTTCAGA-3′; KProx mut, 5′-TCAGCATACTTTTGTACGATCAGCTTCAGA-3′; KDist, 5′-AATTTGAGTCCTTCGCCAATCCGGTTACTG-3′; NF-Y, 5′-AGACCGTACGTGATTGGTTAATCTCTT-3′; NF-Y mut, 5′-AGACCGTACGAAATACGGGAATCTCTT-3′; CDP/Cut, 5′-ACCCAATGATTATTAGCCAATTTCTGA-3′.
Expression of the NF-Y complex
Cloning
The coding regions of the respective proteins were first amplified from plasmid DNA (gift from R. Mantovani, Dipartimento di Biologia Animale, Università di Modena e Reggio Emilia, Modena, Italy) using specific primer pairs with appropriate restriction sites. The various coding regions were cloned as follows: mouse NF-YA as an NdeI–SacI fragment into the NdeI–SacI sites of pJK45 for expression of a fusion protein with a C-terminal His tag. pJK45 is a modified pET21b in which the PstI–NruI fragment, comprising the origin of replication, of pACYC177 was inserted into the PstI–PshAI fragment of pET21b. The coding region of human NF-YB was inserted as a BamHI–SmaI fragment into the BamHI–SmaI sites of pGEX4T-1 to generate a fusion protein with a N-terminal GST (glutathione S-transferase) tag. Human NF-YC was isolated as an SpeI–SacI fragment and cloned into the SpeI–SacI sites of pET41a (N-terminal-GST-tagged construct).
Recombinant protein expression and purification
The NF-Y subunit proteins were expressed in the Escherichia coli strain BL21 (DE3) as follows: cultures were grown at 37 °C to D600∼1.0 and then were shifted to 25 °C. After the temperature attained equilibrium, expression was induced by the addition of isopropyl β-D-thiogalactoside in appropriate concentrations (0.2 mM for the NF-YA, 0.4 mM for the NF-YB/YC co-expression). Cells were further grown for 3 h with shaking at 220 rev./min. Cells were then collected by low-speed centrifugation, resuspended in buffer A [50 mM Tris/HCl (pH 8.0), 400 mM NaCl and 5 mM 2-mercaptoethanol], and lysed by sonication. The recombinant NF-YA protein was immobilized on Ni2+-nitrilotriacetate–agarose (Qiagen) according to the manufacturer's instructions. The immobilized NF-YA was used as a bait to isolate the NF-YB/C dimer from the soluble fraction recovered by high-speed centrifugation of the co-expression lysate. After 3 h of incubation, the supernatant was removed and the resin was washed extensively with buffer A. The formed NF-Y trimeric complex was eluted with imidazole and was applied on to a gel filtration column Hiload 16/60 Superdex 200 (Amersham Bioscience) equilibrated with buffer A. The purified complex was concentrated in Vivaspin ultrafiltration columns (Vivascience, Hannover, Germany) to a final concentration of approx. 1 mg/ml.
RESULTS
Sequence analysis of the histone H3 promoters
The histone H3 promoters are known to be very divergent and contain little sequence homology [15,19]. In the alignment of the first 500 bp of the promoter sequence, the most significant sequence similarity was a composite element consisting of a ‘proximal’ CCAAT box and a TATA box at a fixed distance (22 bp, counted from the start of the motifs), which was located close to the transcriptional start site in the promoters. In seven of the 11 promoters (H3/a, H3/c, H3/f, H3/h, H3/k, H3/m and H3/n), a ‘distal CCAAT box’ was found upstream of the CCAAT/TATA-composite element at a distance of 41/42 bp (counted from the start of the motifs). In two further promoters, a distal CCAAT box was found at a distance of 32 bp (H3/b) or 68 bp (H3/d). Some promoters contain up to two more upstream CCAAT boxes. In addition, the H3/m and the H3/n promoters contain an ATF (activating transcription factor) box, comprising an 8 bp palin-dromic TGACGTCA motif as described by van Wijnen et al. [20].
Promoter deletion experiments of six human histone H3 promoters
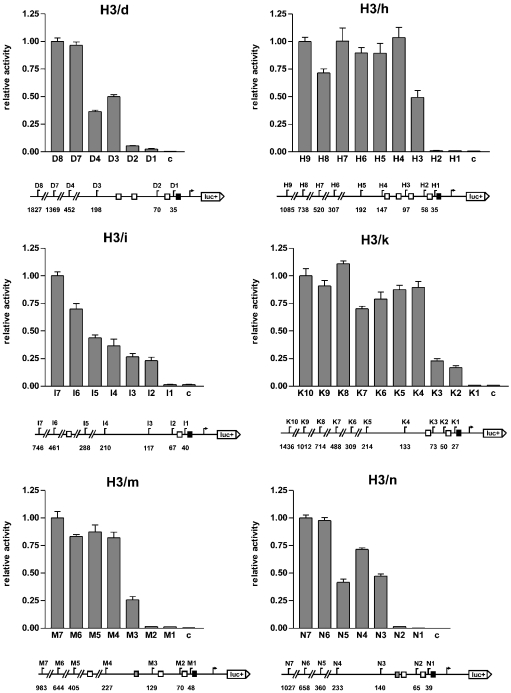
To determine the functional lengths of the human histone H3 promoters, promoter deletion experiments of six human histone H3 genes (H3/d, H3/h, H3/i, H3/k, H3/m and H3/n) were performed (see Figure 1). Promoter fragments of different sizes with a length between 27 and 1827 bp (based on transcriptional start site) were cloned into the reporter gene vector pGL3-basic (fusion site: start codon ATG), and the reporter gene assay was performed as described in the Experimental section.
