Abstract
PKCδ (protein kinase Cδ) is a serine/threonine kinase that plays a key role in growth regulation and tissue remodelling. Traditional models of PKC activation have focused on lipid cofactors and anchoring proteins that localize the active conformation of PKCδ to membranes, in close proximity with its target substrates. However, recent studies identify a distinct mode for PKCδ activation involving tyrosine phosphorylation by Src family kinases. The tyrosine-phosphorylated form of PKCδ (which accumulates in the soluble fraction of cells exposed to oxidant stress) displays lipid-independent kinase activity and is uniquely positioned to phosphorylate target substrates throughout the cell (not just on lipid membranes). This review summarizes (1) recent progress towards understanding structure–activity relationships for PKCδ, with a particular focus on the stimuli that induce (and the distinct functional consequences that result from) tyrosine phosphorylation events in PKCδ's regulatory, hinge and catalytic domains; (2) current concepts regarding the role of tyrosine phosphorylation as a mechanism to regulate PKCδ localization and actions in mitochondrial and nuclear compartments; and (3) recent literature delineating distinct roles for PKCδ (relative to other PKC isoforms) in transcriptional regulation, cell cycle progression and programmed cell death (including studies in PKCδ−/− mice that implicate PKCδ in immune function and cardiovascular remodelling). Collectively, these studies argue that the conventional model for PKCδ activation must be broadened to allow for stimulus-specific differences in PKCδ signalling during growth factor stimulation and oxidant stress.
Keywords: Abl, oxidant stress, phosphorylation, protein kinase Cδ, Src family kinase
Abbreviations: DAG, diacylglycerol; DNA-PK, DNA-dependent protein kinase; EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; IκB, inhibitor κB; IKK, IκB kinase; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; NFκB, nuclear factor κB; PDGF, platelet-derived growth factor; PDK, phosphoinositide-dependent kinase; PKC, protein kinase C; aPKC, atypical PKC; cPKC, conventional PKC; nPKC, novel PKC; PLS, phospholipid scramblase 3; RACK, receptor for activated C-kinase; ROS, reactive oxygen species; SFK, Src family kinase; SH2, Src homology 2; SHIP, SH2-domain-containing inositol 5′-phosphatase; STAT, signal transducer and activator of transcription; WT-PKCδ, wild-type PKCδ
INTRODUCTION
The PKC (protein kinase C) family comprises a multigene family of related serine/threonine kinases that play key roles in growth regulation and programmed cell death. Our particular interest has been in the PKC-activated signalling mechanisms that regulate contraction, the evolution of ischaemic preconditioning, and the pathogenesis of hypertrophy and failure in the heart [1,2]. Traditional models of PKC activation have focused on the role of physiological second messengers [such as calcium and DAG (diacylglycerol)] or tumour-promoting phorbol esters (such as PMA) to anchor PKCs in their active conformations on membranes. However, most cells co-express multiple PKC isoforms that elicit distinct (and occasionally functionally opposing) cellular responses. PKC isoform specificity has been attributed to distinctive compartmentalization patterns for individual PKC isoforms. The prevailing model holds that protein–protein interactions between a particular PKC isoform and its unique membrane-associated anchoring protein (itself localized to a distinct membrane subdomain) serve to recruit the PKC isoform to a distinct subcellular compartment, in close proximity with its unique target substrates. However, this receptor-driven, lipid cofactor-dependent mechanism for PKC activation involving membrane-associated anchoring proteins does not adequately explain the PKC-dependent phosphorylation of proteins in non-membrane compartments. In particular, the well known effects of PKC to alter contractile function by phosphorylating myofibrillar proteins in the sarcomere (which are not associated with lipid membranes) [3] are not readily explained by the traditional model of PKC activation.
This review summarizes recent evidence that PKCδ acts as a lipid-independent enzyme when it is tyrosine-phosphorylated by SFKs (Src family kinases). These newer results suggest that the conventional model, that considers PKCδ as a generic kinase (whose phosphorylations are regulated entirely by translocation to membranes and access to substrate), must be broadened to include additional factors that influence PKCδ's enzymology. According to this revised model, the phosphorylation events triggered by allosterically activated PKCδ in membranes (during cellular activation by receptors that promote DAG accumulation) are likely to be functionally distinct from the events driven by the tyrosine-phosphorylated form of PKCδ that accumulates in the soluble fraction of cells subjected to oxidant stress.
PKC STRUCTURE AND REGULATION: DISTINCTIVE PROPERTIES OF THE PKCδ ISOFORM
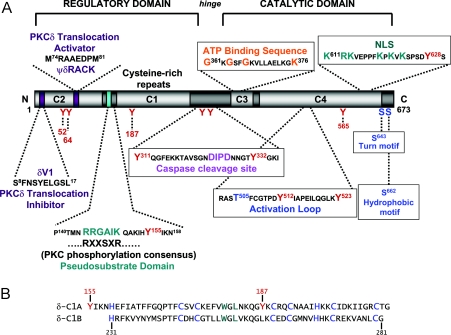
PKC isoforms are single polypeptide chains with N-terminal regulatory domains that contain an autoinhibitory pseudosubstrate domain, two membrane-targeting modules (termed C1 and C2) and a highly conserved C-terminal catalytic domain (that contains the C3 and C4 motifs required for ATP/substrate binding and catalytic activity) (Figure 1A). PKC isoforms are broadly subdivided into three subfamilies based upon their structurally distinct N-terminal regulatory domains. cPKCs (conventional PKCs; α, βI, βII and γ) contain two membrane-targeting modules, designated C1 and C2. The C1 domain consists of tandem ∼50-residue DAG/PMA-binding sequences termed C1A and C1B; each adopts a globular conformation and co-ordinates Zn2+ at a metal ion-binding site formed by three cysteines and one histidine. X-ray crystallographic studies (of PKCδ's C1B domain complexed with PMA) identify C1 domains as hydrophobic switches. Each C1 domain consists of two β-sheets and a short C-terminal α-helix; PMA (or endogenously generated DAG) binds to a hydrophilic cleft situated in an otherwise hydrophobic surface at the tip of the C1 domain (between two ‘unzipped’ β-strands). By capping this polar groove, PMA (or DAG) forms a contiguous hydrophobic surface that promotes PKC binding to membranes [4,5]. The second cPKC membrane targeting motif is the C2 domain, a motif that is found in many proteins that participate in membrane trafficking and signal transduction. C2 domains characteristically consist of eight antiparallel β-strands connected by loops of variable lengths. C2 domains of cPKC isoforms bind anionic phospholipids in a calcium-dependent manner due to the presence of several invariant calcium-binding residues in three loops at one end of the structure.
Figure 1. (A) Structural domains on PKCδ implicated in functional regulation, and (B) sequences of the twin C1A and C1B domains.
(A) See the text for details. Numbering is based on the rat sequence. (B) The histidine and cysteine residues that co-ordinate Zn2+ ions and are a characteristic feature of the C1 motif (H-X12-C-X2-C-X13/14-C-X2-C-X4-H-X2-C-X7-C) are highlighted in blue. Conserved hydrophobic residues, that are not involved in the structural integrity of the protein but rather are believed to be exposed to external solvent and mediate membrane binding, are depicted in green. Tyrosine residues reported to be sites for regulatory phosphorylations (adjacent to or within the C1A domain sequence) are illustrated in red.
nPKCs (novel PKCs; δ, ε, η and θ) also have twin C1 domains and a C2 domain (which, in the case of nPKCs, precedes the C1 domain) in their N-terminal regulatory regions. However, C2 domain-like sequences of nPKCs lack calcium-co-ordinating acidic residue side chains. Hence nPKCs are maximally activated by DAG/PMA, without requiring calcium. Of note, the calcium-binding-like loop of the C2 domain of PKCδ contains an accessible MY52PE sequence that conforms to an optimal SFK substrate. This sequence is unique to PKCδ and is not found in other PKC isoforms that do not become tyrosine phosphorylated in response to PMA [6].
aPKCs (atypical PKCs; ζ and ι/λ) are the third PKC isoform subfamily. aPKCs lack a calcium-sensitive C2 domain and contain only a single cysteine-rich zinc finger structure that does not bind DAG or PMA. As a result, aPKC isoforms are not allosterically regulated by calcium or DAG/PMA. Rather, aPKCs are activated by a distinct set of phospholipid cofactors. aPKCs are also activated via stimulus-induced phosphorylation events (a topic that is beyond the scope of this review, and is described in great detail in recent excellent reviews in the literature [6a,6b]).
Current models of PKC activation are based largely on studies of cPKC isoforms, which reside (in a closed/inactive conformation, with the autoinhibitory pseudosubstrate domain occluding the substrate-binding pocket) in the soluble fraction of quiescent cells. In the absence of calcium or DAG, cPKCs interact weakly/transiently with membranes. Agonists that promote phosphoinositide hydrolysis and Ins(1,4,5)P3 generation lead to the mobilization of intracellular calcium, which binds to the C2 domain and increases its affinity for membranes. This initial association of cPKC with membranes facilitates the interaction of the C1 domain with DAG (the other product of phosphoinositide hydrolysis). C1/C2 domain engagement with membranes promotes a conformational change that expels the autoinhibitory pseudosubstrate domain from the substrate-binding pocket and facilitates the PKC-mediated phosphorylation of membrane substrates. With the exception of the C2 domain-mediated effects of calcium, nPKC isoform activation for the most part follows a similar mechanism. For both cPKC and nPKC isoforms, translocation to membranes generally is considered a hallmark of activation (and frequently is used as a surrogate marker of PKC isoform activation in intact cells).
Most cells co-express multiple PKC isoforms that display only limited substrate specificity in vitro and yet elicit distinct cellular responses in intact cells. PKC isoform specificity in vivo has been attributed to isoform-specific interactions with various anchoring proteins that localize individual PKC isoforms to specific membrane microdomains (in close proximity with their allosteric activators and/or substrates). To date, a relatively large number of PKC-binding partners have been identified, including STICKs (substrates that interact with C-kinase), various cytoskeletal proteins (such as actin or tubulin), true scaffolding proteins [such as caveolin isoforms and AKAPs (A-kinase anchoring proteins)], and RACKs (receptors for activated C-kinase) [7]. The RACK family of membrane-associated PKC-anchoring proteins has figured particularly prominently in the recent literature, since peptides designed to block or promote PKC isoform-selective interactions with their cognate RACKs are currently being evaluated for various cardiovascular indications in humans. RACK proteins consist of a seven-WD40-motif repeat structure, similar to the protein–protein binding motifs found in the β subunits of heterotrimeric G-proteins. The current model holds that cells express a unique RACK, with a distinct subcellular localization, for each PKC isoform. By selectively/saturably binding only the activated conformation of a PKC, each RACK protein recruits its cognate PKC isoform (in an active conformation) to a specific membrane compartment [8]. To date, proteins with characteristics of RACKs for PKCβ (RACK1), PKCε (RACK2 or β-COP) and PKCδ (p32/gC1qBP) have been identified [9–11]. However, it is important to note that RACK proteins also can fulfil functions unrelated to PKC. For example, RACK1 is reported to act as a scaffold to organize signalling complexes containing SFKs, heterotrimeric G-protein βγ subunits, dynamin-1, integrin β subunits, STAT1 (signal transducer and activator of transcription 1), the receptor protein tyrosine phosphatase PTPμ, and phosphodiesterase 4D5 [9,12]. RACK2 (or β-COP, a coated-vesicle protein that participates in intracellular transport and vesicular release) was identified as a binding partner for certain RGS (regulators of G-protein signalling) proteins [9,13].
