Abstract
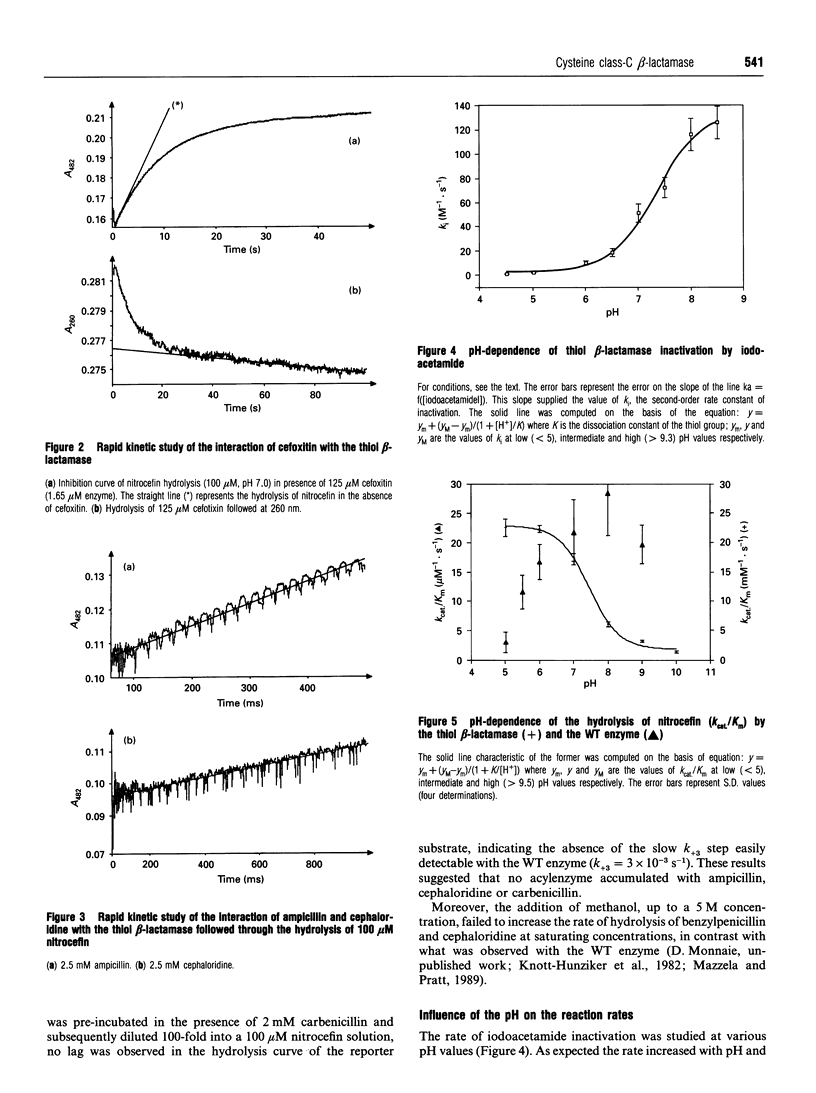
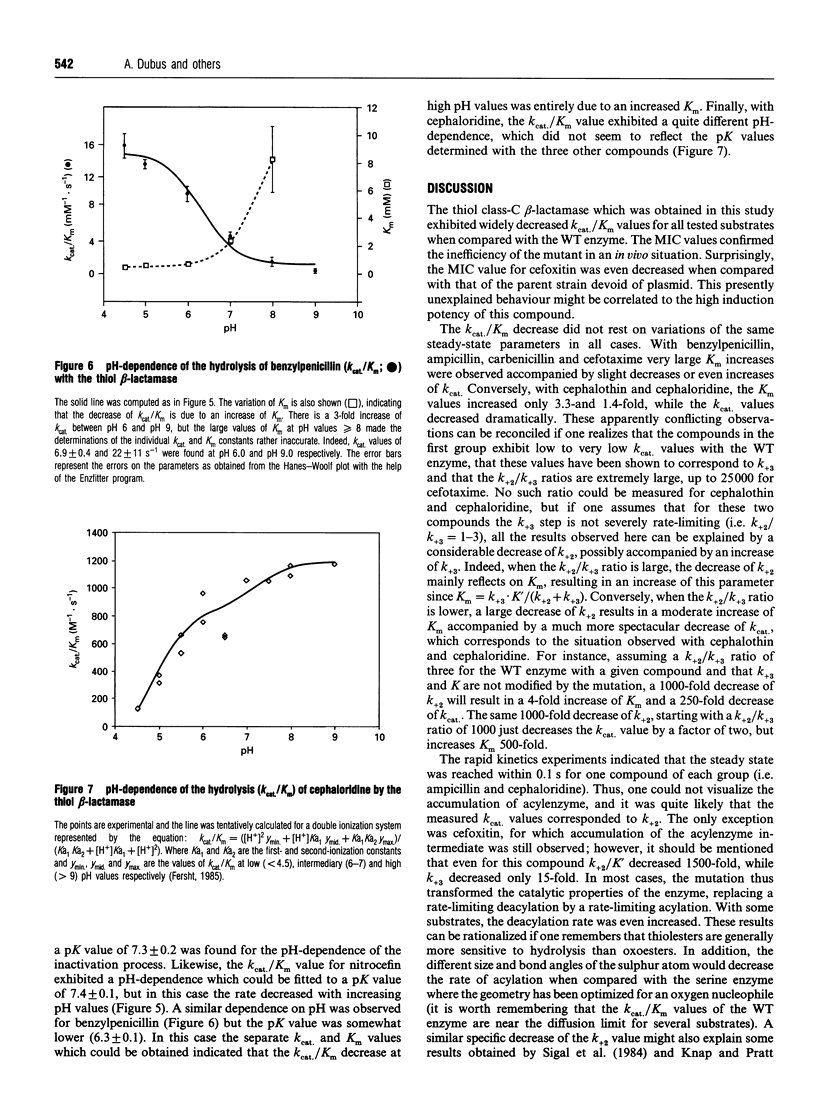
A cysteine residue has been substituted for the active-site serine of the class-C beta-lactamase produced by Enterobacter cloacae 908R by site-directed mutagenesis. The modified protein exhibited drastically reduced kcat./Km values on all tested substrates. However, this decrease was due to increased Km values with some substrates and to decreased kcat. values with others. These apparently contradictory results could be explained by a selective influence of the mutation on the first-order rate constant characteristic of the acylation step, a hypothesis which was confirmed by the absence of detectable acylenzyme accumulation with all the tested substrates, with the sole exception of cefoxitin.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi H., Ishiguro M., Imajoh S., Ohta T., Matsuzawa H. Active-site residues of the transpeptidase domain of penicillin-binding protein 2 from Escherichia coli: similarity in catalytic mechanism to class A beta-lactamases. Biochemistry. 1992 Jan 21;31(2):430–437. doi: 10.1021/bi00117a018. [DOI] [PubMed] [Google Scholar]
- Broome-Smith J. K., Hedge P. J., Spratt B. G. Production of thiol-penicillin-binding protein 3 of Escherichia coli using a two primer method of site-directed mutagenesis. EMBO J. 1985 Jan;4(1):231–235. doi: 10.1002/j.1460-2075.1985.tb02340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 1, 2a, 2b, and 2b'. Antimicrob Agents Chemother. 