Figure 1. Reporter gene analysis of histone H3 promoter deletion series in the HEK-293 cell line.
The reporter gene activity of the indicated constructs is shown as a statistical result (relative to the longest construct, which is set to the value of 1.0; error bars, S.E.M.; c, control construct pGL3-basic) of five independent experiments. A diagram of the promoters with the lengths of the constructs indicated is shown below each graph. Arrow, transcription start site; numbers, distance to transcription start site in bp; luc+, firefly luciferase coding sequence; black boxes, TATA boxes; white boxes, CCAAT boxes; grey boxes, ATF boxes; fusion site, ATG start codon.
Within the first 500 bp upstream of the respective H3 genes, promoter fragments with a length of 133–227 bp were identified as ‘proximal, functional promoters’. They contain the proximal TATA/CCAAT composite element and at least one more upstream CCAAT box. In the H3/m and H3/n genes these fragments also contain an upstream ATF box. Shorter fragments without a second, distal CCAAT box (constructs D2, H2, K3, M2 and N2) were substantially less active, and promoter fragments that contained only the TATA box were only marginally active (constructs D1, H1, I1, K1, M1 and N1). Longer promoter fragments in the H3/h, H3/k and H3/m genes (H5–H9, K5–K10 and M5–M7) were not significantly more active than the proximal, functional promoters (H4, K4 and M4). In the H3/d and H3/n genes, longer fragments of 1369 bp (D7) and 658 bp (N6) respectively were approx. twice as active as the proximal, functional promoters. The H3/i promoter was an exception because it does not contain a second, distal CCAAT box in the vicinity of the proximal CCAAT box. The promoter construct I2, which contains the proximal TATA/CCAAT composite element exhibited an activity of 25% when compared with the 746 bp construct I7. Owing to the gradual increase in activity of the successive longer promoter fragments (I3–I7), no definite proximal, functional promoter could be identified for the H3/i gene.
In conclusion, with the promoter deletion series described above, proximal functional promoters could be defined in five of the six examined histone H3 promoters. These promoters showed full or almost full activity within the first 500 bp upstream of the respective H3 genes: H3/d, 198 bp; H3/h, 147 bp; H3/k, 133 bp; H3/m, 227 bp; H3/n, 140 bp. For the H3/i gene, no proximal, functional promoter could be defined. The proximal, functional promoters contain a TATA box and a proximal CCAAT box, which is just upstream of the TATA box. Furthermore, they contain a second, ‘distal’ CCAAT box, in some cases further CCAAT boxes, and for H3/m and H3/n, an ATF box.
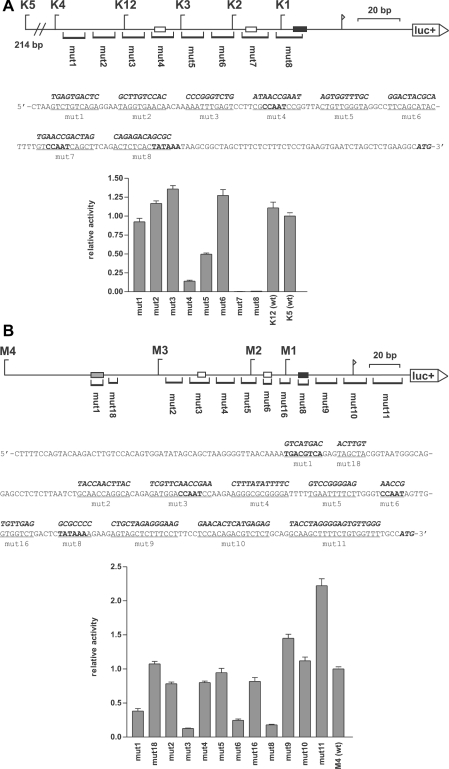
Scanning mutagenesis of the H3/m and H3/k promoters
In addition to the TATA, CCAAT and ATF elements, no further common elements had been found on alignment of the H3 gene promoters. To elucidate the function of the TATA, CCAAT and ATF elements and to find possible additional elements, scanning mutagenesis was performed along the entire length of the histone H3/k and H3/m promoters (see Figure 2). Within the histone H3/k promoter, just three main cis-elements were essential for promoter function: the TATA box and two tandemly arranged CCAAT boxes. One of the twin CCAAT boxes, the ‘proximal CCAAT box’, is located just upstream of the TATA box and the other, the ‘distal CCAAT box’, is located 42 bp upstream of the proximal CCAAT box. Mutation of the TATA box or the proximal CCAAT box in the H3/k promoter (mut7 and mut8) led to an almost complete loss of promoter activity, and the mutation of the distal CCAAT box (mut4) led to an 85% loss of activity. Mutations of other regions, namely the region upstream of the distal CCAAT box (mut1, mut2 and mut3) and the region between the two CCAAT boxes (mut5 and mut6) did not significantly affect promoter activity (mut5 was 50% less active for unknown reasons).