Recent studies indicate that differences in the spatial compartmentalization of individual PKCs may not be the only (or even the prime) mechanism for the regulation of PKC signalling under certain conditions. This is particularly striking for PKCδ, which is dynamically regulated via agonist-induced activation-loop phosphorylation [14], is released from membranes as a lipid-independent enzyme with altered substrate specificity in cells exposed to oxidant stress [15], and displays altered cofactor requirements and substrate specificity as a result of caspase-dependent cleavage in cells undergoing apoptosis [16]. These distinct mechanisms for PKCδ regulation are described in greater detail in the sections that follow.
PKC ISOFORM PHOSPHORYLATION
Serine/threonine phosphorylation at the activation loop and the C-terminus
The traditional model of PKC activation focuses on allosteric activation by calcium and DAG. However, more recent studies have identified a series of sequential ‘priming’ phosphorylations at highly conserved serine/threonine phosphorylation motifs in all PKC isoforms that lock the enzyme in a closed, stabilized, catalytically competent and protease/phosphatase-resistant conformation [17,18]. The first phosphorylation is on an ‘activation-loop’ threonine which plays a critical role to align residues in the catalytic pocket; without activation-loop phosphorylation, cPKC isoforms are effectively catalytically inactive [19–21]. cPKC activation-loop phosphorylation has been attributed to PDK-1 (phosphoinositide-dependent kinase-1), which complexes with the C-terminus of the membrane-localized unphosphorylated enzyme [21]. cPKCs are believed to then autophosphorylate on a conserved proline-flanked ‘turn motif’ and a hydrophobic FXXFS/TF/Y motif (19 residues C-terminal to the turn motif). These priming phosphorylations are completed during the maturation of cPKCs (and are retained during normal culture conditions). cPKC activation is through membrane translocation and the conformational changes induced by calcium and lipid cofactors.
While nPKCs undergo similar priming phosphorylations, the regulation and consequences of these events differ from those described for cPKC isoforms in certain respects. For example, PKCδ retains little phosphorylation at its activation loop (Thr505) in many cell types. PKCδ-Thr505 phosphorylation is induced by PMA or the α1-adrenergic receptor agonist noradrenaline (norepinephrine) in cardiomyocytes, the gastrin receptor in human gastric cancer cells, and thrombin receptors in endothelial cells [14,22,23]. Of note, PMA-(or noradrenaline-) induced PKCδ-Thr505 phosphorylation in cardiomyocytes is blocked by GF109203X (a general inhibitor of cPKCs and nPKCs), but not by Go6976, a selective cPKC inhibitor [14]. This result is surprising, based upon the prevailing notion that PDK-1 (a GF109203X-insensitive enzyme) is the PKCδ-Thr505 kinase. Rather, it suggests that a nPKC isoform, further identified as nPKCε (based upon overexpression studies with kinase-inactive forms of individual nPKCs), plays a role in the control of PKCδ-Thr505 phosphorylation in cardiomyocytes [14]. This could suggest that nPKCε acts directly as the PKCδ-Thr505 kinase (similar to the role of PKCε as the activation-loop kinase for PKCμ in HEK293 cells [24]), although an indirect pathway for the in vivo regulation of PKCδ-Thr505 phosphorylation involving a nPKC-activated kinase or a nPKC-regulated phosphatase also remains possible and deserves further study.
Activation-loop phosphorylation is essential to generate the catalytically competent forms of cPKC/aPKC isoforms. In contrast, PKCδ is a functional kinase even without Thr505 phosphorylation. This has been attributed to an acidic Glu at position 500 in PKCδ that assumes the role of the phosphorylated activation-loop Thr in cPKCs. Nevertheless, the catalytic activity of membrane-associated allosterically activated PKCδ is increased by Thr505 phosphorylation [14,20,25]. The notion that PKCδ activation is via co-ordinate phosphorylation and translocation events deserves emphasis; both mechanisms must be considered when evaluating PKCδ signalling pathways – or the efficacy of pharmacological inhibitors designed to abrogate PKCδ's actions.
Phosphorylation in the hydrophobic motif is important for reversible PKC isoform stimulation by physiological agonists; it releases PKC from membranes and facilitates PKC down-regulation [26]. Hydrophobic motif and turn motif phosphorylations on PKCδ are stable modifications that are not regulated by culture conditions or agonist treatment. In contrast, PKCε retains little phosphorylation at its hydrophobic motif (Ser729) in resting cardiomyocytes. PKCε-Ser729 phosphorylation is induced by PMA or noradrenaline [14]. C-terminal phosphorylations on cPKCs generally have been characterized as intramolecular autophosphorylation events [27]. While some evidence points to a similar intramolecular autophosphorylation mechanism for PKCε-Ser729, other studies are more consistent with a phosphorylation event mediated by a hydrophobic motif kinase in trans [variably characterized as PKCδ or mTOR (mammalian target of rapamycin)] [14,28,29].
Tyrosine phosphorylation
PKCδ is phosphorylated on tyrosine residues in cells transformed with Src or Ras or acutely stimulated with H2O2, PMA, EGF (epidermal growth factor) or PDGF (platelet-derived growth factor). Mouse, rat and human PKCδ contain 19, 21 and 20 tyrosine residues respectively. Multiple sites for tyrosine phosphorylation have been identified in PKCδ's catalytic domain (Tyr512 and Tyr523), regulatory domain (Tyr52, Tyr155 and Tyr187) and hinge region (Tyr311 and Tyr332). In contrast with the sites for Ser/Thr phosphorylation, these tyrosine residues are not conserved across PKC family members. Hence tyrosine phosphorylation is a relatively specific regulatory mechanism for PKCδ, and not a common regulatory mechanism for the entire family of PKC enzymes.
Most studies have relied on in vitro kinase assays to resolve tyrosine phosphorylation-dependent changes in PKCδ function. No uniform pattern or consequence of PKCδ tyrosine phosphorylation can be extracted from the published literature, since the catalytic activity of tyrosine-phosphorylated PKCδ is variably described as decreased, increased, or even altered with regard to substrate specificity and cofactor requirements [30–34]. In fact, it has become increasingly evident that the precise configuration of tyrosine residues phosphorylated on PKCδ depends upon the nature of the inciting stimulus and dictates the functional properties of the enzyme. In general, tyrosine phosphorylation of the catalytic domain (in cells treated with H2O2) increases the kinase activity of PKCδ, whereas phosphotyrosines in PKCδ's regulatory domain (in cells treated with PMA or PDGF) influence the cellular actions of PKCδ without influencing kinase activity. The literature exploring the upstream regulators and downstream consequences of PKCδ tyrosine phosphorylation is reviewed in the sections that follow.
PKCδ-Tyr155
Tyr155 is flanked by the regulatory domain pseudosubstrate motif and the C1A domain (Figures 1A and 1B). Its role has been explored in heterologous overexpression systems, since physiologically relevant stimuli that trigger phosphorylation of Tyr155 have not been identified. Tyr155 phosphorylation is not induced by PMA or PDGF [35,36]; a role for oxidant stress has not been considered. However, Tyr155 phosphorylation profoundly influences growth regulation by PKCδ. Overexpression of WT-PKCδ (wild-type PKCδ) slows proliferation in many cell types [37]. However, cells that overexpress PKCδ with a single Tyr→Phe substitution at Tyr155 grow more rapidly; these cells grow in soft agar and form tumours in nude mice [34,36]. Efforts to define the mechanism(s) for altered growth regulation by PKCδ-Y155F have been uninformative. WT-PKCδ and PKCδ-Y155F display grossly similar [3H]phorbol dibutyrate binding, catalytic activity and subcellular localizations.
PKCδ-Tyr187
Chimaeric PKC constructs (in which the regulatory and catalytic domains of PKCα, PKCδ and PKCε are exchanged at the highly conserved sequence in the hinge region) were used as a strategy to map the PMA- and PDGF-dependent tyrosine phosphorylation(s) to the regulatory domain of PKCδ [36,38]. Further studies with a panel of PKCδ mutants with Tyr→Phe substitutions at conserved tyrosine residues in the regulatory domains of all species (Tyr52, Tyr64, Tyr155 and Tyr187) identified Tyr187 (in the C1A domain; Figure 1B) as the major site for PMA- and PDGF-dependent phosphorylation [35,36]. Tyr187 phosphorylation does not influence the kinase activity of PKCδ (in vitro, using a pseudosubstrate domain peptide). Heterologously overexpressed PKCδ-Y187F also mimics the effect of WT-PKCδ to inhibit growth of NIH 3T3 and C6 glial cells. Like WT-PKCδ, PKCδ-Y187F mediates PMA-dependent induction of the monocyte differentiation programme in 32D myeloid progenitor cells [35,37]. However, PKCδ-Y187F does not mimic the effect of WT-PKCδ to promote differentiation in C6 glial cells [36]. The kinase that phosphorylates PKCδ at Tyr187 has not been identified. There is indirect evidence that phosphorylation of PKCδ-Tyr187 does not involve SFKs (although Fyn is reported to dock on PKCδ via a mechanism that requires Tyr187 phosphorylation [39]).
PKCδ-Tyr52
An accessible Tyr52 (with Glu at the +2 position, conforming to an SFK substrate) is unique to the C2 domain of PKCδ [6]. The C2 domains of other PKC isoforms do not have a tyrosine at this position. Our current understanding of the mechanisms and consequences of PKCδ-Tyr52 phosphorylation come largely from studies in rat basophilic leukaemia cells (RBL-2H3), where engagement of the high-affinity immunoglobulin E receptor (FcεRI) activates PKCδ and leads to phosphorylation of Tyr52 by Lyn [40]. Interestingly, phosphorylated Tyr52 serves as a docking site for the SH2 (Src homology 2) domain of Lyn. Therefore Lyn-dependent PKCδ-Tyr52 phosphorylation amplifies PKCδ–Lyn interactions. A reciprocal regulatory control, involving PKCδ-mediated phosphorylation of Lyn (or of Src at Ser12), also has been identified [41,42], although some have argued that regulation of Src by PKCδ is via an indirect mechanism involving PKCδ-dependent phosphorylation of the protein tyrosine phosphatase PTPα, which activates Src (presumably by dephosphorylating the inhibitory Src-Ser527 site [43]). The functional consequences of SFK phosphorylation by PKCδ are disputed; it variably has been linked to changes (decrease or increase) in Lyn/Src activity as well as dissociation of PKCδ–Src complexes [40,42].
PKCδ-Tyr332
Studies in RBL-2H3 cells have identified Lyn-dependent phosphorylation of PKCδ at Tyr332. While Tyr332 is not the major site for tyrosine phosphorylation (since similar high overall levels of PKCδ tyrosine phosphorylation are detected in cells overexpressing WT-PKCδ or PKCδ-Y332F), Tyr332 is one of two tyrosine residues in PKCδ with an isoleucine at the +3 position (conforming to a consensus binding sequence for the SH2 domain of Shc). PKCδ-Tyr332 phosphorylation creates a docking site for Shc. A single Tyr→Phe substitution at Tyr332 abrogates the PKCδ–Shc interaction; Tyr→Phe substitutions at Tyr372 (the other tyrosine with an isoleucine position +3), Tyr52, Tyr64, Tyr187 or Tyr 565 do not interfere with the PKCδ–Shc interaction [44].
Shc is a scaffold protein that also binds SHIP (SH2-domain-containing inositol 5′-phosphatase), which dephosphorylates PtdIns(3,4,5)P3 to PtdIns(3,4)P2 and thereby negatively regulates Akt phosphorylation. Recent studies indicate that Shc co-ordinates a physiologically important indirect interaction between PKCδ and SHIP, with PKCδ regulating antigen-induced SHIP tyrosine phosphorylation (acting as a negative regulator of mast cell degranulation) and SHIP regulating PKCδ localization and activation [44]. The evidence that phosphotyrosine residues in PKCδ's regulatory and/or hinge region act as docking sites for Src kinases and adapters such as Shc raises the possibility that the biological role of PKCδ might extend beyond its role as a kinase. In support of this notion, kinase-inactive PKCδ mimics the effect of WT-PKCδ to induce apoptosis in vascular smooth muscle cells [45].
