Abstract
A portion of the genetic basis for many common autoimmune disorders has been uncovered by genome-wide association studies (GWAS), but GWAS do not reveal causal variants, effector genes, or the cell types impacted by disease-associated variation. We have generated 3D genomic datasets consisting of promoter-focused Capture-C, Hi-C, ATAC-seq, and RNA-seq and integrated these data with GWAS of 16 autoimmune traits to physically map disease-associated variants to the effector genes they likely regulate in 57 human cell types. These 3D maps of gene cis-regulatory architecture are highly powered to identify the cell types most likely impacted by disease-associated genetic variation compared to 1D genomic features, and tend to implicate different effector genes than eQTL approaches in the same cell types. Most of the variants implicated by these cis-regulatory architectures are highly trait-specific, but nearly half of the target genes connected to these variants are shared across multiple autoimmune disorders in multiple cell types, suggesting a high level of genetic diversity and complexity among autoimmune diseases that nonetheless converge at the level of target gene and cell type. Substantial effector gene sharing led to the common enrichment of similar biological networks across disease and cell types. However, trait-specific pathways representing potential areas for disease-specific intervention were identified. To test this, we pharmacologically validated squalene synthase, a cholesterol biosynthetic enzyme encoded by the FDFT1 gene implicated by our approach in MS and SLE, as a novel immunomodulatory drug target controlling inflammatory cytokine production by human T cells. These data represent a comprehensive resource for basic discovery of gene cis-regulatory mechanisms, and the analyses reported reveal mechanisms by which autoimmune-associated variants act to regulate gene expression, function, and pathology across multiple, distinct tissues and cell types.
INTRODUCTION
Autoimmune diseases are a group of complex disorders arising from dysregulated inflammatory responses that damage self-tissues and exhibit both shared and unique etiologies. Rare, naturally occurring or engineered mutations in patients or animal models have identified a handful of genes and cell types that contribute to the development and progression of disease, but in the majority of cases, common genetic variation between humans underlies autoimmune disease susceptibility. The polygenic and pleiotropic nature of autoimmunity poses serious challenges to comprehensive identification of causal variants, effector genes, effector cell types, novel therapies, and clinical management strategies. Examples of genes implicated by both common and rare genetic variation to date are CTLA4, with known roles in rheumatoid arthritis (RA), type 1 diabetes (T1D), and Hashimoto’s thyroiditis (HT)1-3, PTPN22 in systemic lupus erythematosus (SLE), T1D, RA and Grave’s disease (GD)4,5, and IL23R in ulcerative colitis (UC), Crohn’s disease (CRO), SLE, RA and psoriasis (PSO)6-8. These factors control the activation, proliferation, and function of pathogenic T helper cells and regulatory T cells, which are cell types known to mediate or control inflammation in autoimmune animal models and in humans. Identifying additional effector genes for these traits will be important for understanding disease pathology and pointing to new therapeutic approaches.
The majority of autoimmune disease-associated variants have been uncovered with GWAS. Most GWAS variants are non-coding, commonly located far from their effector genes, and their contribution to disease remains unclear. The human genome is packed and folded tightly into chromatin such that only ~1% of DNA participates in gene regulation9, and these cis-regulatory elements (cRE) can influence gene expression by ‘looping’ to physically contact distant gene promoters10. In this study, we leveraged existing autoimmune GWAS and our own matched ATAC-seq, RNA-seq, and high-resolution chromosome conformation capture data from over 50 diverse human cell types to identify putatively causal regulatory autoimmune single nucleotide polymorphisms (SNPs) in open chromatin and to comprehensively map these accessible variants to the genes they likely control. This analysis revealed shared, trait-specific, and cell type-specific gene regulatory networks likely influenced by autoimmune disease-associated genetic variation, including ‘druggable’ targets with no prior known role in immune regulation or autoimmune disease. An example we took forward to functional validation is the FDFT1 gene encoding a lipid biosynthetic enzyme, which exhibits significantly increased expression in multiple immune cell types from multiple sclerosis and lupus patients compared to healthy subjects and we show regulates inflammatory cytokine production by human T cells.
METHODS
Data and resource:
Table S1 lists all datasets, including new datasets and datasets used in this study that were previously published. The original published studies provided configurations and technical details in ATAC-seq, Hi-C, and Capture-C library generation.
Immune cell isolation:
The pDC, cDC1, and NK cells were purified from the apheresis products obtained from healthy, screened, consented human donors through University of Pennsylvania Human Immunology Core (HIC). Surface markers used for sorting were: pDC = CD3neg CD14neg CD16neg CD19neg CD20neg CD56neg HLADR+ CD11cneg, cDC2 = CD3neg CD14neg CD16neg CD19neg CD20neg CD56neg HLADR+ CD11c+ CD1c+, NK = CD45+ CD3neg CD56+. Gating strategies are included in Supplemental File 1. For T cell subsets, peripheral blood in the form of buffy coat products was obtained from anonymized healthy adults of sex and age ranges indicated in Table S2 via Canadian Blood Services (Vancouver BC, Canada) with informed consent and ethical approval from The University of British Columbia Clinical and Canadian Blood Service Research Ethics Boards (H18-02553). CD4+ T cells were obtained by incubation with CD4+ RosetteSep enrichment cocktail (Stemcell Technologies) before centrifugation over Ficoll-Paque (GE Healthcare). Ex vivo CD4+ T cell subsets were isolated to >95% purity by FACS with the following surface markers: Treg = CD25+ CD127low; Th1 = CD25neg CXCR3+ CCR6neg CCR4neg; Th2 = CD25neg CXCR3neg CCR6neg CCR4+ and Th17 = CD25neg CXCR3neg CCR6+ CCR4+. Tr1 cells (CD4+ CD45RAneg IL10+) were isolated following 16 hours anti-CD3/CD28 stimulation using IL-10 secretion capture assay as previously described11. The mAbs used are listed in Table S2. T cell subsets were expanded as previously described12. Briefly, artificial APCs expressing CD32, CD80, and CD58 were irradiated (7500 rad), loaded with anti-CD3 mAb (OKT3, 100 ng/mL), and used to stimulate a 1:1 ratio of sorted T cells in the presence of recombinant human IL-2 (100U/mL for non-Tregs and 1000U/mL for Tregs) for two weeks, with media and IL-2 replenished every 3-4 days. At day 14, after cells had returned to a resting state, aliquots of 1-2 × 105 cells were stored at −80°C as dry cell pellets (for DNA extraction) or in 100μL RNALater (Sigma-Aldrich, for RNA extraction). Then 10 × 106 cells were set aside for lysis/fixation and remaining cells were cryopreserved. Gating strategies and cell characterization data are included in Supplemental File 1.
Purification and stimulation of CD4+ T cells from human tonsil:
Fresh tonsils were obtained as discarded surgical waste from de-identified immune-competent children undergoing tonsillectomy to address airway obstruction or a history of recurrent tonsillitis. These studies were approved by The Children’s Hospital of Philadelphia Institutional Review Board as non-human subject research. The mean age of donors was 5.6 years (range 3-16 years) and 75% were male. Tonsillar mononuclear cells were isolated from tissues by mechanical disruption (tonsils were minced and pressed through a 70-micron cell screen) followed by Ficoll-Paque centrifugation. Total CD4+ T cells were purified using EasySep™ human CD4+ T cell isolation kit I (STEM cells Technologies, cat#17555) by immunomagnetic negative selection as per manufacturer’s protocol. Isolated untouched, highly purified (93-98%) human CD4 T cells were activated using anti-CD3+anti-CD28 Dynabeads (1:1, Thermofisher scientific, cat # 11161D) for 8-24 hours. IL-2, IL-17A, IL-10, IL-4, TNF, and IFNg in supernatants were measured using LEGENDplex™ human cytokine capture beads (Biolegend) on a Cytoflex flow cytometer (Beckman).
Cell fixation:
Briefly, ~107 cells were suspended in 10 mL of RPMI medium supplemented with 10% FBS. To fix the cells, 270 μL of 37% formaldehyde was added, and the suspension was incubated for 10 minutes at room temperature on a platform rocker. The fixation reaction was then quenched by adding 1.5 mL of cold 1M glycine (4°C). The fixed cells were centrifuged at 210 × g for 5 minutes at 4°C, and the supernatant was removed. The cell pellets were washed with 10 mL of cold PBS (4°C) and centrifuged again under the same conditions. After washing, the cell pellets were resuspended in 5 mL of cold lysis buffer (10 mM Tris pH 8, 10 mM NaCl, 0.2% NP-40/Igepal, supplemented with a protease inhibitor cocktail) and incubated on ice for 20 minutes. Following incubation, the cells were centrifuged at 680 × g, and the lysis buffer was removed. The cell pellets were then resuspended in 1 mL of fresh lysis buffer, transferred to 1.5 mL Eppendorf tubes, and snap frozen in ethanol/dry ice or liquid nitrogen. The frozen cell pellets were stored at −80°C for subsequent 3C library generation.