Figure 2. Scanning mutagenesis of the promoters of histone H3 genes H3/k (A) and H3/m (B).
Top panels: to scale diagram of the promoters. luc+, firefly luciferase coding sequence; arrow, transcription start site; black boxes, TATA box; white boxes, CCAAT boxes; grey box, ATF box. The mutated areas and the names of the mutated constructs are indicated at the bottom of each diagram. The positions of the deletion constructs are shown on top of each diagram for orientation. Middle panels: nucleotide sequences of the promoters. The regions that were mutated in the respective mutation constructs are underlined and the mutated sequence is shown above in bold italic style. The start codon of the firefly luciferase coding sequence, the CCAAT boxes, the TATA boxes and the ATF box are shown in bold. Bottom panels: the reporter gene activity of the constructs in the HEK-293 cell line is shown as a statistical mean for four independent experiments [relative to the wild-type promoter K5 (wt) or M4 (wt) respectively, which is set to the value of 1.0; error bars, S.E.M.].
Similarly, in the H3/m promoter the TATA box, the proximal CCAAT box and the distal CCAAT box were the essential cis-elements. Mutations of these elements (mut8, mut6 and mut3 respectively) led to a severe loss of activity over the range of 80–90%. Additionally, the mutation of the ATF box (mut1) led to a 60% loss of promoter activity. Mutation of the other promoter regions, even the mutation of the transcription initiation site (mut10), did not affect promoter activity. Surprisingly, a mutation within the 5′-untranslated region of the histone H3/m gene (mut11) led to a rise in promoter activity. The reason for this effect remained unsolved.
Thus the scanning mutagenesis of the H3/k and the H3/m promoters revealed that a tandemly arranged pair of CCAAT boxes and a TATA box define the functional framework of the histone H3 promoters. In the H3/m promoter, an ATF box also contributes to the promoter function.
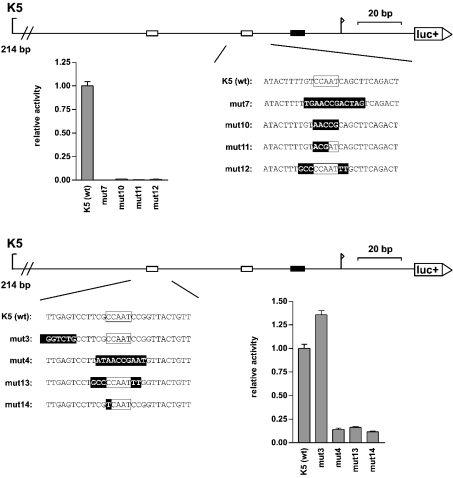
Mutagenesis of the CCAAT boxes of the H3/k promoter
Several different transcription factors can bind to CCAAT boxes. Since their sequence requirements vary slightly, we further characterized the sequence specificities of the CCAAT boxes of the histone H3/k promoter (Figure 3). We tried whether mutations within the ‘CCAAT’-motifs were tolerated. A mutation of the proximal H3/k CCAAT box to ‘ACGAT’ (mut11) led to an almost complete loss of promoter activity. Similarly, a point mutation within the distal CCAAT box to ‘TCAAT’ (mut14) caused the same loss of promoter activity as a mutation of the whole distal CCAAT box (mut4).
Figure 3. Specific mutagenesis of the CCAAT boxes of the histone H3/k promoter.
To scale diagrams of the histone H3/k promoter: luc+, firefly luciferase coding sequence; arrow, transcription start site; black boxes, TATA box; white boxes, CCAAT boxes. At the bottom of each diagram, the wild-type sequence of the region surrounding the proximal CCAAT box (upper panel) and the distal CCAAT box (lower panel) and the mutated sequence of the mutation constructs are shown (mutated bases are inversely shaded). The reporter gene activity of the constructs in the HEK-293 cell line is shown below each diagram as a statistical mean for four independent experiments [relative to the wild-type promoter K5 (wt), which is set to the value of 1.0; error bars, S.E.M.].
The factor NF-Y (also called CBF) is the principal CCAAT-box-binding activator. Bi et al. [21] and Mantovani [22] reported that the NF-Y consensus-binding sequence is YRRCCAATCA (with Y representing the pyrimidine base and R the purine base). The surrounding bases of the CCAAT boxes in the histone H3 promoters correspond loosely to this NF-Y consensus sequence. A mutation of the surrounding bases of the proximal CCAAT box of the histone H3/k promoter (three upstream and two down-stream, mut12) to a non-consensus NF-Y sequence led to an almost complete loss of promoter activity. Similarly, a mutation of the surrounding bases of the ‘CCAAT’ motif of the distal H3/k CCAAT box to a non-consensus NF-Y sequence (mut13) caused the same loss of promoter activity as a mutation of the whole ‘CCAAT’ box (mut4).