PKCδ-Tyr311
Tyr311 in the hinge region of PKCδ is flanked by sequence that conforms to that of an optimal Src substrate. PKCδ-Tyr311 phosphorylation is prominent in cells treated with H2O2 [31,46]. However, the functional ramifications of Tyr311 phosphorylation are still disputed. Tyr311 phosphorylation has been linked to increased kinase activity in cells treated with H2O2 and altered PKCδ trafficking/down-regulation kinetics in cells transformed with Src [47]. However, this second conclusion is based on a single study by the Courtneidge laboratory, which identified a high level of PKCδ tyrosine phosphorylation, reduced PKCδ protein expression and accelerated PKCδ degradation in Src-transformed NIH 3T3 cells (relative to parental NIH 3T3 cells) [47]. Biochemical studies identified Tyr311 as a minor phosphorylation site on PKCδ in Src-transformed cells; most PKCδ phosphorylation was mapped to other tyrosine residues. However, a single Tyr→Phe mutation at Tyr311 was sufficient to completely abrogate Src-dependent PKCδ tyrosine phosphorylation (and prevent the accelerated PKCδ down-regulation kinetics). These results were interpreted as evidence that PKCδ undergoes a highly ordered sequence of tyrosine phosphorylation reactions that are initiated at Tyr311, and that Src-dependent Tyr311 phosphorylation (by itself, or in conjunction with a subsequent modification) accelerates the kinetics of PKCδ down-regulation. It was postulated that PKCδ phosphorylation at Tyr311 and Tyr332 (which flank a caspase cleavage site in PKCδ's hinge region; Figure 1) induces a conformational change that exposes the adjacent (but otherwise hidden) caspase cleavage site. However, it is worth noting that PKCδ cleavage products have never been identified directly under these conditions. It is also noteworthy that the original study provided evidence that PMA-dependent down-regulation of PKCδ-Y311F is not accelerated relative to that of WT-PKCδ [47]. This is consistent with recent studies in cardiomyocytes that identified PMA- and H2O2-dependent increases in PKCδ-Tyr311 phosphorylation, but failed to link Tyr311 phosphorylation to accelerated PMA-induced down-regulation kinetics for PKCδ [14]. Collectively, these results suggest that phosphorylation of PKCδ at Tyr311 may influence PKCδ down-regulation in Src-transformed cells, but it does not modulate physiological PKCδ trafficking/down-regulation mechanisms.
We demonstrated recently that the Tyr311-phosphorylated form of PKCδ accumulates in the particulate fraction of cardiomyocytes treated with PMA [15]. In contrast, H2O2 promotes the redistribution of PKCδ from the particulate to the soluble fraction; in H2O2-treated cardiomyocytes, PKCδ phosphorylated on Tyr311 accumulates in both the soluble and particulate fractions. In both cases, Tyr311 phosphorylation is by one or more SFKs. However, PMA and H2O2 promote PKCδ tyrosine phosphorylation via different mechanisms. SFKs (Src, Fyn, Yes and Lyn) constitutively complex with PKCδ (but not PKCα or PKCε) in cardiomyocytes. H2O2 activates SFKs and increases SFK–PKCδ complex formation; this provides an obvious mechanism to explain the H2O2-dependent increase in Tyr311 phosphorylation. However, the mechanism underlying the PMA-dependent increase in Tyr311 phosphorylation is less obvious, since PMA neither activates SFKs nor increases complex formation between SFKs and PKCδ. A clue to the mechanism for Tyr311 phosphorylation in cardiomyocytes treated with PMA came from in vitro kinase assays. We showed that PKCδ (immunoprecipitated from quiescent cardiomyocyte cultures) undergoes a low level of 32P incorporation when in vitro kinase assays are performed without lipid. 32P incorporation into PKCδ (at Ser/Thr and Tyr residues, including Tyr311) increases dramatically in the presence of phosphatidylserine/PMA. Since phosphatidylserine/PMA does not activate SFKs, we interpret these results as evidence that phosphatidylserine/PMA induces a conformational change in PKCδ that renders the protein a better substrate for one or more pre-complexed SFKs, resulting in increased Tyr311 phosphorylation.
Further studies have begun to expose the functional consequences of PKCδ-Tyr311 phosphorylation in cardiomyocytes [15]. As noted, Tyr311 phosphorylation does not appear to influence the kinetics of PMA-dependent PKCδ down-regulation. Rather, Tyr311 phosphorylation appears to play a more prominent role in regulating PKCδ kinase activity. Using conventional in vitro immune complex kinase assays, we demonstrated that PKCδ was recovered from quiescent cultures with little or no lipid-independent kinase activity; PKCδ-mediated phosphorylation of model substrates, such as ε-peptide (the pseudosubstrate domain sequence of PKCε), δ-peptide (an optimal PKCδ phosphorylation motif based upon the murine eEF-1α sequence) and histone, requires lipid (phosphatidylserine/PMA). In contrast, PKCδ was recovered from H2O2-treated cultures (including from the soluble fraction) as a Tyr311-phosphorylated protein with lipid-independent kinase activity. The activation-dependent change in cofactor requirements is accompanied by an equally striking change in substrate specificity. Histone (which generally is viewed as a model substrate for cPKCs, and a poor substrate for nPKCs) is effectively phosphorylated by PKCδ recovered from resting cardiomyocytes, as long as the assays are performed in the presence of lipid cofactors. However, histone is not significantly phosphorylated (even when lipid cofactors are added to the assays) by PKCδ recovered from PMA- or H2O2-stimulated cardiomyocytes. This striking difference in substrate specificity for the unstimulated and phosphorylated/activated forms of PKCδ raises a significant concern that at least some of the current dogma regarding the selective functions of PKC isoforms (derived largely from studies on overexpressed enzymes that tend to localize aberrantly and become excessively phosphorylated) may not hold for native enzymes in differentiated cell types.
Additional in vitro kinase assays with recombinant PKCδ and active Src identified PKCδ-Tyr311 as a direct target for Src-dependent phosphorylation; tyrosine phosphorylation ‘fine tunes’ the substrate specificity of PKCδ, since the Src-phosphorylated form of PKCδ displays substantial lipid-independent kinase activity towards δ-peptide, but little towards ε-peptide or histone [15]. The identification of the Src-phosphorylated form of PKCδ as a lipid-independent kinase with altered substrate specificity is noteworthy; the enzymology of this modified form of PKCδ (which accumulates in the soluble fraction of cardiomyocytes subjected to oxidative stress) could explain the well recognized effects of PKC to phosphorylate contractile proteins in the sarcomere (which are not associated with lipid membranes).
The distinct enzymology of the tyrosine-phosphorylated form of PKCδ, as well as the potential role of PKCδ as a signal-regulated scaffold (with phospho-Tyr311 – alone or along with Tyr332 – acting as a docking site for the adapter protein Shc, SFKs and/or other signalling proteins), constitute interesting and potential fruitful areas for future research.
PKCδ FUNCTIONS: NON-CARDIAC CELL TYPES
PKCδ has been implicated in cell cycle regulation and programmed cell death in many cell types. Unlike PKCβ (which stimulates growth) and PKCε (which acts as an oncogene when overexpressed in rat fibroblasts and promotes tumours in nude mice), PKCδ generally slows proliferation, induces cell cycle arrest, and/or enhances the differentiation of various undifferentiated cell lines [48–50]. In many cases, the growth-inhibitory effects of PKCδ have been linked to changes in the expression of factors that influence cell cycle progression. For example, PKCδ decreases cyclin D1 and cyclin E expression and up-regulates p27Kip1 in vascular smooth muscle and endothelial cells [48,49].
While PKCδ generally suppresses normal cell proliferation, PKCδ activation is linked to growth-stimulatory responses in certain contexts. The effect of PKCδ to stimulate growth generally has been identified in transformed or cancer cell lines, where it has been attributed to activation of the ERK (extracellular-signal-regulated kinase)/MEK [MAPK (mitogen-activated protein kinase)/ERK kinase] cascade. ERK is activated by the constitutively active PKCδ mutant heterologously overexpressed in COS cells [51], and it is implicated as the effector for the oestrogen-activated ErbB2/PKCδ/Ras autocrine/paracrine pathway that promotes the proliferation of oestrogen receptor-positive MCF-7 human breast cancer cells [52]. Moreover, PKCδ also regulates other biological effectors that are critical for cancer biology. For example, PKCδ inhibits basal transcription of the tumour suppressor protein p53 in human myeloid leukaemia cells [53]. Since p53 is a key element in the surveillance mechanism used by cells to maintain genomic stability by eliminating cells with damaged DNA, inhibition of p53 is permissive for tumour formation. PKCδ activation has also been linked to anchorage-independent tumour cell growth and enhanced tumour cell survival. These mechanisms would influence the metastatic potential of tumour cells, perhaps explaining the observed correlation between elevated PKCδ levels and highly aggressive forms of metastatic breast cancer [54].
PKCδ also activates NFκB (nuclear factor κB). This ubiquitous transcription factor plays a key role in regulating immune and inflammatory responses; it also influences tumorigenesis through the induction of target genes that accelerate transit through the cell cycle, block apoptosis, promote angiogenesis, and enhance tumour cell invasiveness. A PKCδ/NFκB pathway has been implicated in enhanced protein expression of ICAM-1 (intercellular cell-adhesion molecule 1) and increased neutrophil adhesiveness in endothelial cells, the induction of certain IAP (inhibitor of apoptosis) protein family members in human colon cancer cells, and increased expression of pro-inflammatory mediators in airway epithelial cells [22,55,56]. Several potential mechanisms linking PKCδ to increased NFκB-dependent gene expression have been suggested. NFκB is held in the cytoplasm by IκB (inhibitor κB) proteins that mask its nuclear localization sequence. There is evidence that PKCδ itself, or its downstream effector PKCμ (or protein kinase D), can act as IKKs (IκB kinases) to phosphorylate IκB [57,58]. The phosphorylated form of IκB undergoes ubiquitination and degradation by the 26 S proteasome, liberating NFκB, which then translocates to the nucleus and binds κB-regulatory elements. PKCδ is also reported to increase the transactivation potential of NFκB via an IKK/IκB-independent pathway (speculated to involve direct phosphorylation of NFκB by kinases downstream from PKCδ, such as p38 MAPK or Akt [22,59,60]). Interestingly, recent studies identified reciprocal regulation of PKCδ by NFκB, by showing that a NFκB-responsive regulatory element in the PKCδ promoter links tumour necrosis factor-α stimulation to increased PKCδ mRNA and protein expression [61].
PKCδ also plays a role in transcriptional regulation by phosphorylating STAT1, STAT3 and p300 [62–64]. PKCδ-dependent phosphorylation of STAT1 on Ser727 is required for the transcriptional regulation of interferon-sensitive genes, whereas PKCδ-dependent phosphorylation of Ser727 of STAT3 reduces its DNA-binding and transcriptional activity. p300 is a transcriptional co-activator/histone acetyltransferase that is also phosphorylated at Ser89 by PKCδ (and not cPKCs). This modification is reported to repress p300 transcriptional co-activator function by inhibiting its intrinsic histone acetyltransferase activity [64]. Finally, PKCδ is reported to form a complex with the seven-transmembrane-spanning-domain Frizzled receptor, where it plays an essential role in the Wnt/JNK (c-Jun N-terminal kinase) pathway by regulating the localization and activity of Dishevelled [65].