1989 Mar;33(3):264–270. doi: 10.1128/aac.33.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 2c, 2d, 2e, 3, and 4. Antimicrob Agents Chemother. 1989 Mar;33(3):271–276. doi: 10.1128/aac.33.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen H., Martin M. T., Waley S. G. Beta-lactamases as fully efficient enzymes. Determination of all the rate constants in the acyl-enzyme mechanism. Biochem J. 1990 Mar 15;266(3):853–861. [PMC free article] [PubMed] [Google Scholar]
- De Meester F., Joris B., Reckinger G., Bellefroid-Bourguignon C., Frère J. M., Waley S. G. Automated analysis of enzyme inactivation phenomena. Application to beta-lactamases and DD-peptidases. Biochem Pharmacol. 1987 Jul 15;36(14):2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- Fisher J., Charnas R. L., Bradley S. M., Knowles J. R. Inactivation of the RTEM beta-lactamase from Escherichia coli. Interaction of penam sulfones with enzyme. Biochemistry. 1981 May 12;20(10):2726–2731. doi: 10.1021/bi00513a004. [DOI] [PubMed] [Google Scholar]
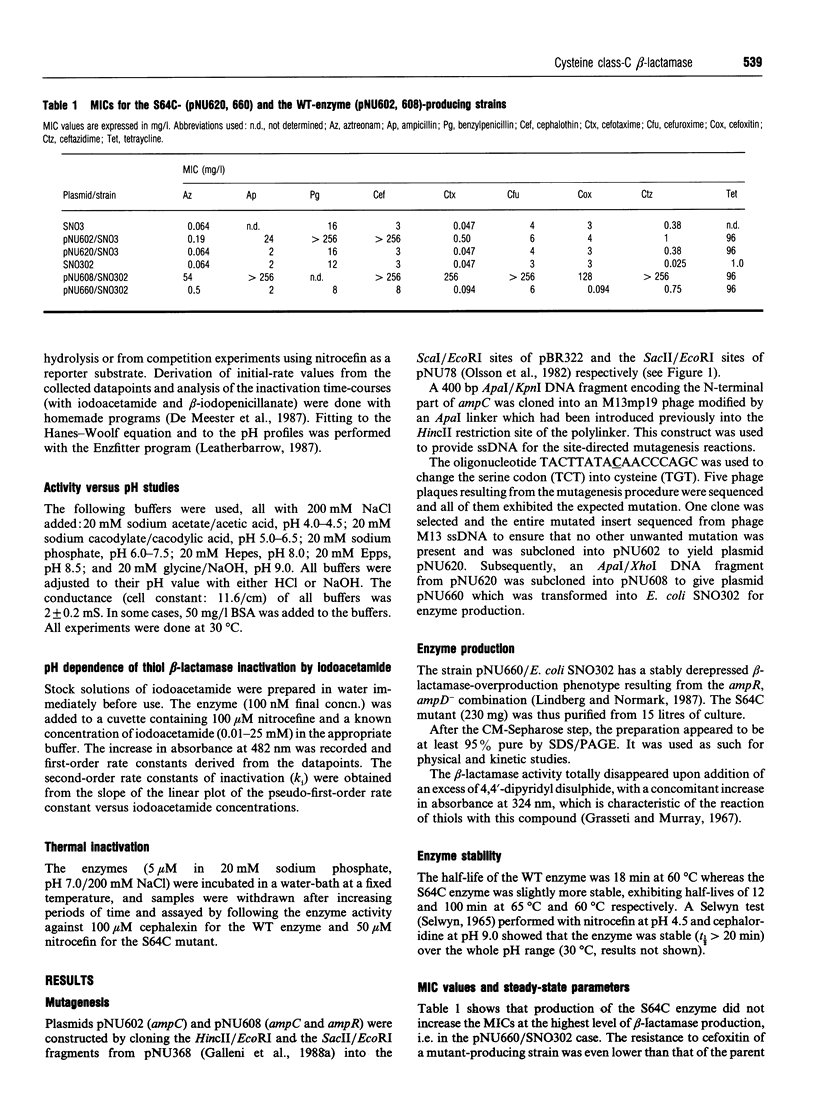
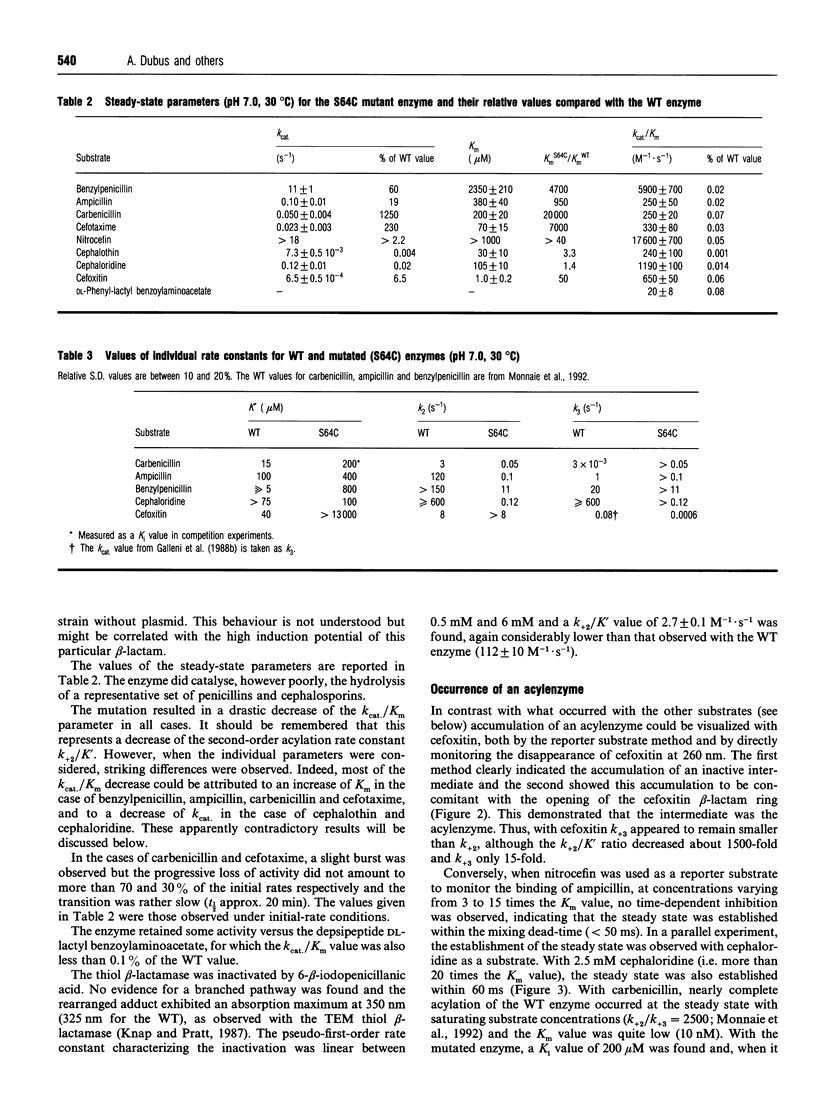