Thus either a change in the ‘CCAAT’ motif or a change in the surrounding bases to a non-consensus NF-Y sequence led to a complete loss of function of both H3/k CCAAT boxes. This points to NF-Y as the relevant transcription factor.
EMSA of the CCAAT boxes of the H3/k promoter
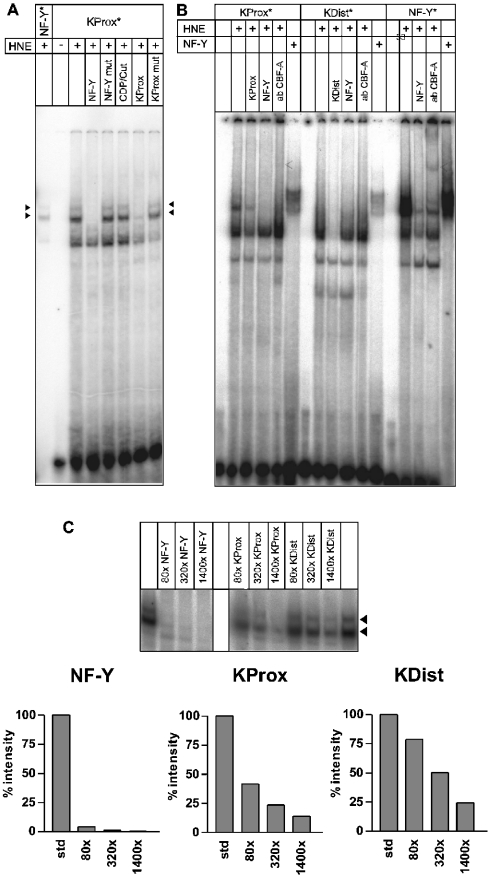
The results described above indicate that the CCAAT boxes are the most important, if not the only, cis-elements besides the TATA box in the histone H3 promoters. Furthermore, the sequences of the CCAAT boxes correspond loosely to an NF-Y consensus-binding sequence and their mutation to a non-consensus NF-Y recognition sequence abolishes the CCAAT-box function. To elucidate the identity of the CCAAT-box-binding factor, EMSAs were performed with HeLa nuclear extract and double-stranded oligonucleotides comprising the CCAAT boxes of the H3/k as probes. The probes were 30 bp long and contained the wild-type CCAAT sequence with the CCAAT motif at positions 16–20.
The proximal CCAAT box of the H3/k promoter ‘KProx’ showed three bands (Figure 4A), the upper two of which formed a double band consisting of a strong lower and a weak upper band. This double band had the same pattern and showed the same shift as the double band of labelled NF-Y consensus oligonucleotide. Furthermore, it was competed by NF-Y consensus oligonucleotide and unlabelled KProx, but not by the ‘KProx mut’ oligonucleotide with a mutated CCAAT box. Thus the upper double band depends on a functional CCAAT box. Moreover, it was not competed by CDP/Cut consensus oligonucleotide, the binding sequence of another possible CCAAT-box-binding protein. In contrast, the lower band was competed by ‘KProx mut’, but not by the NF-Y consensus oligonucleotide. The lower band therefore represents a specific binding activity, but it was not attributed to a functional CCAAT box. Taken together, the upper double band represents, with high probability, an NF-Y bandshift complex.
Figure 4. Autoradiograms of the EMSA experiments.
(A) Competition experiments with the proximal CCAAT box of the histone H3/k promoter (KProx). Composition of the individual incubation mixtures: labelled oligonucleotides are marked with an *, proteins added (HNE, HeLa nuclear extract; NF-Y, isolated NF-Y) are indicated by ±, competitors (80-fold molar excess) and antibody are indicated in the lower part of the heading. The NF-Y double bands are marked by black arrowheads. It may be noted that the specific activity of the NF-Y * probe used was much lower than the specific activity of the KProx* probe. (B) Supershift and complex-binding experiments with HNE and recombinant NF-Y. The experimental set-up and the heading is the same as for (A). To the incubation mixture indicated by ab CBF-A, 1 μg of antibody raised against NF-YB was added. Approx. 100 ng of isolated NF-Y complex was added to the labelled oligonucletides where indicated. The supershift band is indicated by the open arrow. (C) Titration competition experiments. The binding of labelled double-stranded oligonucleotides containing the consensus sequence of the binding site of NF-Y was competed by increasing concentrations of unlabelled double-stranded oligonucleotides (NF-Y, KProx and KDist). The molar excess of unlabelled competitor oligonucleotide is indicated. The intensities of the signals were quantified with respect to the PhosphoImager signals by the Aida 2.2 software from Raytest and expressed as the percentage ratio to the uncompeted signal intensities.