PKCδ has emerged as a common mediator of apoptosis in response to many stimuli. However, the precise mechanism(s) executing PKCδ's pro-apoptotic actions may vary considerably, depending upon the biological context (the cell type, the nature of the pro-apoptotic stimulus, etc. [66]). For example, a PKCδ/p38/MAPK pathway mediates the pro-apoptotic effects of PMA in androgen-dependent LNCaP prostate cancer cells. In this system, allosterically activated PKCδ in membranes appears to be sufficient to induce apoptosis; proteolytic cleavage of PKCδ by caspase (to liberate a constitutively active catalytic fragment) is not detected [67].
PKCδ plays a central role in the genotoxic stress response leading to the induction of apoptosis in cells exposed to DNA-damaging agents. Activation of tyrosine kinases (Abl, Lyn) leads to the tyrosine phosphorylation (activation) of PKCδ and the activation of a MEKK1 (MEK kinase 1)/MKK7 (MAPK kinase 7)/JNK pathway [68]. Full-length PKCδ is detected in the nucleus during the initial phase of the genotoxic stress response in certain cell types. The structural requirements for entry of PKCδ into the nucleus have been mapped to six basic amino acid residues in a functional bipartite nuclear localization sequence in the C-terminus of PKCδ (K611RKVEPPFKPKVK623) [69]. Tyrosine phosphorylation is not required for this initial nuclear targeting of full-length PKCδ [39]. However, tyrosine phosphorylation serves as an amplification step at later stages of the apoptosis signalling response, since tyrosine phosphorylation (at Tyr187 or at the Tyr311/Tyr332 pair of residues that flank the caspase cleavage site) is required for caspase activation, caspase-mediated proteolytic cleavage of PKCδ, and the generation of the 40 kDa catalytic domain fragment of PKCδ that accumulates in the nucleus at an even higher rate than the full-length protein [39,47,70,71]. The 40 kDa catalytic domain fragment of PKCδ is a constitutively active enzyme (since it is freed from the autoinhibitory constraints imposed by the N-terminal regulatory domain); it induces apopotosis when overexpressed in certain cell types [71]. Of note, a recent study identified certain differences in the enzymology of the freed PKCδ catalytic domain fragment and the full-length enzyme: the freed PKCδ catalytic domain fragment acts as a sphingosine-dependent kinase to phosphorylate 14-3-3 proteins, whereas full-length PKCδ is inhibited by sphingosine [16]. These results might suggest that full-length PKCδ and the freed catalytic domain fragment could play distinct roles (phosphorylating distinct nuclear targets) at different stages of apoptosis. Indeed, among the few known nuclear targets for PKCδ, some differences have already surfaced. The nuclear targets for PKCδ identified to date include: (1) nuclear DNA-PK (DNA-dependent protein kinase), an enzyme essential for the repair of double-stranded DNA breaks that is inhibited by PKCδ-dependent phosphorylation [72]; (2) hRad9, a key component of the genotoxin-activated checkpoint signalling complex which also binds anti-apoptosis Bcl-2 family proteins and mediates apoptotic responses to DNA damage when phosphorylated by PKCδ [73]; (3) lamin B, a nuclear structural protein that is cleaved upon phosphorylation, leading to the disassembly of the nuclear lamina [74]; (4) c-Abl, a kinase activated by apoptotic stimuli that forms complexes with Lyn, DNA-PK and PKCδ in the nucleus; and (5) p73β, a structural/functional homologue of p53 that interacts with c-Abl complexes and activates transcription from p53 promoters [75]. p73β and DNA-PK are two examples of nuclear PKCδ substrates that are phosphorylated preferentially by the freed PKCδ catalytic fragment relative to the full-length enzyme.
PKCδ localizes to mitochondria as part of the pro-apoptotic signalling mechanism triggered by PMA or oxidant stress in keratinocytes, U-937 cells and MCF-7 cells. PKCδ translocation to mitochondria has been described as a kinase-dependent process that induces apoptotic cell death by amplifying local ceramide formation, altering the local regulation of calcium signalling events in mitochondria (without influencing the global processing of calcium signals in the cytosol), and mediating the H2O2-dependent loss of membrane potential, release of cytochrome c and activation of caspase 3 [76–79]. Many of the mitochondrial actions of PKCδ have been attributed to a physical and functional interaction with c-Abl. The consensus of most studies is that oxidant stress activates PKCδ via a c-Abl-independent mechanism, since H2O2 promotes equivalent PKCδ activation, PKCδ translocation to mitochondria, and PKCδ tyrosine phosphorylation in wild-type and c-Abl−/− cells [78]. However, c-Abl activation and translocation to the mitochondria requires PKCδ [80,81]. Once activated, c-Abl phosphorylates PKCδ at Tyr512, which further increases the kinase activity of PKCδ. In this fashion, a PKCδ/Abl amplification loop drives the mitochondrial death pathway. Few biologically relevant substrates of PKCδ (or c-Abl) in mitochondria have been identified. One notable exception is PLS3 (phospholipid scramblase 3), a mitochondrial form of PLS that sensitizes cells to PKCδ-dependent apoptosis (Table 1) [82].
Table 1. Substrates of PKCδ.
HAT, histone acetyltransferase.
| Category | Substrate | Consequences | Ref. |
|---|---|---|---|
| Signalling molecules | SFKs | Variable (see text) | [40,42] |
| c-Abl | Increased activity | [80] | |
| SHPTP1 protein tyrosine phosphatase (SHP1) | Decreased phosphatase activity | [118] | |
| Protein tyrosine phosphatase PTPα | Increased phosphatase activity; Src activation | [43] | |
| RasGRP | Uncertain | [110] | |
| PKCε (hydrophobic motif) | Promotes release from membranes and down-regulation | [14] | |
| STAT1 (Ser727) | Required for interferon-mediated gene transcription | [62] | |
| STAT3 (Ser727) | Reduced DNA-binding and transcriptional activity | [63] | |
| p300 | Decreased HAT activity; repressed transcriptional co-activator function | [64] | |
| 14-3-3 (at the putative helix 3 dimer interface) | Presumed to interfere with 14-3-3 dimerization and interactions with binding partners (Bad, Raf, etc.) | [16] | |
| gp130 | Increased gp130–STAT3 interaction | [119] | |
| p47phox subunit of NADPH oxidase | Increased activity | [120] | |
| β4 integrin | Decreased cell attachment to laminin | [121] | |
| Mitochondrial proteins | PLS3 | Increased transbilayer phospholipid movement | [82] |
| Nuclear proteins | DNA-PK | Decreased activity; increased DNA-damage-induced apoptosis | [72] |
| Lamin B | Apoptosis | [74] | |
| hRad4 | Increased hRad9–Bcl-2 interactions; apoptosis | [73] | |
| p73β (Ser289) | p73β activation; apoptosis | [75] |
Many of the predictions regarding PKCδ function – derived from studies in cell culture systems – have been validated in PKCδ−/− mice, which develop and reproduce grossly normally, but display defects that expose critical roles for PKCδ in immune function and vascular biology. PKCδ−/− mice exhibit autonomous hyperproliferation of B cells, leading to the development of immune-complex glomerulonephritis and lymphocyte infiltration in many organs; these results have been taken as evidence that PKCδ is a critically important negative regulator of B-cell proliferation [83]. PKCδ−/− vessels (isografted into PKCδ−/− or PKCδ+/+ recipient mice) are also more prone to vein graft arteriosclerosis, relative to PKCδ+/+ vessels [84]. In vitro studies established that smooth muscle cells isolated from PKCδ−/− vessels are similar to wild type with respect to mitogen-stimulated proliferation in vitro, but PKCδ−/− smooth muscle cell cultures produce significantly less ROS (reactive oxygen species) in response to UV irradiation and are markedly resistant to H2O2-induced cell death [84]. A recent study also identified a critical role for neutrophil PKCδ in brain injury in a murine stroke model [85]. This study linked the defect in PKCδ−/− neutrophil function (impaired adhesion, migration, respiratory burst and degranulation) to reduced neutrophil migration into ischaemic brain tissue and reduced reperfusion tissue injury. Finally, there is evidence that mechanical stress promotes translocation of PKCδ to the cytoskeleton and cell migration; mechanical stress-induced migration is defective in smooth muscle cells cultured from PKCδ−/− mice, which exhibit abnormal cytoskeleton structure and diminished stress-induced phosphorylation of paxillin, focal adhesion kinase and vinculin [86]. Collectively, studies in PKCδ−/− mice support the notion that PKCδ plays a pivotal role in oxidant signalling as well as in mechanisms that maintain the delicate balance between cell proliferation and apoptosis.
PKCδ LOCALIZATION AND FUNCTION IN CARDIOMYOCYTES
Recent studies have identified stimulus-specific differences in the mechanism of activation and spatial distribution of PKCδ (that are predicted to lead to distinct PKCδ-triggered responses) in cardiomyocytes. PKCδ is detected (along with PKCα, PKCε and PKCλ) in most cardiomyocyte preparations. In cultured neonatal rat cardiomyocytes, PMA-sensitive PKC isoforms are targeted to distinct subcellular localizations [87–90]. PKCα and PKCε reside in the cytosol at rest, and translocate to the perinuclear region (PKCα) and sarcomeres (PKCε) on activation [91,92]. Native PKCδ translocates from the nucleus to the fibrillar cytoskeleton, perinucleus and focal contacts [91]. Heterologously overexpressed WT-PKCδ (at levels 6–8-fold over those of endogenous PKCδ) translocates from the cytosol and perinuclear region to the nucleus [92]. The consistent identification of both endogenous and overexpressed PKCδ in the nucleus is intriguing, given the known nuclear targets and actions of PKCδ.
PMA-sensitive PKC isoforms also localize (in their active conformation) to buoyant cholesterol- and sphingolipid/glycosphingolipid-enriched membrane microdomains, termed caveolae or lipid rafts. We demonstrated previously that cardiomyocyte caveolae contain a resident ERK signalling cascade (consisting of A-Raf/c-Raf-1, MEK and ERK1/2); PMA promotes translocation of PKCα, PKCδ and PKCε to caveolae and induces a local increase in ERK1/2 activity [93]. These results suggest that caveolae may nucleate PKC-dependent signalling pathways. Our recent studies also suggest that caveolae may constitute particularly important platforms to facilitate cross-talk between PKCδ and SFKs (which also localize to this compartment).
Hypoxia and H2O2 result in the release of PKCδ from the membrane fraction of neonatal rat cardiomyocytes [15,94]; H2O2 has also been reported to release PKCδ from CHO cell membranes [95]. The effect of H2O2 to promote PKCδ translocation to the soluble fraction is specific; PKCα and PKCε are not released from membranes under these conditions. Importantly, the tyrosine-phosphorylated form of PKCδ that accumulates in the soluble fraction of H2O2-treated cardiomyocytes acts as a lipid-independent kinase, suggesting a novel paradigm for PKCδ's actions during oxidant stress.
Many studies implicate PKC isoforms in mechanisms leading to cardiac hypertrophy and ventricular remodelling. The earliest link between PKC and cardiac hypertrophy came from studies that identified PKC (including PKCδ) activation and/or up-regulation in numerous models of cardiac hypertrophy. However, it has become increasingly evident that individual PKC isoforms exert distinct cardiac actions. The preponderance of the literature has concentrated on the cardiac actions of PKCα, PKCβ and PKCε, which activate ERK, induce cardiomyocyte hypertrophy and mediate ischaemic preconditioning [96–99]. PKCδ (along with PKCε) has been implicated in the down-regulation of SERCA2 [sarco(endo)plasmic reticulum Ca2+-ATPase] expression, a characteristic feature of the hypertrophic phenotype; PKCδ overexpression is also reported to activate JNK and p38 MAPK (but not ERK), promote cell detachment, and induce cardiomyocyte apoptosis [100,101]. However, it is important to note that activation of JNK and p38 MAPK by PKCδ generally has been detected at a time when apoptosis is already well developed (well beyond the initial period of PKCδ expression). Hence, while the p38 MAPK pathway has been implicated in the PKCδ-driven apoptosis pathway in prostate cell lines, the relative importance of the JNK/p38 MAPK pathways as a cause (rather than a consequence) of apoptosis in cardiomyocytes has not yet been resolved [102].