3C library generation:
For each library, 107 fixed cells were thawed at 37°C, followed by centrifugation at room temperature for 5 minutes at 1845 × g. The resulting cell pellet was resuspended in 1 mL of deionized water (dH2O) supplemented with 5 μL of a 200X protease inhibitor cocktail, incubated on ice for 10 minutes, and then centrifuged again. The cell pellet was then resuspended in dH2O to a total volume of 650 μL. A 50 μL aliquot of the cell suspension was set aside for pre-digestion quality control (QC), while the remaining sample was divided into three tubes. Both the pre-digestion control and the samples underwent a pre-digestion incubation in a Thermomixer (BenchMark) with the addition of 0.3% SDS, 1× NEB DpnII restriction buffer, and dH2O, shaking at 1,000 rpm for 1 hour at 37°C. Following this, a 1.7% solution of Triton X-100 was added to each tube, and shaking was continued for another hour. After the pre-digestion incubation, 10 μL of DpnII enzyme (NEB, 50 U/μL) was added to each sample tube (but not the control), and the incubation with shaking continued until the end of the day. An additional 10 μL of DpnII was added to each digestion reaction for an overnight digestion. The next day, a further 10 μL of DpnII was added, and the samples were incubated with shaking for another 2-3 hours. Next, 100 μL from each digestion reaction was pooled into a single 1.5 mL tube and set aside for digestion efficiency QC. The remaining samples were heat inactivated by incubating them at 65°C for 20 minutes in a MultiTherm shaking at 1,000 rpm, followed by cooling on ice for an additional 20 minutes. The digested samples were then ligated by adding 8 μL of T4 DNA ligase (HC ThermoFisher, 30 U/μL) and 1X ligase buffer, followed by overnight incubation at 16°C with shaking at 1,000 rpm in a MultiTherm. The next day, 2 μL more of T4 DNA ligase was added to each sample, and the incubation was continued for a few more hours. The ligated samples were then de-crosslinked overnight at 65°C with Proteinase K (20 mg/mL, Denville Scientific) along with pre-digestion and digestion controls. The following morning, both controls and ligated samples were incubated for 30 minutes at 37°C with RNase A (Millipore), followed by phenol/chloroform extraction and ethanol precipitation at −20°C. The 3C libraries were centrifuged at 1,000 × g for 45 minutes at 4°C to pellet the samples, while the controls were centrifuged at 1845 × g. The pellets were then resuspended in 70% ethanol and centrifuged again as described above. Finally, the pellets of the 3C libraries and controls were resuspended in 300 μL and 20 μL of dH2O, respectively, and stored at −20°C. Sample concentrations were measured using a Qubit fluorometer, and digestion and ligation efficiencies were assessed by gel electrophoresis on a 0.9% agarose gel and quantitative PCR (SYBR Green, Thermo Fisher).
Promoter Capture-C (PCC) pre-processing and interaction calling:
in brief, paired-end reads were pre-processed using HICUP pipeline13 with bowtie2 as aligner and GRCh37/hg19 for reference genome. Significant promoters interactions were called using unique read pairs from all baits promoter in the reference by CHICAGO14 pipeline. In addition to analysis of individual fragments (1frag), we also binned four fragments to improve long-distance sensitivity in interactions calling15. Interactions with CHICAGO score > 5 in either 1-fragment or 4-fragment resolution were considered significant. These interactions were output as ibed format (similar to BEDPE format) in which each line represents one physical contact between fragments. Interactions from both resolutions were merged and their genomic coordinates were lifted from GRCh37/hg19 to GRCh38/hg38.
RNA-seq library generation and sequencing:
RNA was isolated from approximately 1 million cells at each stage using Trizol Reagent (Invitrogen), followed by purification with the Direct-zol RNA Miniprep Kit (Zymo Research). To remove any contaminating genomic DNA, the RNA was treated with DNase I. The quality of the purified RNA was assessed using a Bioanalyzer 2100 with the Nano RNA Chip, and only samples with RIN>7 were selected for RNA-seq library preparation. To prepare the RNA samples for sequencing, rRNA was depleted using the QIAseq FastSelect RNA removal kit (Qiagen). The remaining RNA was then processed into RNA-seq libraries using the SMARTer Stranded Total RNA Sample Prep Kit (Takara Bio USA), following the manufacturer’s instructions. Specifically, the purified first-strand cDNA was amplified into RNA-seq libraries using SeqAmp DNA Polymerase along with Forward and Reverse PCR Primers from the Illumina Indexing Primer Set HT. The quality and quantity of the RNA-seq libraries were assessed using the Agilent 2100 Bioanalyzer system and a Qubit fluorometer (Life Technologies). Sequencing was conducted on the NovaSeq 6000 platform at the CHOP Center for Spatial and Functional Genomics.
RNA-seq preprocessing and expression profiling:
the detail configurations, steps, and technical details for each data set are provided in the original studies. In brief, read fragments from fastq files were mapped to genome assembly GRCh37/hg19 or GRCh38/hg38 using STAR16, independently for each replicate and condition. We used GENCODE annotation files for feature annotation and htseq-count for raw read count calculation at each feature. Read counts were transformed into TPM (transcript per million) and normalized internally between replicates/conditions in each individual study. For comparative measurements, we transformed all the expression values into 0-100 scale.
ATAC-seq library generation and sequencing:
A total of 50,000 to 100,000 sorted cells were centrifuged at 550 g for 5 min at 4 °C. The cell pellet was washed with cold PBS and resuspended in 50 μL cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40/IGEPAL CA-630) and immediately centrifuged at 550 g for 10 min at 4 °C. Nuclei were resuspended in the Nextera transposition reaction mix (25 μL 2× TD Buffer, 2.5 μL Nextera Tn5 transposase (Illumina Cat #FC-121-1030), and 22.5 μL nuclease free H2O) on ice, then incubated for 45 min at 37 °C. The tagmented DNA was then purified using the Qiagen MinElute kit eluted with 10.5 μL Elution Buffer (EB). Ten microliters of purified tagmented DNA was PCR amplified using Nextera primers for 12 cycles to generate each library. PCR reaction was subsequently cleaned up using 1.5× AMPureXP beads (Agencourt), and concentrations were measured by Qubit. Libraries were paired-end sequenced on the Illumina HiSeq 4000 platform (100 bp read length).
ATAC-seq preprocessing and peaks calling:
Open chromatin regions (OCRs) were called using the ENCODE ATAC-seq pipeline (https://github.com/ENCODE-DCC/atac-seq-pipeline). In brief, reads were aligned to GRCh37/hg19 assembly or GRCh38/hg38 assembly genome using bowtie2, duplicates were removed, alignments from all replicates were pooled, narrow peaks were call using MACS2. We lifted all coordinates from GRCh37/hg19 to GRCh38/hg38 to ensure consistency between datasets, using LiftOver (https://genome.sph.umich.edu/wiki/LiftOver).
ATAC-seq transcription factor footprint analysis with RGT toolbox:
bam files of mapped reads from replicates and samples were merged for each cell type. The merged bam files were then used for footprint analysis by RGT-HINT17 with parameters: --atac-seq, –paired-end, –organism=hg38. If bam files were generated on GRCh37/hg19, we performed lift-over to GRCh38/hg38 using CrossMap (v0.1.1)18 and hg19ToHg38.over.chain.gz file. We then used RGT-MOTIFANALYSIS matching to scan each footprint for possible transcription binding sites from HOCOMOCO and JASPAR databases for human only with parameter --filter "species:sapiens;database:hocomoco,jaspar_vertebrates". Parameter –rand-proportion 10 was used to generate random putative binding sites with sizes ten times larger than the input footprints. After performing motif matching, we evaluated which transcription factors were more likely to occur in those footprints than in background regions (generated by the previous command) using RGT-MOTIFANALYSIS enrichment with the same filtered databases and default parameters. Output included all the Motif-Predicted Binding Sites (MPBS) that occurred within the identified footprints in each cell type. We overlapped these sites with the loci of our obesity variants.
Hi-C library preparation:
Hi-C library preparation on FACS-sorted cells was conducted using the Arima-HiC kit (Arima Genomics Inc), following the manufacturer’s instructions. In brief, cells were first crosslinked with formaldehyde. After crosslinking, the cells were processed according to the Arima-HiC protocol, which involves digesting chromatin with multiple restriction enzymes. The sequencing libraries were then prepared by shearing the purified, proximally-ligated DNA and selecting DNA fragments ranging from 200 to 600 bp using AmpureXP beads (Beckman Coulter). These size-selected fragments were enriched using Enrichment Beads provided in the Arima-HiC kit and subsequently converted into Illumina-compatible sequencing libraries using the Swift Accel-NGS 2SPlus DNA Library Kit (Swift, 21024) and Swift 2S Indexing Kit (Swift, 26148). The resulting purified and PCR-amplified DNA was subjected to standard quality control checks, including qPCR, Bioanalyzer analysis, and KAPA Library Quantification (Roche, KK4824). Sequencing was performed with unique single indexes on the Illumina NovaSeq 6000 Sequencing System, generating 200 bp reads.