To reinforce the conclusions from the competition experiments, we added an antibody against the NF-YB (CBF-A) subunit to the reaction mixture to form a complex that should generate a supershift. In fact, addition of the antibody generated an extremely weak supershift with the KProx oligonucleotide complex (Figure 4B). The retardation was the same as for the complex formed with the NF-Y consensus oligonucleotide. The binding of the antibody to the primary complex was better reflected by the observed attenuation of the signal of the primary complex.
In addition to the supershift experiments, we analysed the complex formation with isolated recombinantly expressed NF-Y subunits (Figure 4B). The results of these experiments were the same as for the supershift experiments: the NF-Y consensus oligo formed a strong complex with NF-Y, the KProx oligonucleotide also formed a complex with NF-Y, whereas the complex with the KDist oligonucleotide was weaker (Figure 4B).
This is in line with the results from the initial EMSA experiments with the distal CCAAT box of the H3/k promoter. ‘KDist’ generated an electrophoretic retardation signal with nuclear extract from HeLa cells similar to the characteristic NF-Y band-shift complex (Figure 4B, middle panel) but of weak intensity. This signal was completely quenched by the addition of unlabelled KDist oligonucleotide, but only partially by the addition of unlabelled NF-Y consensus oligonucleotide. These experiments together indicate clearly that NF-Y binds to KProx and KDist but with different affinities.
The differences in the binding intensities of NF-Y transcription factor to the two CCAAT boxes of the H3/k promotor were analysed by titration competition experiments with the double-stranded oligonucleotides from the different regions for NF-Y consensus oligonucleotide binding (Figure 4C). Competition with the same unlabelled NF-Y oligonucleotide was complete with an 80-fold excess, whereas an 80-fold excess of KProx only resulted in a reduction in the signal intensity to 30% of the uncompeted signal. The competition with the KDist oligonucleotide was even weaker; only a 1400-fold excess of the unlabelled oligonucleotide quenched the signal to 25%.
All the experiments point to a binding of NF-Y to the CCAAT boxes within the H3/k promoter but with different affinities.
Change of distance between the elements of the H3/k-promoter
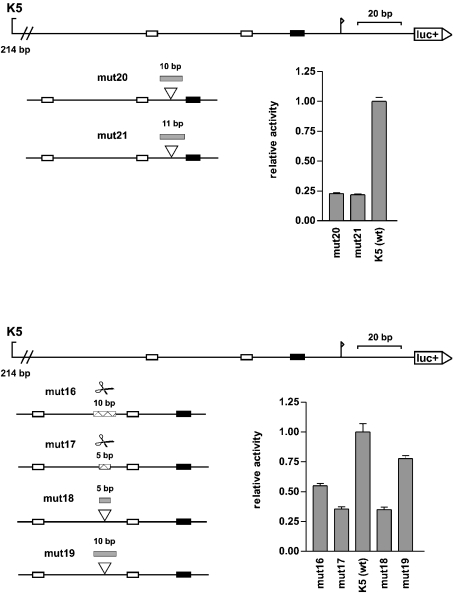
The distances between the proximal CCAAT box and the TATA box on the one hand and the proximal CCAAT box and the distal CCAAT box on the other are very similar in the histone H3 promoters. The proximal CCAAT boxes, which are found in all histone H3 promoters, are 22 bp (in two cases 20 bp) upstream of the TATA boxes (based on the starting points of these motifs), and the distal CCAAT boxes (present in nine of the 11 histone H3 promoters) are in seven histone H3 promoters 41 or 42 bp upstream of the proximal CCAAT boxes (based on the starting points of these motifs). To find whether these conserved distances have a functional relevance, H3/k promoter mutants were constructed with altered distances between these elements (see Figure 5). Insertion of 10 or 11 bases between the proximal CCAAT box and the TATA box of the H3/k promoter caused a 75% reduction in the promoter activity (mut20 and mut21).
Figure 5. Changes in the distances between the relevant cis-elements in the histone H3/k promoter.
To scale diagrams of the histone H3/k promoter: luc+, firefly luciferase coding sequence; arrow, transcription start site; black boxes, TATA boxes; white boxes, CCAAT boxes. At the bottom of each diagram, the positions of inserted or deleted sequences are shown: grey boxes, inserted sequences; white boxes with pair of scissors, deleted sequences. The reporter gene activity of the constructs in the HEK-293 cell line is shown as a statistical mean for four independent experiments [relative to the wild-type promoter K5 (wt), which is set to the value of 1.0; error bars, S.E.M.].
Changing the distance between the two CCAAT boxes of the H3/k promoter also caused a loss of promoter activity. When an entire helix turn (∼10 bp) was deleted (mut16) the loss of activity was 50%. When an entire helix turn was inserted (mut19), the loss of activity was 25%. In contrast, the loss of activity was especially high (65%) when half of the helix turn (5 bp) was deleted or inserted into the spacer sequence (mut17 and mut18), thereby rotating the motifs relative to each other on the double helix by 180°.
Therefore the distances between the cis-elements in the histone H3/k promoter had a functional significance. For optimal promoter activity, the wild-type distances of 41 bp CCAAT-CCAAT and 22 bp CCAAT-TATA are essential.