PKCδ function has been interrogated in the intact heart by overexpressing δV1 (a peptide designed to competitively inhibit docking of PKCδ to its specific membrane-anchoring protein, or RACK) or ψδRACK (a peptide that is believed to destabilize the inactive ‘closed’ conformation of PKCδ by preventing an intramolecular interaction between the ψδRACK sequence and the RACK-binding site; Figure 1A). This strategy is designed to influence PKCδ signalling, without altering the natural stoichiometry of PKCδ in relation to its upstream activators or downstream substrates. Using this approach, PKCδ activation is reported to worsen cell damage during an ischaemic insult, whereas PKCδ inhibition confers cardioprotection [103,104]. Of note, the salutary effects of the PKCδ translocation inhibitor peptide (δV1) are identified only at low levels of overexpression. δV1 overexpression at higher levels results in a lethal cardiomyopathy with contractile dysfunction and histological changes that resemble a desmin (or myofibrillar) cardiomyopathy. The δV1-induced cardiomyopathy is quite distinct from the dilated cardiomyopathy that develops in mice that overexpress the PKCε translocation inhibitor peptide (εV1), further supporting the notion that nPKC isoforms are ‘hard-wired’ to different downstream signalling pathways [105]. Finally, δV1 (PKCδ inhibition) blocks apoptosis/necrosis when infused into the coronary vessels at the time of reperfusion in a porcine model of acute myocardial infarction [106]. This and related studies have provided the rationale to develop PKCδ translocation inhibitors as therapies to prevent irreversible reperfusion injury in humans. However, the precise mechanism(s) for the biological actions of δV1 may not be as straightforward as generally assumed, for several reasons. (1) PKCδ is released from the membranes during oxidant stress. A peptide inhibitor that prevents RACK-driven PKC isoform compartmentation would not be predicted to prevent substrate phosphorylation in the soluble fraction of H2O2-treated cells. (2) PKCδ localizes to multiple subcellular compartments (including surface membranes, lipid rafts, mitochondria and nuclei). The subcellular location of PKCδ's RACK, and the translocation event(s) blocked by δV1, have not been identified. (3) PKCδ translocation events and function can be regulated by tyrosine phosphorylation, with mounting evidence that pools of PKCδ (that differ in their phosphorylation patterns and binding partners) may mediate specialized functions in distinct subcellular compartments. It is perhaps relevant that certain PKCδ tyrosine phosphorylation sites are in the C2 domain (Tyr52 and Tyr64), adjacent to the ψδRACK (or PKCδ activator peptide) sequence. In theory, δV1 might inhibit certain actions of PKCδ by interfering with PKCδ interactions with non-RACK binding partners (such as SFKs) and/or preventing PKCδ phosphorylation by SFKs. This entirely distinct mechanism for PKCδ regulation by translocation inhibitor or activator peptides deserves further study.
Hyperglycaemia has been reported to promote the translocation of PKCδ to membranes, increase ROS production and induce apoptosis in adult cardiomyocytes [107]. While the hyperglycaemia-dependent increase in ROS might be expected to promote tyrosine phosphorylation of PKCδ, a role for PKCδ tyrosine phosphorylation in cell-based or intact tissue models of hyperglycaemia has never been considered. Other recent studies link nPKC (PKCδ and/or PKCε) activation to decreased Akt phosphorylation and blunted Akt activation by EGF receptors [108]. This mechanism is speculated to render cardiomyocytes more vulnerable to stress-induced apoptosis and to contribute to the transition from hypertrophy to cardiac failure.
THE FUTURE CHALLENGE: TO IDENTIFY PKCδ SUBSTRATES
A detailed understanding of the cellular actions of individual PKC isoforms ultimately will require knowledge of their distinct cellular substrates. To date, there has been only limited progress in this area; Table 1 represents a list of proteins identified as PKCδ substrates. PKCδ substrates traditionally have been identified using pharmacological strategies (with activators or inhibitors) or molecular strategies (involving targeted deletion or overexpression of individual PKC isoforms or translocation modifier peptides). Each of these approaches is accompanied by its own distinct set of problems. For example, PMA may not strictly report PKC actions, since high-affinity DAG/PMA-binding C1 domains have been identified in proteins that lack kinase domains (and are unrelated to PKC). The chimaerins (a family of Rac GTPase-activating proteins), RasGRPs (Ras/Rap1 exchange factors) and Munc13 isoforms (scaffolding proteins involved in exocytosis) are examples of proteins that bind PMA with nanomolar affinity and translocate to membranes in response to PMA [109]. The interpretation of studies with PMA can be confounded further by cross-talk between PKCs and other members of the extended phorbol ester binding protein family. For example, recent studies identified RasGRP3 (a guanine nucleotide exchange factor for Ras) as a binding partner and substrate for PKCδ [110]. These proteins co-localize to similar subcellular compartments and interact functionally (in an as yet poorly understood manner) at the level of downstream signalling pathways such as ERK.
Studies with PKC inhibitors can be even more problematic, since many commonly used PKC inhibitors lack the requisite specificity for PKC. For example, chelerythrine (which has been used widely as a general PKC inhibitor) induces apoptosis through a mitochondrial mechanism that is unrelated to PKC inhibition; chelerythrine is also reported to inhibit interactions of Bcl-XL with BH3 (Bcl-2 homology domain 3)-containing proteins such as Bax [111–113]. Rottlerin (which has been touted to be a selective PKCδ inhibitor) uncouples mitochondrial respiration from oxidative phosphorylation and exerts inhibitory actions in PKCδ−/− cells; some studies even challenge the efficacy of rottlerin as an in vitro PKCδ inhibitor [114,115]. Finally, even Ro318220 and GF109203X (agents considered to be relatively selective PKC inhibitors) are reported to inhibit RSK (p90 ribosomal S6 kinase) and p70 S6 kinase [116]. These caveats emphasize that studies using PKC inhibitors as tools to interrogate PKC function must be interpreted with caution, and that newer strategies to identify PKCδ targets are imperative.
A novel chemical genetic approach developed by Shokat and colleagues [117] represents a very exciting methodological breakthrough that holds tremendous promise for the identification of endogenous PKC substrates in future studies. Their approach involves engineering the ATP-binding site in a kinase of interest so that it accepts a structurally modified γ-32P-labelled ATP analogue with a bulky substitution attached at the N6 position [for example N6-(benzyl)ATP or N6-(phenethyl)ATP]. Since the chemically modified γ-32P-labelled ATP analogue binds only the mutated enzyme's active site (which carries an alanine or glycine in place of a conserved bulky residue), only unique substrates of the kinase of interest are labelled in vivo in cells. This strategy has been used to identify substrates of JNK, CDK2 (cyclin-dependent kinase 2) and v-Src. It represents a viable strategy to identify the distinct substrates of allosterically activated PKCδ in membranes as well as tyrosine-phosphorylated PKCδ in the soluble fractions of cells exposed to oxidant stress.
Acknowledgments
Work in the author's laboratory is supported by USPHS-NHLBI grants HL-64639 and HL-77860.
References
- 1.Dempsey E. C., Newton A. C., Mochly-Rosen D., Fields A. P., Reyland M. E., Insel P. A., Messing R. O. Protein kinase C isozymes and the regulation of diverse cell responses. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000;279:L429–L438. doi: 10.1152/ajplung.2000.279.3.L429. [DOI] [PubMed] [Google Scholar]
- 2.Sabri A., Steinberg S. F. Protein kinase C isoform-selective signals that lead to cardiac hypertrophy and the progression of heart failure. Mol. Cell. Biochem. 2003;251:97–101. [PubMed] [Google Scholar]
- 3.Solaro R. J., Burkart E. M. Functional defects in troponin and the systems biology of heart failure. J. Mol. Cell. Cardiol. 2002;34:689–693. doi: 10.1006/jmcc.2002.2028. [DOI] [PubMed] [Google Scholar]
- 4.Cho W. Membrane targeting by C1 and C2 domains. J. Biol. Chem. 2001;276:32407–32410. doi: 10.1074/jbc.R100007200. [DOI] [PubMed] [Google Scholar]
- 5.Hurley J. H., Misra S. Signaling and subcellular targeting by membrane-binding domains. Annu. Rev. Biophys. Biomol. Struct. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pappa H., Murray-Rust J., Dekker L. V., Parker P. J., McDonald N. Q. Crystal structure of the C2 domain from protein kinase C-δ. Structure. 1998;6:885–894. doi: 10.1016/s0969-2126(98)00090-2. [DOI] [PubMed] [Google Scholar]
- 6a.Hirai T., Chida K. Protein kinase Czeta: activation mechanisms and cellular functions. J. Biochem. (Tokyo) 2003;133:1–7. doi: 10.1093/jb/mvg017. [DOI] [PubMed] [Google Scholar]
- 6b.Farese R. V. Function and dysfunction of aPKC isoforms for glucose transport in insulin-sensitive and insulin-resistant states. Am. J. Physiol. Endocrinol. Metab. 2002;283:E1–E11. doi: 10.1152/ajpendo.00045.2002. [DOI] [PubMed] [Google Scholar]
- 7.Jaken S., Parker P. J. Protein kinase C binding partners. BioEssays. 2000;22:245–254. doi: 10.1002/(SICI)1521-1878(200003)22:3<245::AID-BIES6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 8.Mackay K., Mochly-Rosen D. Localization, anchoring, and functions of protein kinase C isozymes in the heart. J. Mol. Cell. Cardiol. 2001;33:1301–1307. doi: 10.1006/jmcc.2001.1400. [DOI] [PubMed] [Google Scholar]
- 9.Schechtman D., Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- 10.Csukai M., Chen C. H., De Matteis M. A., Mochly-Rosen D. The coatomer protein β'-COP, a selective binding protein (RACK) for protein kinase Cε. J. Biol. Chem. 1997;272:29200–29206. doi: 10.1074/jbc.272.46.29200. [DOI] [PubMed] [Google Scholar]