Hi-C pre-processing and interaction calling:
follows the pipeline as a recent study described 19. Paired-end reads from each replicate were pre-processed using the HICUP pipeline v0.7.413, aligned by bowtie2 with hg38 as the reference genome. The alignments files were parsed to pairtools v0.3.0 to process and pairix v0.3.7 to index and compress, then converted to Hi-C matrix binary format .cool by cooler v0.8.11 at multiple resolutions (500bp, 1, 2, 4, 10, 40, 500kbp and 1Mbp) and normalized with ICE method20. The matrices from different replicates were merged at each resolution using cooler. Mustache v1.0.121 and Fit-Hi-C2 v2.0.722 were used to call significant intra-chromosomal interaction loops from merged replicates matrices at three resolutions 1kb, 2kb, and 4kb, with significance threshold at q-value < 0.1 and FDR < 1 × 10−6, respectively. The identified interaction loops were merged between both tools at each resolution. Lastly, interaction loops from all three resolutions were merged with preference for smaller resolution if overlapped.
Definition of cis-Regulatory Elements (cREs):
We intersected ATAC-seq open chromatin regions (OCRs) of each cell type with chromatin conformation capture data determined by Hi-C/Capture-C of the same cell type, and with promoters (−1,500/+500bp of TSS) defined by GENCODE v30.
16 autoimmune traits’ GWAS summary statistics reformatted:
Table S3 lists the studies and accessions from which we drew GWAS summary statistics for each trait. LDSC (https://github.com/bulik/ldsc) recommends reformatting the summary statistics files using munge_sumstats.py. The table shows the number of total variants reported for each trait from the original studies, and the numbers after being filtered by munge_sumstat.py. This step also produced basic metadata about the summary statistics, such as mean chi2, max chi2, lambda GC, and mean value of the signed summary statistic column (beta, odd ratio - OR). Due to the inability of LDSC to produce meaningful heritability h2 for traits with low numbers of variants, we applied --merge-alleles with the list of HapMap3 variants that LDSC used to estimate LD Scores (https://data.broadinstitute.org/alkesgroup/LDSCORE/w_hm3.snplist.bz2). This standardized all the GWAS sumstats files to the 1,217,311 variants from HapMap, discarded all the unmatched, and imputed the missing variants into the sumstats with NULL beta value.
Cell type specific partitioned heritability of each autoimmune trait:
We used LDSC v.1.0.1 with --h2 flag to estimate SNP-based heritability of each trait within each cell type’s 5 defined sets of input genomic regions: (1) OCRs, (2) OCRs at gene promoters, (3) cREs, (4) cREs with an expanded window of ±500 bp and (5) OCRs that were not cREs. Each set of input regions from each cell type was used to create the annotation, which in turn was used to compute annotation-specific LD scores for each cell types regions of interest set. These annotation-specific LD scores were used with 53 categories of the full baseline model. (v2.2) as control compute partitioned heritability. To ensure comparability between traits from different studies, we used --samp-prev and --pop-prev to flag the sample and population prevalence for each trait, so that LDSC estimates SNP-heritability on the liability scale. The number of cases and controls were used to compute sample prevalence of each trait. We used the most recently reported trait prevalence for the European population for each trait, noted in Table S3.
Genetic loci included in variant-to-genes mapping:
For three autoimmune traits whose studies performed fine-mapping analysis, we used the significant variants within their provided credible sets. For the other traits, we leveraged their reported variants from associated loci which reached genome-wide significance within original studies. Proxies for each lead variant were queried using TopLD23 and LDlinkR tool24 with the GRCh38 Genome assembly, 1000 Genomes phase 3 v5 variant set, European population, and LD threshold of r2≥0.8 (results in Table S3, column “N proxies”).
Differential analysis and clustering of correlated genes:
Normalized transcripts per million (TPM) of all measured genes in 46 of 57 cell types was used to perform differential analysis using DEseq2 package25, where cell type and system (immune, metabolic, neural and other) were used as variables for the modeling contrast. Because many of the genes we gathered from the variant-to-gene mapping were lowly expressed in corresponding cell types or others, causing relatively high levels of variability, we used apeglm method for effect size (logarithmic fold change estimates) shrinkage26 to alleviate this phenomenon during the genes ranking. Weighted correlation network analysis (WGCNA) package27 was used to cluster genes from the variant-to-genes mapping process. WGCNA network construction power was chosen based on the analysis of scale-free topology for soft-thresholding. blockwiseModules with a power of 10 was used to create the correlation network and cluster genes into modules of similarly expressed genes.
Pathways enrichment analyses:
we performed two types of analyses on the set of genes from the variant-to-genes mapping process:
Gene set over-representation analysis (ORA)
was performed using clusterProfiler package28 to identify GO biology process terms (org.Hs.eg.db databese) enriched within our genes set in each cell type. A relaxed P-value cutoff was set at 0.1, and the minimum including genes was set at 2 to ensure the capture of all possible enriched terms. An adjusted P-value of 0.05 was later used to filter the significant terms.
Active-subnetwork-oriented gene set enrichment analysis:
we used the pathfindR package29 to identify active subnetworks in protein-protein interaction networks from Biogrid, KEGG, STRING, GeneMania, and IntAct databases, using the list of genes from the variant-to-genes mapping process. Then we provided the statistics from the differential expression analysis for pathfinder, fold-changes were computed for cell type versus mean of other cell types, with cell system as a variable. We performed enrichment analyses on the identified subnetworks, discovering enriched KEGG pathways and GO biology process terms.
Prediction of variant’s effect on transcription factor binding:
genomic positions (0-based coordinates) and allele alternatives of each proxy (from SNPlocs.Hsapiens.dbSNP155.GRCh38 package with matching reference sequence from BSgenome package) were used to scan all position frequency matrix databases (from MotifDb package) for potential transcription factor binding disruptive effects. The motifbreakR function from motifBreakR package was used, with filterp=TRUE and setting a p-value threshold=0.0005, information content methods method='ic' with even background probabilities of the four nucleotides bkg = c(A=0.25, C=0.25, G=0.25, T=0.25) and BPPARAM = BiocParallel::SerialParam() to allow serial evaluation.
GWAS-eQTL colocalization:
We used the same summary statistics for autoimmune disorders as in the LDSC analysis. Variants from the 1000 Genomes Project v3 samples were used as a reference panel. For all the lead SNPs that had proxies identified in variant-to-gene mapping analysis, we clumped and selected their median genomic positions for each lead. We extended genomic windows of ±250,000 bases in both directions around these median positions, or to the most distant proxies of each clump, to create 325 non-overlapped loci. We used eQTLs from 3 databases: eQTL Catalogue30 (available at https://github.com/eQTL-Catalogue), the Database of Immune Cell EQTLs (DICE eQTL)31 available at https://dice-database.org in GRCh37 reference genome, lifted to GRCh38 using UCSC LiftOver tool and LiftRsNumber.py), and the OneK1K database32 (available at https://onek1k.org). We overlapped 325 immune GWAS loci with the significant gene expression eQTLs associations (FDR > 0.05) for all 108 tissues of eQTL Catalogue, the 15 eGenes sets of DICE and 14 eSNP sets of OneK1K to narrow the list of genes that are regulated in response to immune diseases associated variants. For each gene in this list, we extract available eQTL summary statistics within the overlapping locus: association P values, effect size estimate (β), and standard error of β; minor allele frequencies were included for OneK1K, and inferred from dnSNP155 for DICE. We used coloc.abf function from R package coloc33 to test colocalization for each gene between eQTLs of each cell types and all GWAS of each disease included in each overlapping locus.
Single-cell RNA-seq analysis:
We collected immune single-cell RNA-seq for the autoimmune diseases, focused on the data with healthy control and no comorbidity and prioritized processed data from peripheral blood samples and with cell identity labels. The collected datasets are in Table S3. Origin tissues from which the immune cell types differentiated from were indicated in the table. We applied differential gene expression analysis comparing disease versus control on each separate cell type population of each data set, using the default Wilcoxon Rank Sum test of the Seurat package34; p-values were FDR adjusted.
RESULTS
Large-scale survey of 3D cis-regulatory architectures identifies target tissues enriched for autoimmune disease heritability
Genes are frequently controlled by regulatory elements that are far away in the linear sequence of the genome, but due to the highly folded structure of chromatin in the nucleus, are in close proximity in three-dimensional space. The importance of this long-range mode of gene regulation was first appreciated at the Hbb and IFNg loci35-37, and has been confirmed at a whole genome scale by studies showing that the set of genes contacted by distant regulatory regions exhibit 10- to 20-fold higher levels of expression than genes not in contact with distant cRE38. Therefore, a spatial map of genome structure in a given cell type represents a crucial tool for understanding any gene’s full cis-regulatory architecture and how genetic variants might contribute to its expression. We used ATAC-seq and chromosome conformation capture – promoter Capture-C (PCC) and Hi-C – to map the promoter-connected open chromatin (putative cRE) landscapes of 57 human cell types representing immune, metabolic, neuronal, and pluripotent stem cell lineages (Table S1). All datasets were generated by our laboratory using our standardized wet- and dry-bench pipeline and include 9 immune cell types not previously published (EBV-B, T helper 1, T helper 2, T helper 17, T regulatory 1, T regulatory, plasmacytoid dendritic, CD1c+ dendritic, and natural killer cells).