DISCUSSION
The aim of the present study was to elucidate the transcriptional regulation of the histone H3 genes. We started by mapping the promoters of six human histone H3 genes (H3/d, H3/h, H3/i, H3/k, H3/m and H3/n) using promoter deletion series in reporter gene assays (Figure 1). The transcripts of the genes chosen represented 68% of the total histone H3 mRNA in fetal human tissues and a majority of the total histone H3 mRNA in eight human cell lines [23] and were therefore a representative group of the total set of histone H3 genes. The experiments revealed that the functional lengths of the promoters were over the range of 133–227 bp (with the exception of H3/i, where no clear functional length could be defined): H3/d, 198 bp; H3/h, 147 bp; H3/k, 133 bp; H3/m, 227 bp; H3/n, 140 bp.
To identify the essential cis-elements within these functional promoters, the promoters of the histone H3/k and the histone H3/m genes were further analysed using scanning mutagenesis (Figure 2). The scanning mutagenesis revealed, in both promoters, that a TATA box and two CCAAT boxes were essential for promoter activity. In addition, an ATF box contributed to the activity of the H3/m promoter. No further cis-elements were identified. Thus the specific promoter activity of the histone H3 genes is mediated essentially by a pair of CCAAT boxes.
Mantovani [22] found that many cell-cycle-regulated genes, such as Cdc2 (cell division cycle), Cdc6, CDC25A/B/C, cyclin A2, cyclin B1/B2, E2F1 (E2 promoter-binding factor 1) or topoisomerase IIα contain one or more CCAAT boxes in their promoters. Also, the promoters of the other histone classes, with the exception of histone H4, contain at least one CCAAT box. The bipartite histone H2A/H2B promoters contain multiple CCAAT boxes [18], and the histone H1 promoters contain a single CCAAT box the same distance from the TATA box as the proximal CCAAT box of the histone H3 promoters [24]. Therefore these promoters might be cell-cycle-regulated in the same way by means of one or more CCAAT boxes. The principle CCAAT-box-binding activator is NF-Y (also called CBF) [25,26]. NF-Y is composed of three subunits (NF-YA, NF-YB and NF-YC), all of them being necessary for DNA binding. The activity of NF-Y in the nucleus has been shown to be cell-cycle-regulated: Bolognese et al. [27] have reported that the concentration of NF-YA in the nucleus peaks in mid-S phase and Frontini et al. [28] have shown that the localization of NF-YC is mostly cytoplasmic in G0/G1 phase, but that it targets the nucleus with the onset of S phase. Therefore the histone H3 genes might be activated in a cell-cycle-regulated way by NF-Y in concert with other cell-cycle-regulated genes.
To test this hypothesis, we analysed the CCAAT boxes of the H3/k promoter for NF-Y-binding specificity. Either a change in the central ‘CCAAT’ motif or a change in the surrounding bases to a non-consensus NF-Y-binding sequence caused a complete loss of the activating effects of the CCAAT boxes in reporter gene assays (see Figure 3). Furthermore, we were able to show that the CCAAT boxes of the H3/k promoter bound to NF-Y in EMSA experiments with HeLa nuclear extract and with the isolated recombinantly expressed trimeric NF-Y complex. The affinity of NF-Y for binding the two CCAAT boxes seems to be very different. Binding to the proximal CCAAT box is much stronger than binding to the distal CCAAT box. In contrast, mutation of the distal CCAAT box resulted in a sharp decrease in the promoter activity to approx. 15%. This discrepancy can be explained by binding of factors other than NF-Y to the distal CCAAT box or by a co-operative binding of NF-Y to the two CCAAT boxes. The results of the spacing experiments also point to the synergistic interaction of NF-Y in the H3 promoter. Binding of NF-Y to the distal CCAAT box of H3/m and to the histone H1.5 promoter was also reported by van Wijnen et al. [20]. Therefore NF-Y is very probably the ATF in histone H3 gene regulation that causes the cis-activating effect of the two CCAAT boxes in a cell-cycle-regulated fashion.
The basic functional promoter structure of the 11 replication-dependent histone H3 genes is quite similar, with a TATA box and two CCAAT boxes as the main elements. This explains why the isolated promoter activities compared with each other were basically similar in five different human cell lines [23]. Furthermore, it supports the notion that the aberrant expression pattern of the histone H3 genes that we have observed in human tumour cell lines is not due to soluble transcription factors but may depend on epigenetic effects within the chromatin context of the histone genes [23].
van Wijnen et al. [20] reported that the H3/m promoter yielded an EMSA gel shift band that was caused by a complex of the factor HiNF-D. The binding of HiNF-D, which has been described as a complex of cdc2 (Cdk1), cyclin A, pRb (p105) and CDP/Cut [29,30], required almost the full functional H3/m promoter including the upstream region around the ATF box. Recently, HiNF-D has been described as a late S phase repressor of histone H4 gene expression [31]. Probably, HiNF-D exerts a similar function in histone H3 gene regulation. However, so far no data about the function of HiNF-D in histone H3 promoter regulation has been published. Furthermore, most of the histone H3 promoters do not contain an ATF box, nor do they contain any significant sequence homology in the respective region. Thus it is not known whether HiNF-D has a general role in histone H3 promoter regulation also.