- Galleni M., Amicosante G., Frère J. M. A survey of the kinetic parameters of class C beta-lactamases. Cephalosporins and other beta-lactam compounds. Biochem J. 1988 Oct 1;255(1):123–129. doi: 10.1042/bj2550123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleni M., Frère J. M. A survey of the kinetic parameters of class C beta-lactamases. Penicillins. Biochem J. 1988 Oct 1;255(1):119–122. doi: 10.1042/bj2550119. [DOI] [PMC free article] [PubMed] [Google Scholar]
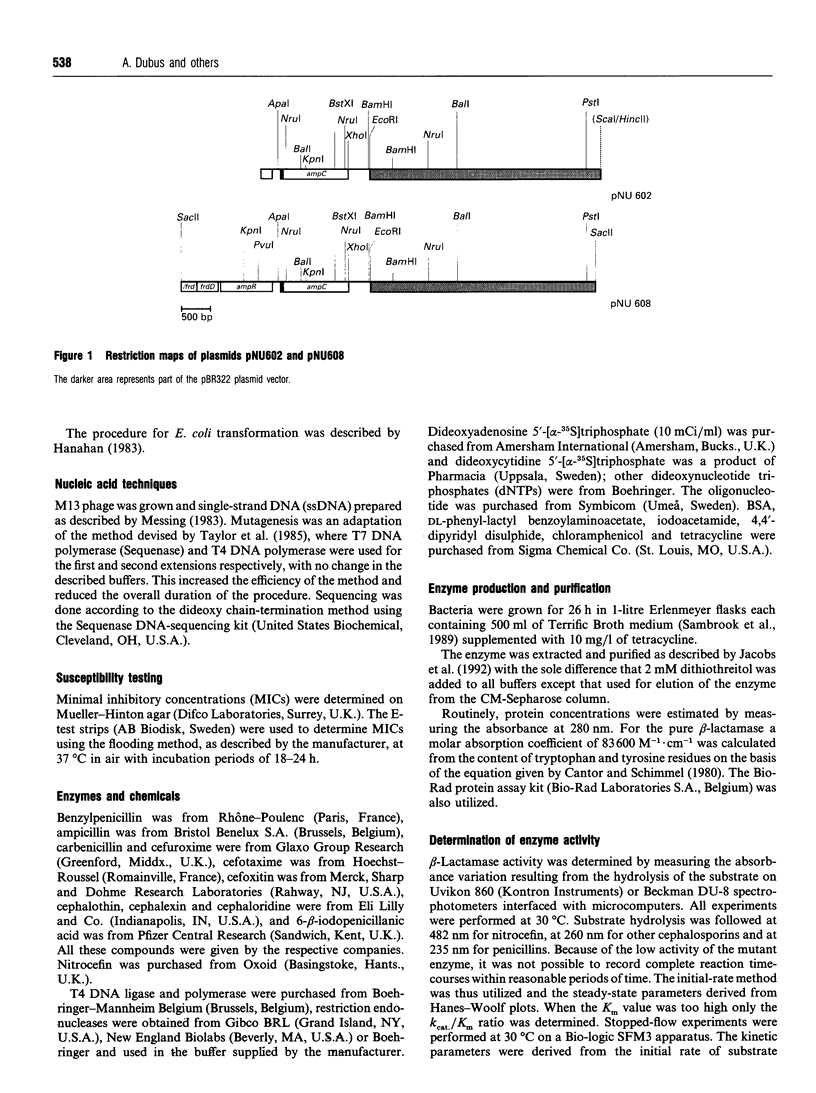
- Galleni M., Lindberg F., Normark S., Cole S., Honore N., Joris B., Frere J. M. Sequence and comparative analysis of three Enterobacter cloacae ampC beta-lactamase genes and their products. Biochem J. 1988 Mar 15;250(3):753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R. M., Christensen H., Waley S. G. Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73. Biochem J. 1990 Dec 15;272(3):613–619. doi: 10.1042/bj2720613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassetti D. R., Murray J. F., Jr Determination of sulfhydryl groups with 2,2'- or 4,4'-dithiodipyridine. Arch Biochem Biophys. 1967 Mar;119(1):41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- Hadonou A. M., Wilkin J. M., Varetto L., Joris B., Lamotte-Brasseur J., Klein D., Duez C., Ghuysen J. M., Frère J. M. Site-directed mutagenesis of the Streptomyces R61 DD-peptidase. Catalytic function of the conserved residues around the active site and a comparison with class-A and class-C beta-lactamases. Eur J Biochem. 