- 11.Robles-Flores M., Rendon-Huerta E., Gonzalez-Aguilar H., Mendoza-Hernandez G., Islas S., Mendoza V., Ponce-Castaneda M. V., Gonzalez-Mariscal L., Lopez-Casillas F. p32 (gC1qBP) is a general protein kinase C (PKC)-binding protein. J. Biol. Chem. 2002;277:5247–5255. doi: 10.1074/jbc.M109333200. [DOI] [PubMed] [Google Scholar]
- 12.Chen S., Dell E. J., Lin F., Sai J., Hamm H. E. RACK1 regulates specific functions of Gβγ. J. Biol. Chem. 2004;279:17861–17868. doi: 10.1074/jbc.M313727200. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan B. M., Harrison-Lavoie K. J., Marshansky V., Lin H. Y., Kehrl J. H., Ausiello D. A., Brown D., Druey K. M. RGS4 and RGS2 bind coatomer and inhibit COPI association with Golgi membranes and intracellular transport. Mol. Biol. Cell. 2000;11:3155–3168. doi: 10.1091/mbc.11.9.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rybin V. O., Sabri A., Short J., Braz J. C., Molkentin J. D., Steinberg S. F. Cross regulation of nPKC isoform function in cardiomyocytes: Role of PKCε in activation loop phosphorylations and PKCδ in hydrophobic motif phosphorylations. J. Biol. Chem. 2003;278:14555–14564. doi: 10.1074/jbc.M212644200. [DOI] [PubMed] [Google Scholar]
- 15.Rybin V. O., Guo J., Sabri A., Elouardighi H., Schaefer E., Steinberg S. F. Stimulus-specific differences in PKCδ localization and activation mechanisms in cardiomyocytes. J. Biol. Chem. 2004;279:19350–19361. doi: 10.1074/jbc.M311096200. [DOI] [PubMed] [Google Scholar]
- 16.Hamaguchi A., Suzuki E., Murayama K., Fujimura T., Hikita T., Iwabuchi K., Handa K., Withers D. A., Masters S. C., Fu H., Hakomori S. Sphingosine-dependent protein kinase-1 (SDK1), directed to 14-3-3, is identified as the kinase domain of PKCδ. J. Biol. Chem. 2003;278:41557–41565. doi: 10.1074/jbc.M305294200. [DOI] [PubMed] [Google Scholar]
- 17.Parekh D. B., Ziegler W., Parker P. J. Multiple pathways control protein kinase C phosphorylation. EMBO J. 2000;19:496–503. doi: 10.1093/emboj/19.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton A. C., Johnson J. E. Protein kinase C: a paradigm for regulation of protein function by two membrane-targeting modules. Biochim. Biophys. Acta. 1998;1376:155–172. doi: 10.1016/s0304-4157(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 19.Marshall C. J. Signal transduction. Hot lips and phosphorylation of protein kinases. Nature (London) 1994;367:686. doi: 10.1038/367686a0. [DOI] [PubMed] [Google Scholar]
- 20.Le Good J. A., Ziegler W. H., Parekh D. B., Alessi D. R., Cohen P., Parker P. J. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 21.Sonnenburg E. D., Gao T., Newton A. C. The phosphoinositide-dependent kinase, PDK-1, phosphorylates conventional protein kinase C isozymes by a mechanism that is independent of phosphoinositide 3-kinase. J. Biol. Chem. 2001;276:45289–45297. doi: 10.1074/jbc.M107416200. [DOI] [PubMed] [Google Scholar]
- 22.Rahman A., Anwar K. N., Uddin S., Xu N., Ye R. D., Platanias L. C., Malik A. B. Protein kinase C-δ regulates thrombin-induced ICAM-1 gene expression in endothelial cells via activation of p38 mitogen-activated protein kinase. Mol. Cell. Biol. 2001;21:5554–5565. doi: 10.1128/MCB.21.16.5554-5565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogasa M., Miyazaki Y., Hiraoka S., Kitamura S., Nagasawa Y., Kishida O., Miyazaki T., Kiyohara T., Shinomura Y., Matsuzawa Y. Gastrin activates nuclear factor kappaB (NFκB) through a protein kinase C dependent pathway involving NFκB inducing kinase, inhibitor kappaB (IκB) kinase, and tumour necrosis factor receptor associated factor 6 (TRAF6) in MKN-28 cells transfected with gastrin receptor. Gut. 2003;52:813–819. doi: 10.1136/gut.52.6.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brändlin I., Eiseler T., Salowsky R., Johannes F. J. Protein kinase Cμ regulation of the JNK pathway is triggered via phosphoinositide-dependent kinase 1 and protein kinase Cε. J. Biol. Chem. 2002;277:45451–45457. doi: 10.1074/jbc.M205299200. [DOI] [PubMed] [Google Scholar]
- 25.Stempka L., Schnolzer M., Radke S., Rincke G., Marks F., Gschwendt M. Requirements of protein kinase C δ for catalytic function. J. Biol. Chem. 1999;274:8886–8892. doi: 10.1074/jbc.274.13.8886. [DOI] [PubMed] [Google Scholar]
- 26.Feng X., Becker K. P., Stribling S. D., Peters K. G., Hannun Y. A. Regulation of receptor-mediated protein kinase C membrane trafficking by autophosphorylation. J. Biol. Chem. 2000;275:17024–17034. doi: 10.1074/jbc.275.22.17024. [DOI] [PubMed] [Google Scholar]
- 27.Behn-Krappa A., Newton A. C. The hydrophobic phosphorylation motif of conventional protein kinase C is regulated by autophosphorylation. Curr. Biol. 1999;9:728–737. doi: 10.1016/s0960-9822(99)80332-7. [DOI] [PubMed] [Google Scholar]
- 28.Cenni V., Doppler H., Sonnenburg E. D., Maraldi N., Newton A. C., Toker A. Regulation of novel protein kinase Cε by phosphorylation. Biochem. J. 2002;363:537–545. doi: 10.1042/0264-6021:3630537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parekh D., Ziegler W., Yonezawa K., Hara K., Parker P. J. Mammalian TOR controls one of two kinase pathways acting upon nPKCδ and nPKCε. J. Biol. Chem. 1999;274:34758–34764. doi: 10.1074/jbc.274.49.34758. [DOI] [PubMed] [Google Scholar]
- 30.Denning M. F., Dlugosz A. A., Threadgill D. W., Magnuson T., Yuspa S. H. Activation of the epidermal growth factor receptor signal transduction pathway stimulates tyrosine phosphorylation of protein kinase Cδ. J. Biol. Chem. 1996;271:5325–5331. doi: 10.1074/jbc.271.10.5325. [DOI] [PubMed] [Google Scholar]
- 31.Konishi H., Tanaka M., Takemura Y., Matsuzaki H., Ono Y., Kikkawa U., Nishizuka Y. Activation of protein kinase C by tyrosine phosphorylation in response to H2O2. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11233–11237. doi: 10.1073/pnas.94.21.11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., Mischak H., Yu J. C., Wang L. M., Mushinski J. F., Heidaran M. A., Pierce J. H. Tyrosine phosphorylation of protein kinase C-δ in response to its activation. J. Biol. Chem. 1994;269:2349–2352. [PubMed] [Google Scholar]
- 33.Benes C., Soltoff S. P. Modulation of PKCδ tyrosine phosphorylation and activity in salivary and PC-12 cells by Src kinases. Am. J. Physiol. Cell. Physiol. 2001;280:C1498–C1510. doi: 10.1152/ajpcell.2001.280.6.C1498. [DOI] [PubMed] [Google Scholar]
- 34.Acs P., Beheshti M., Szallasi Z., Li L., Yuspa S. H., Blumberg P. M. Effect of a tyrosine 155 to phenylalanine mutation of protein kinase C-δ on the proliferative and tumorigenic properties of NIH 3T3 fibroblasts. Carcinogenesis. 2000;21:887–891. doi: 10.1093/carcin/21.5.887. [DOI] [PubMed] [Google Scholar]
- 35.Li W., Chen X. H., Kelley C. A., Alimandi M., Zhang J., Chen Q., Bottaro D. P., Pierce J. H. Identification of tyrosine 187 as a protein kinase C-δ phosphorylation site. J. Biol. Chem. 1996;271:26404–26409. doi: 10.1074/jbc.271.42.26404. [DOI] [PubMed] [Google Scholar]
- 36.Kronfeld I., Kazimirsky G., Lorenzo P. S., Garfield S. H., Blumberg P. M., Brodie C. Phosphorylation of protein kinase Cδ on distinct tyrosine residues regulates specific cellular functions. J. Biol. Chem. 2000;275:35491–35498. doi: 10.1074/jbc.M005991200. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe T., Ono Y., Taniyama Y., Hazama K., Igarashi K., Ogita K., Kikkawa U., Nishizuka Y. Cell division arrest induced by phorbol ester in CHO cells overexpressing protein kinase C-δ subspecies. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10159–10163. doi: 10.1073/pnas.89.21.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brodie C., Bogi K., Acs P., Lorenzo P. S., Baskin L., Blumberg P. M. PKCδ inhibits the expression of glutamine synthetase in glial cells via the PKCδ regulatory domain and its tyrosine phosphorylation. J. Biol. Chem. 1998;273:30713–30718. doi: 10.1074/jbc.273.46.30713. [DOI] [PubMed] [Google Scholar]
- 39.Blass M., Kronfeld I., Kazimirsky G., Blumberg P. M., Brodie C. Tyrosine phosphorylation of protein kinase Cδ is essential for its apoptotic effect in response to etoposide. Mol. Cell. Biol. 2002;22:182–195. doi: 10.1128/MCB.22.1.182-195.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J. S., Swann P. G., Szallasi Z., Blank U., Blumberg P. M., Rivera J. Tyrosine phosphorylation-dependent and -independent associations of protein kinase C-δ with Src family kinases in the RBL-2H3 mast cell line. Oncogene. 1998;16:3357–3368. doi: 10.1038/sj.onc.1201886. [DOI] [PubMed] [Google Scholar]
- 41.Gould K. L., Woodgett J. R., Cooper J. A., Buss J. E., Shalloway D., Hunter T. Protein kinase C phosphorylates pp60src at a novel site. Cell. 1985;42:849–857. doi: 10.1016/0092-8674(85)90281-8. [DOI] [PubMed] [Google Scholar]
- 42.Zang Q., Lu Z., Curto M., Barile N., Shalloway D., Foster D. A. Association between v-Src and protein kinase Cδ in v-Src-transformed fibroblasts. J. Biol. Chem. 1997;272:13275–13280. doi: 10.1074/jbc.272.20.13275. [DOI] [PubMed] [Google Scholar]
- 43.Brandt D. T., Goerke A., Heuer M., Gimona M., Leitges M., Kremmer E., Lammers R., Haller H., Mischak H. Protein kinase Cδ induces Src kinase activity via activation of the protein tyrosine phosphatase PTPα. J. Biol. Chem. 2003;278:34073–34078. doi: 10.1074/jbc.M211650200. [DOI] [PubMed] [Google Scholar]
- 44.Leitges M., Gimborn K., Elis W., Kalesnikoff J., Hughes M. R., Krystal G., Huber M. Protein kinase C-δ is a negative regulator of antigen-induced mast cell degranulation. Mol. Cell. Biol. 2002;22:3970–3980. doi: 10.1128/MCB.22.12.3970-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goerke A., Sakai N., Gutjahr E., Schlapkohl W. A., Mushinski J. F., Haller H., Kolch W., Saito N., Mischak H. Induction of apoptosis by protein kinase C-δ is independent of its kinase activity. J. Biol. Chem. 2002;277:32054–32062. doi: 10.1074/jbc.M203734200. [DOI] [PubMed] [Google Scholar]