To determine which of these tissues are most impacted by autoimmune disease-associated genetic variation, we measured the enrichment of heritability across the 3D cRE landscapes of each cell type by stratified linkage disequilibrium score regression (S-LDSR)39 (Fig. 1A) using recent European-population GWAS summary statistics of sixteen autoimmune diseases: allergy (ALG), eczema (ECZ), ankylosing spondylitis (AS), psoriasis (PSO), Crohn's disease (CRO), ulcerative colitis (UC), celiac disease (CEL), inflammatory bowel disease (IBD), rheumatoid arthritis (RA), juvenile idiopathic arthritis (JIA), multiple sclerosis (MS), systemic lupus erythematosus (SLE), type I diabetes mellitus (T1D), Hashimoto's thyroiditis (HT), Graves' disease (GD) and vitiligo (VIT) (Table S3). Genetic heritability for the risk of the majority of autoimmune diseases showed statistically significant enrichment in the cRE landscapes of most or all of the 22 analyzed immune cell types and states, and was largely not enriched in metabolic, neuronal, and pluripotent cell types (Fig. 1A, detail in Table S4). Heritability for JIA, AS and PSO was significantly enriched in the least number of immune cell types (one, two and three, respectively). Grave’s disease displayed very strong enrichment for the most cell types, 34 in total, including immune, metabolic, neural and tumor cell types (Fig. S1A).
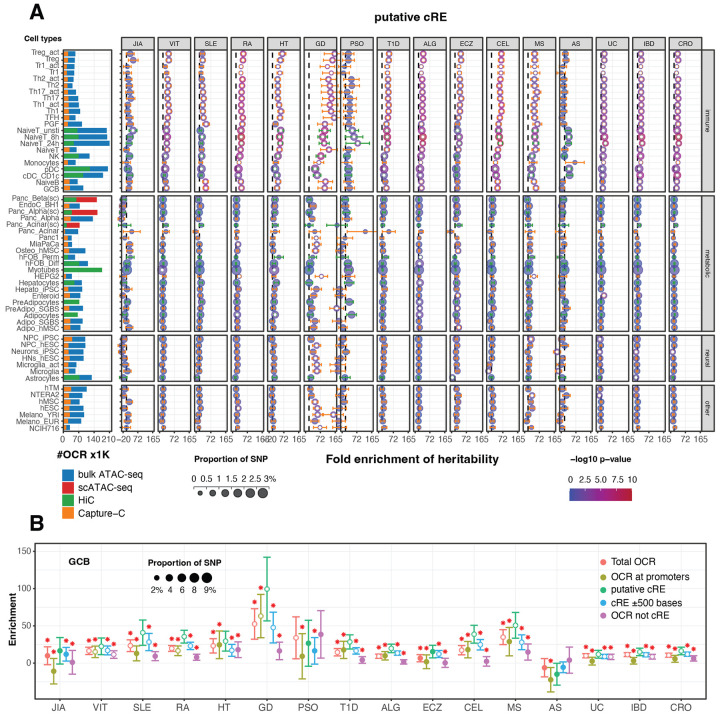
Figure 1 – Autoimmune disease heritability enrichment across diverse cell types.
A. Bar plots (left) depict total number of open chromatin regions (OCR x1000) identified by bulk (blue) or single-cell (red) ATAC-seq and the proportion of those OCR contacting a gene promoter (putative cRE) as measured by HiC (green) or promoter capture-C (orange). The dot plots depict heritability enrichment for each cell type across 16 autoimmune traits as determined by LDSC analysis. Whiskers represent enrichment standard errors, with colors matched for HiC vs. capture-C. The colors of the dots correspond to p-values in −log10, with dots featuring a white asterisk indicating a significant p-value ≤ 0.05. The size of the dots corresponds to the proportion of SNP contribution to heritability. A dashed line at 1 indicates no enrichment. B. Dot plot depicting heritability enrichment for germinal center B cells (GCB) for each immune trait, grouped by all OCR (red), promoter OCR (light green), promoter-contacting OCR (cRE, green), cRE expanded to 500 bp flanking regions (blue), and OCR not contacting a promoter (violet). Red asterisks indicate a significant p-value<0.05 in one-side (lesser) T-test comparing other categories with the green putative cRE.
Statistically significant enrichment (P<0.05) was observed across all 22 immune cell types for at least one immune trait (Fig. S1B). The cRE landscapes of naïve and activated CD4+ helper T cells exhibited enrichment for 15 out of 16 immune traits, whereas monocytes showed enrichment for only 7 traits. The regulatory landscapes of naïve and GCB cells were most highly enriched for SLE heritability and demonstrated strong enrichment for RA and CEL, consistent with the clear role of autoantibodies in driving these conditions44. Conversely, ECZ, AS, JIA, and PSO variants were not enriched in B cells. Differentiated effector T cell (Th1, Th2, Th17) landscapes were highly enriched in most autoimmune traits, but not enriched for AS, JIA, PSO. Th17 regulatory landscapes were particularly enriched in ECZ and MS, diseases known to be driven by Th17-derived cytokines40-42. TR1 and TFH cells exhibited moderate heritability enrichment across many diseases, with TFH landscapes exhibiting particularly strong enrichment in RA heritability. NK regulatory architectures were most significantly enriched for UC, CRO, and IBD heritability. The cRE landscapes of monocytes, CD1c+ DC (cDC2), and interferon-producing pDC cells were enriched for heritability for multiple diseases, but were not enriched for ECZ- or PSO-associated variants. Treg cells stood out as the most highly enriched cell type for T1D GWAS loci, and featured prominently in VIT, RA, HT, ECZ, UC, CRO, and IBD heritability. This suggests widespread involvement of autoimmune susceptibility variants in the regulation of gene expression in Treg, and aligns with their established role in self-tolerance and regulation of inflammation43,44. The EBV-transformed PGF B cell line was the only immune cell type significantly enriched for JIA heritability, was strongly enriched for MS heritability, and was one of just three cell types enriched for PSO variants. Interestingly, the incidence and pathobiology of these diseases have been linked to EBV infection45-48, suggesting possible genetic interactions between EBV and autoimmune variants in infected B cells, however, heritability for another EBV-linked disease, SLE, was not enriched in PGF cells49,50. These findings underscore the diverse and complex interplay of immune cell types in autoimmune disease susceptibility.
Interestingly, the 3D regulatory architectures of some non-immune cell types also exhibited enrichment for several immune traits (Fig.1A, Table S4). For instance, adipocyte lineage cells showed significant enrichment for ECZ, ALG, HT and RA heritability, and enrichment for ALG and ECZ heritability was found on the adipocyte/osteoblast axis, including hFOB (fetal osteoblasts), hMSC-derived osteoblasts, pre-adipocytes, adipocytes, hMSC-derived adipocytes, hepatocytes and colon-derived enteroids. Significantly, neural progenitor cell cRE landscapes are enriched in heritability for MS, an autoimmune disease targeting nervous tissue in the CNS, and the cRE landscape of intestinal organoids (enteroids) was very strongly enriched for UC heritability, suggesting that UC variants also operate in non-immune intestinal cells to control UC risk51. GD heritability was enriched in the greatest number of non-immune cell types, including Panc1, MiaPaCa (pancreatic ductal cancer cell lines), osteoblast (hMSC-derived), HEPG2, NPC-hESC, hypothalamic neurons (hES-derived), microglia, activated microglia, hMSC, hESC, melanocyte-YRI, NCIH716 (colorectal adenocarcinoma).
3D cRE architectures are enriched for autoimmune disease heritability compared to 1D epigenomic feature maps
Importantly, these LDSC analyses show that 3D promoter-contacted open chromatin (putative cRE) landscapes specifically and consistently exhibit the strongest heritability enrichment across all cell types compared to open chromatin landscapes not informed by Hi-C/PCC or compared to open chromatin proximal to gene promoters (Fig. 1B). The magnitude of heritability enrichment for each immune trait using cell type cRE landscapes was significantly greater in 72% (2616 of 3648) of pairwise comparisons (red stars in Fig. 1B). Moreover, the subset of open chromatin regions not detected as interacting with a gene promoter by Hi-C/PCC consistently exhibited the lowest level of heritability enrichment and generally failed to reach statistical significance compared to other biological constraints tested. The originally reported LDSR method extended the analysis to the ±500 bp regions flanking their regulatory element categories39. Expanding our analysis similarly to include ±500 bp flanking our defined cRE resulted in the inclusion of more weighted variants (represented by larger blue dots in all panels in Fig. 1B), more enriched cell types (Fig. S1), decreased enrichment range across cell types, decreased 95% CI, and decreased level of significance. These comparative metrics underscore the enhanced predictive value of 3D chromatin-based maps of putative gene regulatory architecture for assessing the heritability of autoimmune traits across tissue types.
Chromatin-based V2G maps reveal trait-specific genetic influence on disease risk via genes shared across traits and cell types
Having shown that GWAS-informed open chromatin-promoter interactomes are highly enriched for autoimmune heritability in multiple immune cell types, we next determined which genes are in contact with, and therefore likely regulated by, accessible autoimmune variants by systematically mapping the genomic positions of linkage disequilibrium (LD) proxies within the cRE landscape of each cell type. This approach cannot confidently identify co-inherited proxies in the highly polymorphic and extended LD landscape of the MHC region on chromosome 6, so in line with other variant-to-gene efforts we excluded this chromosomal region from our analysis. These 3D autoimmune variant-gene mappings revealed 5,488 genes targeted by chromatin contacts with 3,642 proxies linked to all immune traits in all cell types (full result in Table S5), leading to combinatorial complexity of over 25,000 variant-gene pairs (Fig. 2A). The total number of variants vs. target genes implicated per trait (Fig. 2B) or per cell type (Fig. 2C) were highly concordant, however, individual variants and genes generally showed cell type- and trait-specific distributions. Most of the implicated variants were trait-specific, with only 18% (679 proxies) shared between traits (Fig. 2D, details in Fig. S2A). Outliers were variants rs34536443, rs35350651 and rs653178 on chromosomes 12 and 19 that were associated with seven autoimmune traits. On average, a single variant contacted 4 gene promoters in a specific cell type through chromatin contact loops. Immune cells exhibited a statistically significantly higher number of autoimmune variant-gene contacts, and thus higher total number of implicated genes compared to non-immune cell types (Fig. S2B), with pDC, cDC2, and NK cells exhibiting significantly more variant-gene contacts than other immune cell types.