1992 Jul 1;207(1):97–102. doi: 10.1111/j.1432-1033.1992.tb17025.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Houba-Hérin N., Hara H., Inouye M., Hirota Y. Binding of penicillin to thiol-penicillin-binding protein 3 of Escherichia coli: identification of its active site. Mol Gen Genet. 1985;201(3):499–504. doi: 10.1007/BF00331346. [DOI] [PubMed] [Google Scholar]
- Jacob F., Joris B., Frère J. M. Active-site serine mutants of the Streptomyces albus G beta-lactamase. Biochem J. 1991 Aug 1;277(Pt 3):647–652. doi: 10.1042/bj2770647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C., Dubus A., Monnaie D., Normark S., Frère J. M. Mutation of serine residue 318 in the class C beta-lactamase of Enterobacter cloacae 908R. FEMS Microbiol Lett. 1992 Apr 1;71(1):95–100. doi: 10.1016/0378-1097(92)90548-3. [DOI] [PubMed] [Google Scholar]
- Joris B., Dusart J., Frere J. M., van Beeumen J., Emanuel E. L., Petursson S., Gagnon J., Waley S. G. The active site of the P99 beta-lactamase from Enterobacter cloacae. Biochem J. 1984 Oct 1;223(1):271–274. doi: 10.1042/bj2230271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knap A. K., Pratt R. F. Chemical modification of the RTEM-1 thiol beta-lactamase by thiol-selective reagents: evidence for activation of the primary nucleophile of the beta-lactamase active site by adjacent functional groups. Proteins. 1989;6(3):316–323. doi: 10.1002/prot.340060314. [DOI] [PubMed] [Google Scholar]