- 46.Konishi H., Yamauchi E., Taniguchi H., Yamamoto T., Matsuzaki H., Takemura Y., Ohmae K., Kikkawa U., Nishizuka Y. Phosphorylation sites of protein kinase Cδ in H2O2-treated cells and its activation by tyrosine kinase in vitro. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6587–6592. doi: 10.1073/pnas.111158798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blake R. A., Garcia-Paramio P., Parker P. J., Courtneidge S. A. Src promotes PKCδ degradation. Cell Growth Differ. 1999;10:231–241. [PubMed] [Google Scholar]
- 48.Fukumoto S., Nishizawa Y., Hosoi M., Koyama H., Yamakawa K., Ohno S., Morii H. Protein kinase C-δ inhibits the proliferation of vascular smooth muscle cells by suppressing G1 cyclin expression. J. Biol. Chem. 1997;272:13816–13822. doi: 10.1074/jbc.272.21.13816. [DOI] [PubMed] [Google Scholar]
- 49.Ashton A. W., Watanabe G., Albanese C., Harrington E. O., Ware J. A., Pestell R. G. Protein kinase Cδ inhibition of S-phase transition in capillary endothelial cells involves the cyclin-dependent kinase inhibitor p27(Kip1) J. Biol. Chem. 1999;274:20805–20811. doi: 10.1074/jbc.274.30.20805. [DOI] [PubMed] [Google Scholar]
- 50.Braun M. U., Mochly-Rosen D. Opposing effects of δ- and ζ-protein kinase C isozymes on cardiac fibroblast proliferation: use of isozyme-selective inhibitors. J. Mol. Cell. Cardiol. 2003;35:895–903. doi: 10.1016/s0022-2828(03)00142-1. [DOI] [PubMed] [Google Scholar]
- 51.Ueda Y., Hirai S. I., Osada S. I., Suzuki A., Mizuno K., Ohno S. Protein kinase C-δ activates the MEK-ERK pathway in a manner independent of ras and dependent on raf. J. Biol. Chem. 1996;271:23512–23519. doi: 10.1074/jbc.271.38.23512. [DOI] [PubMed] [Google Scholar]
- 52.Keshamouni V. G., Mattingly R. R., Reddy K. B. Mechanism of 17-β-estradiol-induced Erk1/2 activation in breast cancer cells; a role for HER2 and PKC-δ. J. Biol. Chem. 2002;277:22558–22565. doi: 10.1074/jbc.M202351200. [DOI] [PubMed] [Google Scholar]
- 53.Abbas T., White D., Hui L., Yoshida K., Foster D. A., Bargonetti J. Inhibition of human p53 basal transcription by down-regulation of protein kinase Cδ. J. Biol. Chem. 2004;279:9970–9977. doi: 10.1074/jbc.M306979200. [DOI] [PubMed] [Google Scholar]
- 54.Kiley S. C., Clark K. J., Duddy S. K., Welch D. R., Jaken S. Increased protein kinase Cδ in mammary tumor cells: relationship to transformation and metastatic progression. Oncogene. 1999;18:6748–6757. doi: 10.1038/sj.onc.1203101. [DOI] [PubMed] [Google Scholar]
- 55.Wang Q., Wang X., Evers B. M. Induction of cIAP-2 in human colon cancer cells through PKCδ/NF-κB. J. Biol. Chem. 2003;278:51091–51099. doi: 10.1074/jbc.M306541200. [DOI] [PubMed] [Google Scholar]
- 56.Page K., Li J., Zhou L., Iasvovskaia S., Corbit K. C., Soh J. W., Weinstein I. B., Brasier A. R., Lin A., Hershenson M. B. Regulation of airway epithelial cell NF-κB-dependent gene expression by protein kinase C-δ. J. Immunol. 2003;170:5681–5689. doi: 10.4049/jimmunol.170.11.5681. [DOI] [PubMed] [Google Scholar]
- 57.Vancurova I., Miskolci V., Davidson D. NF-κB activation in tumor necrosis factor α-stimulated neutrophils is mediated by protein kinase Cδ. J. Biol. Chem. 2001;276:19746–19752. doi: 10.1074/jbc.M100234200. [DOI] [PubMed] [Google Scholar]
- 58.Storz P., Döppler H., Toker A. Protein kinase Cδ selectively regulates protein kinase D-dependent activation of NF-κB in oxidative stress signaling. Mol. Cell. Biol. 2004;24:2614–2626. doi: 10.1128/MCB.24.7.2614-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanden Berghe W., Plaisance S., Boone E., De Bosscher K., Schmitz M. L., Fiers W., Haegeman G. p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for NFκB p65 transactivation mediated by tumor necrosis factor. J. Biol. Chem. 1998;273:3285–3290. doi: 10.1074/jbc.273.6.3285. [DOI] [PubMed] [Google Scholar]
- 60.Rahman A., True A. L., Anwar K. N., Ye R. D., Voyno-Yasenetskaya T. A., Malik A. B. Gαq and Gβγ regulate PAR-1 signaling of thrombin-induced NF-κB activation and ICAM-1 transcription in endothelial cells. Circ. Res. 2002;91:398–405. doi: 10.1161/01.res.0000033520.95242.a2. [DOI] [PubMed] [Google Scholar]
- 61.Suh K. S., Tatunchak T. T., Crutchley J. M., Edwards L. E., Marin K. G., Yuspa S. H. Genomic structure and promoter analysis of PKC-δ. Genomics. 2003;82:57–67. doi: 10.1016/s0888-7543(03)00072-7. [DOI] [PubMed] [Google Scholar]
- 62.Uddin S., Sassano A., Deb D. K., Verma A., Majchrzak B., Rahman A., Malik A. B., Fish E. N., Platanias L. C. Protein kinase C-δ is activated by type I interferons and mediates phosphorylation of Stat1 on serine 727. J. Biol. Chem. 2002;277:14408–14416. doi: 10.1074/jbc.M109671200. [DOI] [PubMed] [Google Scholar]
- 63.Jain N., Zhang T., Kee W. H., Li W., Cao X. Protein kinase Cδ associates with and phosphorylates Stat3 in an interleukin-6-dependent manner. J. Biol. Chem. 1999;274:24392–24400. doi: 10.1074/jbc.274.34.24392. [DOI] [PubMed] [Google Scholar]
- 64.Yuan L. W., Soh J. W., Weinstein I. B. Inhibition of histone acetyltransferase function of p300 by PKCδ. Biochim. Biophys. Acta. 2002;1592:205–211. doi: 10.1016/s0167-4889(02)00327-0. [DOI] [PubMed] [Google Scholar]
- 65.Kinoshita N., Iioka H., Miyakoshi A., Ueno N. PKCδ is essential for Dishevelled function in a noncanonical Wnt pathway that regulates Xenopus convergent extension movements. Genes Dev. 2003;17:1663–1676. doi: 10.1101/gad.1101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brodie C., Blumberg P. M. Regulation of cell apoptosis by protein kinase C-δ. Apoptosis. 2003;8:19–27. doi: 10.1023/a:1021640817208. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka Y., Gavrielides M. V., Mitsuuchi Y., Fuji T., Kazanietz M. G. Protein kinase C promotes apoptosis in LNCaP prostate cancer cells through activation of p38 MAPK and inhibition of the AKT survival pathway. J. Biol. Chem. 2003;278:33753–33762. doi: 10.1074/jbc.M303313200. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida K., Miki Y., Kufe D. Activation of SAPK/JNK signaling by protein kinase Cδ in response to DNA damage. J. Biol. Chem. 2002;277:48372–48378. doi: 10.1074/jbc.M205485200. [DOI] [PubMed] [Google Scholar]
- 69.DeVries T. A., Neville M. C., Reyland M. E. Nuclear import of PKCδ is required for apoptosis: identification of a novel nuclear import sequence. EMBO J. 2002;21:6050–6060. doi: 10.1093/emboj/cdf606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghayur T., Hugunin M., Talanian R. V., Ratnofsky S., Quinlan C., Emoto Y., Pandey P., Datta R., Huang Y., Kharbanda S., et al. Proteolytic activation of protein kinase C δ by an ICE/CED 3-like protease induces characteristics of apoptosis. J. Exp. Med. 1996;184:2399–2404. doi: 10.1084/jem.184.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emoto Y., Manome Y., Meinhardt G., Kisaki H., Kharbanda S., Robertson M., Ghayur T., Wong W. W., Kamen R., Weichselbaum R., Kufe D. Proteolytic activation of protein kinase C δ by an ICE-like protease in apoptotic cells. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bharti A., Kraeft S. K., Gounder M., Pandey P., Jin S., Yuan Z. M., Lees-Miller S. P., Weichselbaum R., Weaver D., Chen L. B., et al. Inactivation of DNA-dependent protein kinase by protein kinase Cδ: implications for apoptosis. Mol. Cell. Biol. 1998;18:6719–6728. doi: 10.1128/mcb.18.11.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoshida K., Wang H. G., Miki Y., Kufe D. Protein kinase Cδ is responsible for constitutive and DNA damage-induced phosphorylation of Rad9. EMBO J. 2003;22:1431–1441. doi: 10.1093/emboj/cdg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cross T., Griffiths G., Deacon E., Sallis R., Gough M., Watters D., Lord J. M. PKC-δ is an apoptotic lamin kinase. Oncogene. 2000;19:2331–2337. doi: 10.1038/sj.onc.1203555. [DOI] [PubMed] [Google Scholar]
- 75.Ren J., Datta R., Shioya H., Li Y., Oki E., Biedermann V., Bharti A., Kufe D. p73β is regulated by protein kinase Cδ catalytic fragment generated in the apoptotic response to DNA damage. J. Biol. Chem. 2002;277:33758–33765. doi: 10.1074/jbc.M110667200. [DOI] [PubMed] [Google Scholar]
- 76.Sumitomo M., Ohba M., Asakuma J., Asano T., Kuroki T., Asano T., Hayakawa M. Protein kinase Cδ amplifies ceramide formation via mitochondrial signaling in prostate cancer cells. J. Clin. Invest. 2002;109:827–836. doi: 10.1172/JCI14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Majumder P. K., Pandey P., Sun X., Cheng K., Datta R., Saxena S., Kharbanda S., Kufe D. Mitochondrial translocation of protein kinase C-δ in phorbol ester-induced cytochrome c release and apoptosis. J. Biol. Chem. 2000;275:21793–21796. doi: 10.1074/jbc.C000048200. [DOI] [PubMed] [Google Scholar]
- 78.Majumder P. K., Mishra N. C., Sun X., Bharti A., Kharbanda S., Saxena S., Kufe D. Targeting of protein kinase C-δ to mitochondria in the oxidative stress response. Cell Growth Differ. 2001;12:465–470. [PubMed] [Google Scholar]
- 79.Pinton P., Leo S., Wieckowski M. R., Di Benedetto G., Rizzuto R. Long term modulation of mitochondrial Ca2+ signals by protein kinase C isozymes. J. Cell Biol. 2004;165:223–232. doi: 10.1083/jcb.200311061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun X., Wu F., Datta R., Kharbanda S., Kufe D. Interaction between protein kinase Cδ and the c-Abl tyrosine kinase in the cellular response to oxidative stress. J. Biol. Chem. 2000;275:7470–7473. doi: 10.1074/jbc.275.11.7470. [DOI] [PubMed] [Google Scholar]
- 81.Kumar S., Bharti A., Mishra N. C., Raina D., Kharbanda S., Saxena S., Kufe D. Targeting of the c-Abl tyrosine kinase to mitochondria in the necrotic cell death response to oxidative stress. J. Biol. Chem. 2001;276:17281–17285. doi: 10.1074/jbc.M101414200. [DOI] [PubMed] [Google Scholar]
- 82.Liu J., Chen J., Dai Q., Lee R. M. Phospholipid scramblase 3 is the mitochondrial target of protein kinase Cδ-induced apoptosis. Cancer Res. 2003;63:1153–1156. [PubMed] [Google Scholar]
- 83.Miyamoto A., Nakayama K., Imaki H., Hirose S., Jiang Y., Abe M., Tsukiyama T., Nagahama H., Ohno S., Hatakeyama S., Nakayama K. I. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cδ. Nature (London) 2002;416:865–869. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]