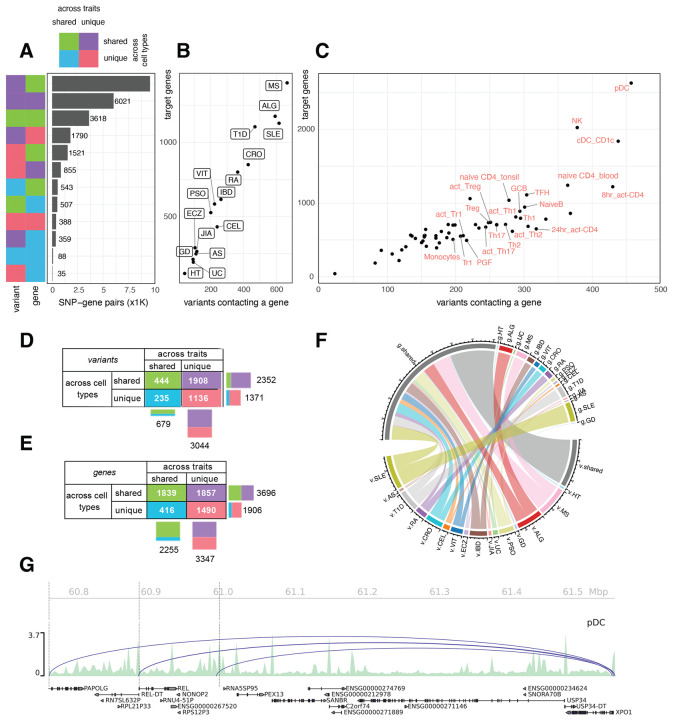
Figure 2 – 3D chromatin-based variant-to-gene mapping across autoimmune diseases and cell types.
A. Bar plot depicting the number of variant-gene pairs in which the variant (left column) or the gene (right column) is unique vs. shared across traits and cell types (green - shared across traits and cell types, purple - unique to trait but shared across cell types, cyan - unique to cell type but shared across traits, red – unique to trait and cell type). B. Scatter-plot depicting target gene number versus variant number for each trait. C. Scatter-plot depicting target gene number versus variant number for each cell type with immune cell types indicated. D. Detail of variants unique vs. shared across traits and cell types. E. Detail of genes unique vs. shared across traits and cell types. F. Chord diagram shows proportions of genes (g.) and variant (v.) of each trait are trait-specific (color-coded) or shared (grey). G. Genome-view of locus containing XPO1 gene, contacted by 3 variants from 5 different traits, uniquely detected in pDC.
While implicated variants were largely trait-specific, 40% of target genes (2,255 of 5,602) were commonly implicated across autoimmune disorders (Fig. 2E, detail in Fig. S3A), with 11 genes implicated across 9 traits (e.g., TYK2, ICAM1, ICAM3, TNFIP3, CLEC16A, BACH2). Genes implicated in B cells tended to be widely shared among other immune cell types (Fig. S3B), while T cell subsets and dendritic cell types showed the most exclusive pattern of V2G-implicated genes. Among the traits, SLE had the highest number of exclusively implicated genes (569), followed by ALG (456) and MS (465) (Fig. 2F). The lower degree of exclusivity for target genes compared to that of variants across traits is due to the fact that nearly half of the trait-specific variants end up targeting the same genes (Fig. 2G). An example is the XPO1 gene, whose promoter was contacted by multiple variants from various immune diseases, but only in pDC (Fig. 2G). The degree of overlap of implicated variants and target genes across cell types was much higher than across traits, with 63% of variants (2,305 out of 3,642 Fig. 2D and Fig. S4A) and 66% of genes (3,612 out of 5,488, Fig. 2E and Fig. S4B) implicated across two or more cell types. Only 388 variant-gene pairs were unique to both trait and cell type. Overall, these analyses show that the majority of variant-gene pairs are composed of target genes shared across traits and cell types and implicated variants that are largely trait-unique but shared across multiple cell types. These results suggest that functional variants that are uniquely associated with single autoimmune traits often converge on the same genes across multiple cell types and therefore may ultimately share common mechanisms of influence on disease risk.
Chromatin-based V2G maps implicate both common and unique biological pathways targeted by autoimmune-associated genetic variation
To explore how complex patterns of 3D autoimmune variant-gene contacts across traits and cell types may drive defined biological pathways to influence disease risk, we conducted pathway analysis for genes implicated in each cell type for each trait. This analysis focused on pathways supported by a minimum of five implicated genes and enrichment was weighted by protein-to-protein interaction subnetworks and by the degree to which pathway genes show higher or lower expression in a given cell type compared to their average expression across all cell types. This analysis yielded 2240 significantly enriched GO terms and 310 KEGG pathways (Table S6 and Table S7). KEGG pathways enriched across all or a majority of autoimmune traits were cytokine-cytokine receptor interactions, bacterial infection (Salmonella, Yersinia), viral infection (HTLV-1, Herpes, COVID-19 disease), NFkB and MAPK signaling, C-type lectin and Toll-like receptor signaling; and top shared GO terms were transcription factor binding and ubiquitin protein ligase binding. These processes have experimentally and clinically established roles in innate and adaptive immunity, immune tolerance, and autoimmunity, further validating the biological relevance of the 3D cRE architectures reported here.
The 66 genes within the cytokine-cytokine receptor interaction pathway were enriched in 21 immune, 14 metabolic, and 3 neural cell types (Fig. S5), 36 genes involved in the Salmonella infection pathway were enriched in 21 immune, 11 metabolic, and 6 neural cell types (Fig. S6). Genes involved in DNA transcription factor activity were enriched in 18 immune, 8 metabolic, and 5 neural cell types, and the ubiquitin ligase pathway was enriched across 21 immune, 10 metabolic, and 5 neural cell types. Colon-derived enteroids from IBD patients52 offer an interesting example: the 3D regulatory architectures of these intestinal organoids showed enrichment for UC heritability at genome-scale (Fig. 1A), and a large number of genes (>700) in these cell types were connected to UC, IBD and CRO variants. However, the implicated genes are not all involved in the same biological processes, as pathway analysis shows that only one-third of the pathways enriched for all three traits in these organoids are shared between CRO and UC, while another third are only enriched in IBD, and the final third are specific to UC (Fig. S7).
While most of the enriched pathways are shared across traits, 31 KEGG pathways show trait-specific enrichment (Fig. S8), and many of these point to processes known to influence disease pathology. For example, the ‘maturity onset of diabetes in the young’ pathway is enriched in our T1D gene set, but as might be expected, not in any other gene sets implicated by other autoimmune traits. Nicotinamide and selenium levels are associated with SLE pathology and as supplements have been shown to ameliorate symptoms in patients53-57, and both ‘nicotinamide metabolism’ and ‘selenocompound metabolism’ were uniquely enriched in the SLE-implicated gene set. One pathway uniquely enriched in the MS-implicated gene set is ‘pyrimidine metabolism’, and levels of pyrimidine metabolites in cerebral-spinal fluid is associated with symptom severity in MS patients, and therapeutic inhibition of pyrimidine biosynthesis ameliorates disease58,59. The only pathway specifically enriched in JIA is ‘nicotine addiction’, and interestingly, maternal smoking and second-hand smoke exposure is a known significant risk factor in JIA60-62.
Comparison of 3D chromatin-based vs. eQTL-based effector gene nomination approaches
To determine whether our GWAS variant-gene contacts correlate with variant-gene quantitative trait effects, we used the COLOC colocalization framework33 to compare the set of genes implicated for each trait in each cell type reported in this study with the set of eGenes identified in eQTL studies in the same or similar cell types from GTEX whole blood, DICE, OneK1K and eQTL_Catalogue datasets. The genetic locations of all variants and target gene promoters from our chromatin-based V2G analysis were clustered to construct 325 loci (see Methods), and colocalization analysis was performed between each immune trait with each cell type eQTL from each resource. GTEX implicated 2190 eGenes from whole blood, the DICE, OneK1K, and eQTL_Catalogue scRNA-seq datasets together implicated 1330 unique eGenes across various immune cell types, compared to 5556 genes implicated by our 3D chromatin-based approach.