- Knap A. K., Pratt R. F. Inactivation of the RTEM-1 cysteine beta-lactamase by iodoacetate. The nature of active-site functional groups and comparisons with the native enzyme. Biochem J. 1991 Jan 1;273(Pt 1):85–91. doi: 10.1042/bj2730085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knap A. K., Pratt R. F. Inactivation of the thiol RTEM-1 beta-lactamase by 6-beta-bromopenicillanic acid. Identity of the primary active-site nucleophile. Biochem J. 1987 Oct 1;247(1):29–33. doi: 10.1042/bj2470029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott-Hunziker V., Petursson S., Waley S. G., Jaurin B., Grundström T. The acyl-enzyme mechanism of beta-lactamase action. The evidence for class C Beta-lactamases. Biochem J. 1982 Nov 1;207(2):315–322. doi: 10.1042/bj2070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F., Lindquist S., Normark S. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J Bacteriol. 1987 May;169(5):1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F., Normark S. Common mechanism of ampC beta-lactamase induction in enterobacteria: regulation of the cloned Enterobacter cloacae P99 beta-lactamase gene. J Bacteriol. 1987 Feb;169(2):758–763. doi: 10.1128/jb.169.2.758-763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzella L. J., Pratt R. F. Effect of the 3'-leaving group on turnover of cephem antibiotics by a class C beta-lactamase. Biochem J. 1989 Apr 1;259(1):255–260. doi: 10.1042/bj2590255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Monnaie D., Virden R., Frère J. M. A rapid-kinetic study of the class C beta-lactamase of Enterobacter cloacae 908R. FEBS Lett. 1992 Jul 20;306(2-3):108–112. doi: 10.1016/0014-5793(92)80979-q. [DOI] [PubMed] [Google Scholar]
- Normark S., Burman L. G. Resistance of Escherichia coli to penicillins: fine-structure mapping and dominance of chromosomal beta-lactamase mutations. J Bacteriol. 1977 Oct;132(1):1–7. doi: 10.1128/jb.132.1.1-7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson O., Bergström S., Normark S. Identification of a novel ampC beta-lactamase promoter in a clinical isolate of Escherichia coli. EMBO J. 1982;1(11):1411–1416. doi: 10.1002/j.1460-2075.1982.tb01331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn M. J. A simple test for inactivation of an enzyme during assay. Biochim Biophys Acta. 1965 Jul 29;105(1):193–195. doi: 10.1016/s0926-6593(65)80190-4. [DOI] [PubMed] [Google Scholar]
- Sigal I. S., DeGrado W. F., Thomas B. J., Petteway S. R., Jr Purification and properties of thiol beta-lactamase. A mutant of pBR322 beta-lactamase in which the active site serine has been replaced with cysteine. J Biol Chem. 1984 Apr 25;259(8):5327–5332. [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]