- 84.Leitges M., Mayr M., Braun U., Mayr U., Li C., Pfister G., Ghaffari-Tabrizi N., Baier G., Hu Y., Xu Q. Exacerbated vein graft arteriosclerosis in protein kinase Cδ-null mice. J. Clin. Invest. 2001;108:1505–1512. doi: 10.1172/JCI12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chou W. H., Choi D. S., Zhang H., Mu D., McMahon T., Kharazia V. N., Lowell C. A., Ferriero D. M., Messing R. O. Neutrophil protein kinase Cδ as a mediator of stroke-reperfusion injury. J. Clin. Invest. 2004;114:49–56. doi: 10.1172/JCI21655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li C., Wernig F., Leitges M., Hu Y., Xu Q. Mechanical stress-activated PKCδ regulates smooth muscle cell migration. FASEB J. 2003;17:2106–2108. doi: 10.1096/fj.03-0150fje. [DOI] [PubMed] [Google Scholar]
- 87.Rybin V. O., Buttrick P. M., Steinberg S. F. PKC-λ is the atypical protein kinase C isoform expressed by immature ventricle. Am. J. Physiol. 1997;272:H1636–H1642. doi: 10.1152/ajpheart.1997.272.4.H1636. [DOI] [PubMed] [Google Scholar]
- 88.Rybin V. O., Steinberg S. F. Protein kinase C isoform expression and regulation in the developing rat heart. Circ. Res. 1994;74:299–309. doi: 10.1161/01.res.74.2.299. [DOI] [PubMed] [Google Scholar]
- 89.Rybin V. O., Steinberg S. F. Do adult rat ventricular myocytes express protein kinase Cα? Am. J. Physiol. 1997;272:H2485–H2491. doi: 10.1152/ajpheart.1997.272.5.H2485. [DOI] [PubMed] [Google Scholar]
- 90.Mochly-Rosen D., Henrich C. J., Cheever L., Khaner H., Simpson P. C. A protein kinase C isozyme is translocated to cytoskeletal elements on activation. Cell Regul. 1990;1:693–706. doi: 10.1091/mbc.1.9.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Disatnik M. H., Buraggi G., Mochly-Rosen D. Localization of protein kinase C isozymes in cardiac myocytes. Exp. Cell Res. 1994;210:287–297. doi: 10.1006/excr.1994.1041. [DOI] [PubMed] [Google Scholar]
- 92.Braz J. C., Bueno O. F., De Windt L. J., Molkentin J. D. PKCα regulates the hypertrophic growth of cardiomyocytes through extracellular signal-regulated kinase1/2 (ERK1/2) J. Cell Biol. 2002;156:905–919. doi: 10.1083/jcb.200108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rybin V. O., Xu X., Steinberg S. F. Activated protein kinase C isoforms target to cardiomyocyte caveolae. Circ. Res. 1999;84:980–988. doi: 10.1161/01.res.84.9.980. [DOI] [PubMed] [Google Scholar]
- 94.Goldberg M., Zhang H. L., Steinberg S. F. Hypoxia alters the subcellular distribution of protein kinase C isoforms in neonatal rat ventricular myocytes. J. Clin. Invest. 1997;99:55–61. doi: 10.1172/JCI119133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ohmori S., Shirai Y., Sakai N., Fujii M., Konishi H., Kikkawa U., Saito N. Three distinct mechanisms for translocation and activation of the δ subspecies of protein kinase C. Mol. Cell. Biol. 1998;18:5263–5271. doi: 10.1128/mcb.18.9.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wakasaki H., Koya D., Schoen F. J., Jirousek M. R., Ways D. K., Hoit B. D., Walsh R. A., King G. L. Targeted overexpression of protein kinase CβII isoform in myocardium causes cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9320–9325. doi: 10.1073/pnas.94.17.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bowman J. C., Steinberg S. F., Jiang T., Gennan D., Fishman G. I., Buttrick P. M. Expression of protein kinase C-β in the heart causes hypertrophy in adult mice and sudden death in neonates. J. Clin. Invest. 1997;100:2189–2195. doi: 10.1172/JCI119755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takeishi Y., Ping P., Bolli R., Kirkpatrick D. L., Hoit B. D., Walsh R. A. Transgenic overexpression of constitutively active protein kinase Cε causes concentric cardiac hypertrophy. Circ. Res. 2000;86:1218–1223. doi: 10.1161/01.res.86.12.1218. [DOI] [PubMed] [Google Scholar]
- 99.Gray M. O., Zhou H. Z., Schafhalter-Zoppoth I., Zhu P., Mochly-Rosen D., Messing R. O. Preservation of base-line hemodynamic function and loss of inducible cardioprotection in adult mice lacking protein kinase C-ε. J. Biol. Chem. 2004;279:3596–3604. doi: 10.1074/jbc.M311459200. [DOI] [PubMed] [Google Scholar]
- 100.Porter M. J., Heidkamp M. C., Scully B. T., Patel N., Martin J. L., Samarel A. M. Isoenzyme-selective regulation of SERCA2 gene expression by protein kinase C in neonatal rat ventricular myocytes. Am. J. Physiol. Cell. Physiol. 2003;285:C39–C47. doi: 10.1152/ajpcell.00461.2002. [DOI] [PubMed] [Google Scholar]
- 101.Heidkamp M. C., Bayer A. L., Martin J. L., Samarel A. M. Differential activation of mitogen-activated protein kinase cascades and apoptosis by protein kinase C ε and δ in neonatal rat ventricular myocytes. Circ. Res. 2001;89:882–890. doi: 10.1161/hh2201.099434. [DOI] [PubMed] [Google Scholar]
- 102.Clerk A., Sugden P. H. Untangling the Web: Specific signaling from PKC isoforms to MAPK cascades. Circ. Res. 2002;89:847–849. [PubMed] [Google Scholar]
- 103.Chen L., Hahn H., Wu G., Chen C. H., Liron T., Schechtman D., Cavallaro G., Banci L., Guo Y., Bolli R., et al. Opposing cardioprotective actions and parallel hypertrophic effects of δ PKC and ε PKC. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11114–11119. doi: 10.1073/pnas.191369098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hahn H. S., Yussman M. G., Toyokawa T., Marreez Y., Barrett T. J., Hilty K. C., Osinska H., Robbins J., Dorn G. W. Ischemic protection and myofibrillar cardiomyopathy: dose-dependent effects of in vivo δPKC inhibition. Circ. Res. 2002;91:741–748. doi: 10.1161/01.res.0000037091.64492.69. [DOI] [PubMed] [Google Scholar]
- 105.Mochly-Rosen D., Wu G., Hahn H., Osinska H., Liron T., Lorenz J. N., Yatani A., Robbins J., Dorn G. W. Cardiotrophic effects of protein kinase Cε. Circ. Res. 2000;86:1173–1179. doi: 10.1161/01.res.86.11.1173. [DOI] [PubMed] [Google Scholar]
- 106.Inagaki K., Chen L., Ikeno F., Lee F. H., Imahashi K., Bouley D. M., Rezaee M., Yock P. G., Murphy E., Mochly-Rosen D. Inhibition of δ-protein kinase C protects against reperfusion injury of the ischemic heart in vivo. Circulation. 2003;108:2304–2307. doi: 10.1161/01.CIR.0000101682.24138.36. [DOI] [PubMed] [Google Scholar]
- 107.Shizukuda Y., Reyland M. E., Buttrick P. M. Protein kinase C-δ modulates apoptosis induced by hyperglycemia in adult ventricular myocytes. Am. J. Physiol. Heart Circ. Physiol. 2002;282:H1625–H1634. doi: 10.1152/ajpheart.00783.2001. [DOI] [PubMed] [Google Scholar]
- 108.Sabri A., Wilson B. A., Steinberg S. F. Dual actions of the Gαq agonist Pasteurella multocida toxin to promote cardiomyocyte hypertrophy and enhance apoptosis susceptibility. Circ. Res. 2002;90:850–857. doi: 10.1161/01.RES.0000016165.23795.1F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kazanietz M. G. Novel “nonkinase” phorbol ester receptors: the C1 domain connection. Mol. Pharmacol. 2002;61:759–767. doi: 10.1124/mol.61.4.759. [DOI] [PubMed] [Google Scholar]
- 110.Brodie C., Steinhart R., Kazimirsky G., Rubinfeld H., Hyman T., Ayres J. N., Hur G. M., Tothy A., Yang D., Garfield S. H., et al. PKCδ associates with and is involved in the phosphorylation of RasGRP3 in response to phorbol esters. Mol. Pharmacol. 2004;66:76–84. doi: 10.1124/mol.66.1.76. [DOI] [PubMed] [Google Scholar]
- 111.Chan S. L., Lee M. C., Tan K. O., Yang L. K., Lee A. S., Flotow H., Fu N. Y., Butler M. S., Soejarto D. D., Buss A. D., Yu V. C. Identification of chelerythrine as an inhibitor of BclXL function. J. Biol. Chem. 2003;278:20453–20456. doi: 10.1074/jbc.C300138200. [DOI] [PubMed] [Google Scholar]
- 112.Clerk A. Death by protein kinase C inhibitor. J. Mol. Cell. Cardiol. 2001;33:1773–1776. doi: 10.1006/jmcc.2001.1458. [DOI] [PubMed] [Google Scholar]
- 113.Yamamoto S., Seta K., Morisco C., Vatner S. F., Sadoshima J. Chelerythrine rapidly induces apoptosis through generation of reactive oxygen species in cardiac myocytes. J. Mol. Cell. Cardiol. 2001;33:1829–1848. doi: 10.1006/jmcc.2001.1446. [DOI] [PubMed] [Google Scholar]
- 114.Soltoff S. P. Rottlerin is a mitochondrial uncoupler that decreases cellular ATP levels and indirectly blocks PKCδ tyrosine phosphorylation. J. Biol. Chem. 2001;276:37986–37992. doi: 10.1074/jbc.M105073200. [DOI] [PubMed] [Google Scholar]
- 115.Leitges M., Elis W., Gimborn K., Huber M. Rottlerin-independent attenuation of pervanadate-induced tyrosine phosphorylation events by protein kinase C-δ in hemopoietic cells. Lab. Invest. 2001;81:1087–1095. doi: 10.1038/labinvest.3780321. [DOI] [PubMed] [Google Scholar]
- 116.Alessi D. R. The protein kinase C inhibitors Ro 318220 and GF 109203X are equally potent inhibitors of MAPKAP kinase-1β (Rsk-2) and p70 S6 kinase. FEBS Lett. 1997;402:121–123. doi: 10.1016/s0014-5793(96)01510-4. [DOI] [PubMed] [Google Scholar]
- 117.Shah K., Shokat K. M. A chemical genetic approach for the identification of direct substrates of protein kinases. Methods Mol. Biol. 2003;233:253–271. doi: 10.1385/1-59259-397-6:253. [DOI] [PubMed] [Google Scholar]
- 118.Yoshida K., Kufe D. Negative regulation of the SHPTP1 protein tyrosine phosphatase by protein kinase C-δ in response to DNA damage. Mol. Pharmacol. 2001;60:1431–1438. doi: 10.1124/mol.60.6.1431. [DOI] [PubMed] [Google Scholar]
- 119.Novotny-Diermayr V., Zhang T., Gu L., Cao X. Protein kinase C-δ associates with the interleukin-6 receptor subunit glycoprotein (gp) 130 via Stat3 and enhances Stat3-gp130 interaction. J. Biol. Chem. 2002;277:49134–49142. doi: 10.1074/jbc.M206727200. [DOI] [PubMed] [Google Scholar]
- 120.Fontayne A., Dang P. M., Gougerot-Pocidalo M. A., El Benna J. Phosphorylation of p47phox sites by PKC α, β II, δ, and ζ: effect on binding to p22phox and on NADPH oxidase activation. Biochemistry. 2002;41:7743–7750. doi: 10.1021/bi011953s. [DOI] [PubMed] [Google Scholar]
- 121.Alt A., Ohba M., Li L., Gartsbein M., Belanger A., Denning M. F., Kuroki T., Yuspa S. H., Tennenbaum T. Protein kinase Cδ-mediated phosphorylation of α6β4 is associated with reduced integrin localization to the hemidesmosome and decreased keratinocyte attachment. Cancer Res. 2001;61:4591–4598. [PubMed] [Google Scholar]