GTEX and 3D chromatin V2G implicated 757 eGenes in common across all 15 cell types and 16 disease traits (Table S8), meaning that our 3D chromatin V2G implicated 35% of the GTEX whole blood eQTL, while the GTEX whole blood eQTL dataset detected 14% of the genes implicated by our 3D chromatin V2G approach. The single-cell eQTL datasets and our 3D chromatin V2G implicated 309 eGenes in common, such that 3D chromatin V2G implicated 23% of the scRNA-seq eQTL datasets, while the the scRNA-seq eQTL datasets detected 6% of the genes implicated by our 3D chromatin V2G approach (Table S9, Fig. 3). SLE yielded the highest number of commonly implicated eGenes across all cell types (51), followed by RA, PSO and ECZ, and naive CD4+ T cells yielded the highest number of commonly implicated eGenes across all autoimmune traits (149), followed by NK cells and monocytes. These data indicate that 3D chromatin V2G is better powered to implicate eQTL-based eGenes than eQTL are at implicating 3D chromatin-based target genes, and in general the overlap between these two effector gene nomination approaches is relatively low.
Figure 3 – Comparison of eGenes implicated by 3D chromatin-based V2G vs. eQTL.
Center panel. Collage of pie plots each depicting the proportion of eGenes identified by eQTL that match (purple) or do not match (light orange) the gene identified by 3D chromatin V2G for each cell type-trait pairing after co-localization. ND=too few variants/cells. Left panels: bar plots depicting the total number of eQTL eGenes for each immune cell type that match or differ across all autoimmune traits. Bottom panels: bar plots depicting the total number of eQTL eGenes for each trait that match or differ across all immune cell types.
Functional validation of a novel immunomodulatory drug target implicated by 3D chromatin-based V2G
Autoimmune GWAS represents the genetics of immune tolerance and immunoregulation; therefore, understanding the mechanisms by which autoimmune variants-target genes and biological pathways provide important disease insights. These could then be translated into novel immunomodulatory therapies tailored to specific autoimmune diseases or targeted at inflammation across multiple disorders including cancer and organ transplant rejection. The V2G maps reported here implicate a large array of genes and pathways in and across various immune cell types and autoimmune diseases. Many of these genes/pathways represent well-known biology, and some are currently the targets of FDA-approved therapies for immune-mediated disease. However, a large number of implicated genes, many of which are readily ‘druggable’, are novel in that they have not been studied in the context of immunology or assessed as an immunomodulatory target in an inflammatory context. Indeed, we have used this V2G approach in previous studies to nominate and functionally validate 12 novel immune targets with previously unappreciated roles in human T cell activation63, follicular helper T cell function64 and T cell help for B cell differentiation38. We have also validated 3 novel non-immune targets that regulate bone mineralization65,66, sleep behavior67, and type 2 diabetes68.
An example of a novel autoimmune drug target implicated in this study is FDFT1, a farnesyl-diphosphate farnesyltransferase (aka, squalene synthase) that catalyzes the first committed step of isoprenoid synthesis for cholesterol production. The FDFT1 gene interacts with 7 MS-associated variants (Fig. 4A, top panel), 7 SLE-associated variants (Fig. 4A, bottom panel), and one variant associated with both RA and SLE (Fig. 4A, middle panel). These variants are part of a diverse regulatory landscape at this locus, with SLE variants contacting FDFT1 in naïve B cells, germinal center B cells, and microglia, and MS variants contacting FDFT1 in pancreatic cells, iPSC-derived neural progenitor cells, TFH cells and Th17 cells (Fig. 4A). Prior studies showed that FDFT1 is downregulated in astrocytes in both mice and humans with multiple sclerosis69,70, and neuronal induction of FDFT1 and squalene synthase activity contributes to myelination in mouse models of chronic demyelinating disease69, however, a role for FDFT1 in the function of any of the immune cell types studied here has not been established.
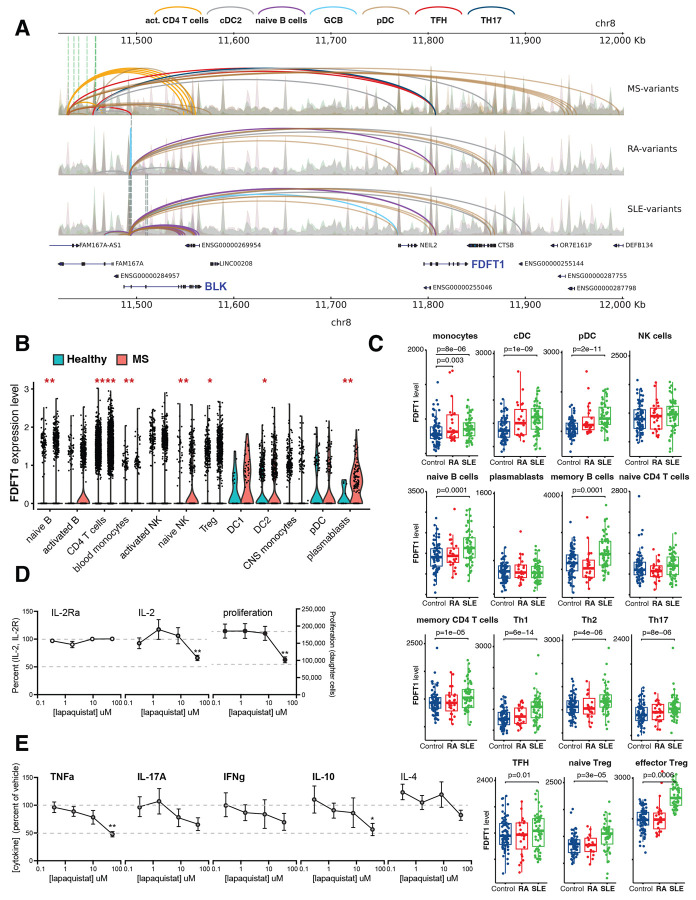
Figure 4 -. Functional validation of the 3D chromatin-based V2G target FDFT1.
A. Diagram depicts loop contacts (arcs) between accessible proxies associated with MS (7, green vertical lines), SLE (7, grey vertical lines) and SLE+RA (1, blue vertical line) and gene promoters at the FAM167A-BLK locus in activated CD4+ T cells (orange), cDC2 (grey), naïve B cells (purple), germinal center B cells (light blue), pDC (brown), TFH cells (red), and Th17 cells (dark blue). MS variants contact FDFT1 in TFH and Th17 cells (top panels) and SLE/RA variants contact FDFT1 in naïve B cells, germinal center B cells, and pDC (bottom panels). B. Differential scRNA-seq65 analysis of FDFT1 expression in immune cell clusters from healthy donors (blue) or MS patients (red). *P<0.01, **P<0.001, ***P<0.0001, ****P<0.00001. C. Differential bulk RNA-seq66 analysis of FDFT1 expression in sorted immune cells from healthy donors (blue), RA patients (red), or SLE patients (green). P values are shown. Effect of the FDFT1 inhibitor lapaquistat on (D) CD25/IL-2R expression, IL-2 secretion, proliferation, and (E) secretion of TNF, IL-17A, IFNg, IL-10, and IL-4 by human tonsillar CD4+ T cells (N=4 donors). P values in D and E are: **, <0.01 and * <0.05.
To determine whether autoimmune disease is associated with altered FDFT1 gene expression in immune cells, we performed a differential analysis of FDFT1 expression in immune cells from SLE patients, MS patients, RA patients, and healthy subjects using single-cell or bulk RNA-seq data (GSE13826671, E-GEAD-39772). Consistent with our chromatin-based MS V2G results, FDTF1 is significantly upregulated in CD4+ T cells from MS patients, as well as in naïve B cells, circulating GCB cells (plasmablasts), NK cells, monocytes, and Treg (Fig. 4B). Similarly, FDFT1 is significantly upregulated in naïve and memory B cells from SLE patients, as well as in innate immune cell types, memory CD4+ T cells, Th1, Th2, Th17, TFH and Treg (Fig. 4C). The RA and SLE variants are located in the first intron of the BLK gene encoding the B cell Src family tyrosine kinase, therefore we also performed a differential analysis of BLK expression. BLK was expressed highly in B cell subsets and exhibited differential expression in SLE patients, but unlike FDFT1, BLK was expressed a low levels in T cells and did not show disease-associated expression in memory or differentiated CD4+ T cells (Fig. S9).
To test whether FDFT1 regulates inflammatory T cell function, we used the squalene synthase inhibitor lapaquistat, a drug historically used to treat hypercholesterolemia, to target FDFT1 activity in CD4+ T cells purified from human tonsil, a rich source of differentiated helper T cell types including TFH and Th17 cells. Lapaquistat had no negative impact on CD4+ T cell activation (as measured by the expression of IL-2 and it’s receptor) or proliferation except at the highest dose (Fig. 4D). However, inhibition of squalene synthase activity resulted in a dose-dependent decrease in activation-induced production of TNF and IL-17A, cytokines produced by human TFH and Th17 cells, as well as decreased secretion of IFNg and IL-10 (Fig. 4E). The reduction in TNF and IL-10 levels was statistically significant. The Th2 cytokine IL-4 was not inhibited by lapaquistat. TNF, IL-17A, IFNg, and IL-10 produced by CD4+ T cell subsets have important roles in the immunopathology characteristic of MS and SLE. These data validate our 3D chromatin-based autoimmune variant-to-gene mapping approach and establish a previously unappreciated role for FDFT1 in the inflammatory function of CD4+ T cells.