The factor p220 (NPAT) has been reported to be necessary for full activity and cell-cycle regulation of human histone genes [32–34]. It is phosphorylated by cyclin E/Cdk2 and is a component of a subset of Cajal bodies that localize to the histone gene clusters in S phase. It remains to be established how p220 (NPAT) activates histone H3 gene transcription.
Liberati et al. [35] showed that NF-Y can bind co-operatively to two neighbouring CCAAT boxes. Therefore binding of NF-Y to the two CCAAT boxes of the histone H3 minimal promoters might be co-operative and require the two neighbouring CCAAT boxes to compensate a modest NF-Y affinity of the single CCAAT boxes. Co-operativity could have the function to enhance sensitivity for cell-cycle-dependent variations in NF-Y concentrations.
This is supported by the analysis of the role of the distances between the CCAAT boxes and between the proximal CCAAT box and the TATA box in the H3/k promoter. Increasing the distance between the proximal CCAAT box and the TATA box by a complete helix turn of 10 and 11 bp respectively caused a loss of promoter activity in reporter gene assays by 75%. Changing the distance between the two CCAAT boxes also led to loss of promoter activity. In the present study, the loss of promoter activity was especially high when only half of the helix turn (5 bp) was inserted or deleted, causing the two CCAAT boxes to rotate relative to each other on the double helix by 180°. These results show that histone H3 promoter activity requires a well-defined set of three cis-elements, namely two CCAAT boxes and a TATA box with only limited flexibility in the distances. Obviously an activation complex forms at the histone H3 promoters that has a defined structure and limited flexibility. Bellorini et al. [36] and Frontini et al. [37] showed that NF-Y interacts with the general, multisubunit transcription factor TF-IID that binds to the TATA box and initiates the assembly of the RNA polymerase II initiation complex. An activation complex probably forms at the histone H3 promoters with two NF-Y factors, bound to the neighbouring CCAAT boxes in a co-operative way, and TF-IID. A future analysis of the molecular interactions between the involved factors could elucidate how the two molecules of NF-Y interact with each other and with TF-IID.
Acknowledgments
We thank R. Mantovani for providing the plasmid DNA containing the coding region of the three NF-Y subunits. This work was supported by the Deutsche Forschungsgemeinschaft. J. K. is a member of the GK 521.
References
- 1.Albig W., Doenecke D. The human histone gene cluster at the D6S105 locus. Hum. Genet. 1997;101:284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- 2.Frank D., Doenecke D., Albig W. Differential expression of human replacement and cell cycle dependent H3 histone genes. Gene. 2003;312:135–143. doi: 10.1016/s0378-1119(03)00609-7. [DOI] [PubMed] [Google Scholar]
- 3.Vasserot A. P., Schaufele F. J., Birnstiel M. L. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc. Natl. Acad. Sci. U.S.A. 1989;86:4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schümperli D., Albrecht U., Koning T. W., Melin L., Soldati D., Stauber C., Luhrmann R. Biochemical studies of U7 snRNPs and of histone RNA 3′ processing. Mol. Biol. Rep. 1990;14:205–206. doi: 10.1007/BF00360475. [DOI] [PubMed] [Google Scholar]
- 5.Dominski Z., Marzluff W. F. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 6.Williams A. S., Ingledue T. C., III, Kay B. K., Marzluff W. F. Changes in the stem-loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res. 1994;22:4660–4666. doi: 10.1093/nar/22.22.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey N. B., Marzluff W. F. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen B. S., Stein J. L., Stein G. S., Ostrer H. Single-copy flanking sequences in human histone gene clusters map to chromosomes 1 and 6. Genomics. 1991;10:486–488. doi: 10.1016/0888-7543(91)90337-e. [DOI] [PubMed] [Google Scholar]
- 9.Albig W., Kioschis P., Poustka A., Meergans K., Doenecke D. Human histone gene organization: nonregular arrangement within a large cluster. Genomics. 1997;40:314–322. doi: 10.1006/geno.1996.4592. [DOI] [PubMed] [Google Scholar]
- 10.Ahn J., Gruen J. R. The genomic organization of the histone clusters on human 6p21.3. Mamm. Genome. 1999;10:768–770. doi: 10.1007/s003359901089. [DOI] [PubMed] [Google Scholar]
- 11.Tripputi P., Emanuel B. S., Croce C. M., Green L. G., Stein G. S., Stein J. L. Human histone genes map to multiple chromosomes. Proc. Natl. Acad. Sci. U.S.A. 1986;83:3185–3188. doi: 10.1073/pnas.83.10.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumperli D. Cell-cycle regulation of histone gene expression. Cell (Cambridge, Mass.) 1986;45:471–472. doi: 10.1016/0092-8674(86)90277-1. [DOI] [PubMed] [Google Scholar]