DISCUSSION
Deciphering the complex interplay between genetic risk factors, cell types, and immune dysregulation is crucial for understanding the pathogenesis of autoimmune disease, and such insights have the potential to revolutionize diagnostics and treatments tailored for improved patient outcomes. Our multi-omic approach provided 3D maps of gene cis-regulatory architecture that powered LDSC analyses to identify the cell types most likely impacted by autoimmune disease-associated genetic variation, followed by identification of those active variants in specific cell types and mapping them to the genes they likely regulate. These results implicated different immune cell types in distinct autoimmune diseases, as well as some non-immune cell types like enteroids, neuronal cells, and pancreatic cells in diseases such as IBD and MS.
Several cell types exhibited negative heritability enrichment for several autoimmune diseases. However, these events were not persistent across those cell types or traits. Thus, it is unlikely the negative enrichments have resulted from misclassification of the alleles in the traits summary statistics, or from particularly biased genetic regions of interest in the cell types. Given that this phenomenon was more pronounced in AS, JIA and PSO, it is likely that the effect of relatively few variants due to sample size employed in the given GWAS contributed to statistical power limitations with the estimation of LDSC, resulting in negative regression slopes in LDSC models.
The convergence of functional disease-specific variants on the same genes and biological networks across different cell types revealed by our 3D chromatin-based variant-to-gene mapping suggests that autoimmune diseases, despite their diverse manifestations, might share common pathways and mechanisms of influence. This places emphasis on the need for better predictive diagnostics with changes in specific gene expression potentially resulting from multiple autoimmune conditions. Understanding that different autoimmune diseases may converge on the same genes and networks implies that therapies targeting these shared pathways could be effective across multiple diseases. This opens up the possibility of developing broad-spectrum treatments that can address the root causes of various autoimmune disorders simultaneously. Conversely, the aspect of cell type-trait specific variants provides the opportunity for precision, personalized medicine, where therapies can be designed to modulate and target only a specific cell type of interest.
We also compared our 3D chromatin-based effector gene identification approach to eQTL approaches. We find that only 10% of the genes nominated by both 3D chromatin V2G and GTEX were implicated by both approaches. The GTEX immune-related dataset is limited to whole blood, and is therefore not powered to determine which individual cell types are driving any of the detected eQTL. Approximately 30% of the nucleated cells in human whole blood are lymphocytes (B cells, T cells, NK cells), ~5% are monocytes, and ~60% are neutrophils, therefore neutrophils, a cell type not included in our analyses, are likely a major driver of GTEX immune eQTLs. The DICE and OneK1K scRNA-seq datasets identified eQTL in immune cell types comparable to those included in our study, however, only 4.5% of the eGenes nominated were implicated by both approaches. The limited overlap could be due in part to the fact that these scRNA-seq datasets implicated less than one quarter the number of eGenes as our study, but crucially, is also consistent with a recent study by Mostafavi et al. suggesting that eQTL and GWAS variants have evolved to control different types of loci73.
The genes implicated by contact with autoimmune-associated variants include several that are part of known disease biology, as well as many that have not been studied in the context of the immunity or autoimmunity. These genes could represent novel therapeutic targets for controlling inflammatory disease. One such gene that we chose to follow up on is FDFT1, encoding a squalene synthase in the cholesterol pathway, which was captured in contact with an MS-associated variant in TFH cells and Th17 cells. While this enzyme has been studied extensively in the context of neuronal myelination, little is known about its specific role in immune cells. We show that FDFT1 antagonism using a direct, small molecule inhibitor of squalene synthase activity reduced production of IL-17, IFNg, and TNF by activated human CD4+ T cells. We did not establish the mechanism by which FDFT1 contributes to pro-inflammatory cytokine production in this study, however, a potential link to IL-17 is that the nuclear receptor RORgt, which is required for Th17 cell differentiation and function, is activated by oxysterols derived from cholesterol catabolism, and presumably would depend on squalene synthase activity74,75. The link to TNF and IFNg is not clear, but a recent in vivo CRISPR screen implicated FDFT1 in the development of memory CD8+ T cells, which are major producers of TNF and IFNg. Other study found that FDFT1 activity disfavors mitochondrial oxidative phosphorylation by diverting lipid synthesis away from CoQ toward cholesterol synthesis76. While more research is needed, our data suggest that FDFT1 is a novel immunomodulator, and validates the power of 3D chromatin-based autoimmune variant-to-gene mapping strategies for identifying therapeutic targets for immune disease.
This study's comprehensive exploration of genetic correlations, tissue-specificity, and shared pathways provides invaluable insights into the complex landscape of autoimmune diseases. By elucidating the underlying mechanisms and highlighting avenues for further research and therapeutic development, the study contributes to advancing our understanding and treatment of autoimmune disorders.
Supplementary Material
ACKNOWLEDGMENTS:
National Institutes of Health, National Institute of Allergy and Infectious Diseases AI146026 (N.R., A.D.W., S.F.A.G.)
Canadian Institutes of Health Research FDN-154304, award from the BC Children's Hospital Research Institute and Canada Research Chair in Engineered Immune Tolerance (M.K.L.)
BC Children’s Hospital Foundation Bertram Hoffmeister Postdoctoral Fellowship (L.C.)
REFERENCEs
- 1.Chiou J., Geusz R.J., Okino M.-L., Han J.Y., Miller M., Melton R., Beebe E., Benaglio P., Huang S., and Korgaonkar K. (2021). Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., and Yoshida S. (2014). Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu X., Pan C.-M., Zhao S.-X., Liang J., Gao G.-Q., Zhang X.-M., Yuan G.-Y., Li C.-G., Xue L.-Q., Shen M., et al. (2011). A genome-wide association study identifies two new risk loci for Graves' disease. Nature Genetics 43, 897–901. 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- 4.Siminovitch K.A. (2004). PTPN22 and autoimmune disease. Nature Genetics 36, 1248–1249. 10.1038/ng1204-1248. [DOI] [PubMed] [Google Scholar]
- 5.Brownlie R.J., Zamoyska R., and Salmond R.J. (2018). Regulation of autoimmune and anti-tumour T-cell responses by PTPN22. Immunology 154, 377–382. 10.1111/imm.12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng L.-L., Wang Y., Zhu F.-L., Xu W.-D., Ji X.-L., and Ni J. (2016). IL-23R mutation is associated with ulcerative colitis: A systemic review and meta-analysis. Oncotarget 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu W.-D., Xie Q.-B., Zhao Y., and Liu Y. (2015). Association of Interleukin-23 receptor gene polymorphisms with susceptibility to Crohn’s disease: A meta-analysis. Scientific Reports 5, 18584. 10.1038/srep18584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong D.-K., Shi X., Han M.-M., Zhang X.-M., Wu N.-N., Sheng X.-Y., and Wang J.-N. (2022). The regulatory mechanism and potential application of IL-23 in autoimmune diseases. Frontiers in Pharmacology 13. 10.3389/fphar.2022.982238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenfelder S., and Fraser P. (2019). Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet 20, 437–455. 10.1038/s41576-019-0128-0. [DOI] [PubMed] [Google Scholar]
- 11.Cook L., Stahl M., Han X., Nazli A., MacDonald K.N., Wong M.Q., Tsai K., Dizzell S., Jacobson K., Bressler B., et al. (2019). Suppressive and Gut-Reparative Functions of Human Type 1 T Regulatory Cells. Gastroenterology 157, 1584–1598. 10.1053/j.gastro.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Cook L., Reid K.T., Häkkinen E., de Bie B., Tanaka S., Smyth D.J., White M.P., Wong M.Q., Huang Q., Gillies J.K., et al. (2021). Induction of stable human FOXP3(+) Tregs by a parasite-derived TGF-β mimic. Immunol Cell Biol 99, 833–847. 10.1111/imcb.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wingett S., Ewels P., Furlan-Magaril M., Nagano T., Schoenfelder S., Fraser P., and Andrews S. (2015). HiCUP: pipeline for mapping and processing Hi-C data. F1000Res 4, 1310. 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cairns J., Freire-Pritchett P., Wingett S.W., Várnai C., Dimond A., Plagnol V., Zerbino D., Schoenfelder S., Javierre B.-M., Osborne C., et al. (2016). CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biology 17, 1–17. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.C S., MC P., SFA G., and AD W. (2021). Restriction enzyme selection dictates detection range sensitivity in chromatin conformation capture-based variant-to-gene mapping approaches. Human genetics 140. 10.1007/s00439-021-02326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z., Schulz M.H., Look T., Begemann M., Zenke M., and Costa I.G. (2019). Identification of transcription factor binding sites using ATAC-seq. Genome Biology 20, 45. 10.1186/s13059-019-1642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H., Sun Z., Wang J., Huang H., Kocher J.P., and Wang L. (2014). CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007. 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su C., Gao L., May C.L., Pippin J.A., Boehm K., Lee M., Liu C., Pahl M.C., Golson M.L., Naji A., et al. (2022). 