- 14.Marzluff W. F., Pandey N. B. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem. Sci. 1988;13:49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- 15.Osley M. A. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 16.Stein G. S., Stein J. L., van Wijnen A. J., Lian J. B. Regulation of histone gene expression. Curr. Opin. Cell Biol. 1992;4:166–173. doi: 10.1016/0955-0674(92)90028-b. [DOI] [PubMed] [Google Scholar]
- 17.van der Meijden C. M., Vaughan P. S., Staal A., Albig W., Doenecke D., Stein J. L., Stein G. S., van Wijnen A. J. Selective expression of specific histone H4 genes reflects distinctions in transcription factor interactions with divergent H4 promoter elements. Biochim. Biophys. Acta. 1998;1442:82–100. doi: 10.1016/s0167-4781(98)00147-x. [DOI] [PubMed] [Google Scholar]
- 18.Trappe R., Doenecke D., Albig W. The expression of human H2A-H2B histone gene pairs is regulated by multiple sequence elements in their joint promoters. Biochim. Biophys. Acta. 1999;1446:341–351. doi: 10.1016/s0167-4781(99)00107-4. [DOI] [PubMed] [Google Scholar]
- 19.Heintz N. The regulation of histone gene expression during the cell cycle. Biochim. Biophys. Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- 20.van Wijnen A. J., Lian J. B., Stein J. L., Stein G. S. Protein/DNA interactions involving ATF/AP1-, CCAAT-, and HiNF-D-related factors in the human H3-ST519 histone promoter: cross-competition with transcription regulatory sites in cell cycle controlled H4 and H1 histone genes. J. Cell. Biochem. 1991;47:337–351. doi: 10.1002/jcb.240470408. [DOI] [PubMed] [Google Scholar]
- 21.Bi W., Wu L., Coustry F., de Crombrugghe B., Maity S. N. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J. Biol. Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- 22.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koessler H., Doenecke D., Albig W. Aberrant expression pattern of replication-dependent histone h3 subtype genes in human tumor cell lines. DNA Cell Biol. 2003;22:233–241. doi: 10.1089/104454903321908629. [DOI] [PubMed] [Google Scholar]
- 24.Doenecke D., Albig W., Bouterfa H., Drabent B. Organization and expression of H1 histone and H1 replacement histone genes. J. Cell. Biochem. 1994;54:423–431. doi: 10.1002/jcb.240540409. [DOI] [PubMed] [Google Scholar]
- 25.Maity S. N., de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 26.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 27.Bolognese F., Wasner M., Dohna C. L., Gurtner A., Ronchi A., Muller H., Manni I., Mossner J., Piaggio G., Mantovani R., et al. The cyclin B2 promoter depends on NF-Y, a trimer whose CCAAT-binding activity is cell-cycle regulated. Oncogene. 1999;18:1845–1853. doi: 10.1038/sj.onc.1202494. [DOI] [PubMed] [Google Scholar]
- 28.Frontini M., Imbriano C., Manni I., Mantovani R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle. 2004;3:217–222. [PubMed] [Google Scholar]
- 29.van Wijnen A. J., Aziz F., Grana X., De Luca A., Desai R. K., Jaarsveld K., Last T. J., Soprano K., Giordano A., Lian J. B., et al. Transcription of histone H4, H3, and H1 cell cycle genes: promoter factor HiNF-D contains CDC2, cyclin A, and an RB-related protein. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12882–12886. doi: 10.1073/pnas.91.26.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Wijnen A. J., van Gurp M. F., de Ridder M. C., Tufarelli C., Last T. J., Birnbaum M., Vaughan P. S., Giordano A., Krek W., Neufeld E. J., et al. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta S., Luong M. X., Bleuming S. A., Miele A., Luong M., Young D., Knudsen E. S., van Wijnen A. J., Stein J. L., Stein G. S. Tumor suppressor pRB functions as a co-repressor of the CCAAT displacement protein (CDP/cut) to regulate cell cycle controlled histone H4 transcription. J. Cell. Physiol. 2003;196:541–556. doi: 10.1002/jcp.10335. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J., Kennedy B. K., Lawrence B. D., Barbie D. A., Matera A. G., Fletcher J. A., Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 33.Ma T., Van Tine B. A., Wei Y., Garrett M. D., Nelson D., Adams P. D., Wang J., Qin J., Chow L. T., Harper J. W. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye X., Wei Y., Nalepa G., Harper J. W. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberati C., di Silvio A., Ottolenghi S., Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J. Mol. Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- 36.Bellorini M., Lee D. K., Dantonel J. C., Zemzoumi K., Roeder R. G., Tora L., Mantovani R. CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Res. 1997;25:2174–2181. doi: 10.1093/nar/25.11.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frontini M., Imbriano C., diSilvio A., Bell B., Bogni A., Romier C., Moras D., Tora L., Davidson I., Mantovani R. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J. Biol. Chem. 2002;277:5841–5848. doi: 10.1074/jbc.M103651200. [DOI] [PubMed] [Google Scholar]