3D chromatin maps of the human pancreas reveal lineage-specific regulatory architecture of T2D risk. Cell Metabolism 34. 10.1016/j.cmet.2022.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., and Mirny L.A. (2012). Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nature Methods 9, 999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roayaei Ardakany A., Gezer H.T., Lonardi S., and Ay F. (2020). Mustache: multi-scale detection of chromatin loops from Hi-C and Micro-C maps using scale-space representation. Genome Biology 21, 1–17. doi: 10.1186/s13059-020-02167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A K., S B., and F A. (2020). Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nature protocols 15. 10.1038/s41596-019-0273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L., Rosen J.D., Sun Q., Chen J., Wheeler M.M., Zhou Y., Min Y.-I., Kooperberg C., Conomos M.P., Stilp A.M., et al. (2022). TOP-LD: A tool to explore linkage disequilibrium with TOPMed whole-genome sequence data. The American Journal of Human Genetics 109, 1175–1181. 10.1016/j.ajhg.2022.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers T.A., Chanock S.J., and Machiela M.J. (2020). LDlinkR: An R Package for Rapidly Calculating Linkage Disequilibrium Statistics in Diverse Populations. Front Genet 11, 157. 10.3389/fgene.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M.I., Huber W., and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu A., Ibrahim J.G., and Love M.I. (2018). Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092. 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P., and Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2, 100141. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulgen E., Ozisik O., and Sezerman O.U. (2019). pathfindR: An R Package for Comprehensive Identification of Enriched Pathways in Omics Data Through Active Subnetworks. Frontiers in Genetics 10. 10.3389/fgene.2019.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerimov N., Hayhurst J.D., Peikova K., Manning J.R., Walter P., Kolberg L., Samoviča M., Sakthivel M.P., Kuzmin I., Trevanion S.J., et al. (2021). A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nature Genetics 53, 1290–1299. 10.1038/s41588-021-00924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmiedel B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G., et al. (2018). Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 175, 1701–1715.e1716. 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazar S., Alquicira-Hernandez J., Wing K., Senabouth A., Gordon M.G., Andersen S., Lu Q., Rowson A., Taylor T.R.P., Clarke L., et al. (2022). Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 376, eabf3041. 10.1126/science.abf3041. [DOI] [PubMed] [Google Scholar]
- 33.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., and Plagnol V. (2014). Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLOS Genetics 10, e1004383. 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao Y., Stuart T., Kowalski M.H., Choudhary S., Hoffman P., Hartman A., Srivastava A., Molla G., Madad S., Fernandez-Granda C., and Satija R. (2024). Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nature Biotechnology 42, 293–304. 10.1038/s41587-023-01767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palstra R.J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., and de Laat W. (2003). The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35, 190–194. 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 36.Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., and Flavell R.A. (2005). Interchromosomal associations between alternatively expressed loci. Nature 435, 637–645. 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 37.Deng W., Lee J., Wang H., Miller J., Reik A., Gregory P.D., Dean A., and Blobel G.A. (2012). Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244. 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pahl M.C., Le Coz C., Su C., Sharma P., Thomas R.M., Pippin J.A., Cruz Cabrera E., Johnson M.E., Leonard M.E., Lu S., et al. (2022). Implicating effector genes at COVID-19 GWAS loci using promoter-focused Capture-C in disease-relevant immune cell types. Genome Biol 23, 125. 10.1186/s13059-022-02691-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., et al. (2015). Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature Genetics 47, 1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann M.A., Fluhr J.W., Ruwwe-Glösenkamp C., Stevanovic K., Bergmann K.C., and Zuberbier T. (2021). Role of IL-17 in atopy-A systematic review. Clin Transl Allergy 11, e12047. 10.1002/clt2.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauffer F., Jargosch M., Baghin V., Krause L., Kempf W., Absmaier-Kijak M., Morelli M., Madonna S., Marsais F., Lepescheux L., et al. (2020). IL-17C amplifies epithelial inflammation in human psoriasis and atopic eczema. J Eur Acad Dermatol Venereol 34, 800–809. 10.1111/jdv.16126. [DOI] [PubMed] [Google Scholar]
- 42.Moser T., Akgün K., Proschmann U., Sellner J., and Ziemssen T. (2020). The role of TH17 cells in multiple sclerosis: Therapeutic implications. Autoimmun Rev 19, 102647. 10.1016/j.autrev.2020.102647. [DOI] [PubMed] [Google Scholar]
- 43.Sakaguchi S., Yamaguchi T., Nomura T., and Ono M. (2008). Regulatory T Cells and Immune Tolerance. Cell 133, 775–787. 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Goswami T.K., Singh M., Dhawan M., Mitra S., Emran T.B., Rabaan A.A., Mutair A.A., Alawi Z.A., Alhumaid S., and Dhama K. (2022). Regulatory T cells (Tregs) and their therapeutic potential against autoimmune disorders - Advances and challenges. Hum Vaccin Immunother 18, 2035117. 10.1080/21645515.2022.2035117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Opoka-Winiarska V., Grywalska E., Sobiesiak A., and Roliński J. (2020). The Impact of Epstein-Barr Virus Infection on Juvenile Idiopathic Arthritis Activity and Patient's Response to Treatment. J Clin Med 9. 10.3390/jcm9113453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rigante D., Bosco A., and Esposito S. (2015). The Etiology of Juvenile Idiopathic Arthritis. Clin Rev Allergy Immunol 49, 253–261. 10.1007/s12016-014-8460-9. [DOI] [PubMed] [Google Scholar]
- 47.Thrastardottir T., and Love T.J. (2018). Infections and the risk of psoriatic arthritis among psoriasis patients: a systematic review. Rheumatol Int 38, 1385–1397. 10.1007/s00296-017-3873-4. [DOI] [PubMed] [Google Scholar]
- 48.Soldan S.S., and Lieberman P.M. (2023). Epstein-Barr virus and multiple sclerosis. Nat Rev Microbiol 21, 51–64. 10.1038/s41579-022-00770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jog N.R., and James J.A. (2020). Epstein Barr Virus and Autoimmune Responses in Systemic Lupus Erythematosus. Front Immunol 11, 623944. 10.3389/fimmu.2020.623944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong T., Parameswaran S., Donmez O.A., Miller D., Forney C., Lape M., Saint Just Ribeiro M., Liang J., Edsall L.E., Magnusen A.F., et al. (2021). Epstein-Barr virus nuclear antigen 2 extensively rewires the human chromatin landscape at autoimmune risk loci. Genome Res 31, 2185–2198. 10.1101/gr.264705.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lasconi C., Pahl M.C., Cousminer D.L., Doege C.A., Chesi A., Hodge K.M., Leonard M.E., Lu S., Johnson M.E., Su C., et al. (2021). Variant-to-Gene-Mapping Analyses Reveal a Role for the Hypothalamus in Genetic Susceptibility to Inflammatory Bowel Disease. Cell Mol Gastroenterol Hepatol 11, 667–682. 10.1016/j.jcmgh.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karakasheva T.A., Zhou Y., Xie H.M., Soto G.E., Johnson T.D., Stoltz M.A., Roach D.M., Nema N., Umeweni C.N., Naughton K., et al. (2023). Patient-derived Colonoids From Disease-spared Tissue Retain Inflammatory Bowel Disease-specific Transcriptomic Signatures. Gastro Hep Adv 2, 830–842. 10.1016/j.gastha.2023.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imaruoka K., Oe Y., Fushima T., Sato E., Sekimoto A., Sato H., Sugawara J., Ito S., and Takahashi N. (2019). Nicotinamide alleviates kidney injury and pregnancy outcomes in lupus-prone MRL/lpr mice treated with lipopolysaccharide. Biochem Biophys Res Commun 510, 587–593. 10.1016/j.bbrc.2019.01.110. [DOI] [PubMed] [Google Scholar]
- 54.Campbell A.M., Kashgarian M., and Shlomchik M.J. (2012). NADPH oxidase inhibits the pathogenesis of systemic lupus erythematosus. Sci Transl Med 4, 157ra141. 10.1126/scitranslmed.3004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M., Lai Y., Chen B., Guo C., Zhou M., Zhao S., Wang S., Li J., Yang N., and Zhang H. (2023). NAMPT is a metabolic checkpoint of IFNγ-producing CD4(+) T cells in lupus nephritis. Mol Ther 31, 193–210. 10.1016/j.ymthe.2022.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye D., Sun X., Guo Y., Shao K., Qian Y., Huang H., Liu B., Wen C., and Mao Y. (2021). Genetically determined selenium concentrations and risk for autoimmune diseases. Nutrition 91-92, 111391. 10.1016/j.nut.2021.111391. [DOI] [PubMed] [Google Scholar]
- 57.Abrishamkar H., Helli B., Zilaei M., Rajaei E., Sheikhi A., & Hosseini S. A. (2024). Selenium and systemic lupus erythematosus (sle): a double- blind randomised controlled trial. [Preprint]. 10.21203/rs.3.rs-3956860/v1. [DOI] [Google Scholar]
- 58.Herman S., Åkerfeldt T., Spjuth O., Burman J., and Kultima K. (2019). Biochemical Differences in Cerebrospinal Fluid between Secondary Progressive and Relapsing-Remitting Multiple Sclerosis. Cells 8. 10.3390/cells8020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vermersch P., Czlonkowska A., Grimaldi L.M., Confavreux C., Comi G., Kappos L., Olsson T.P., Benamor M., Bauer D., Truffinet P., et al. (2014). Teriflunomide versus subcutaneous interferon beta-1a in patients with relapsing multiple sclerosis: a randomised, controlled phase 3 trial. Mult Scler 20, 705–716. 10.1177/1352458513507821. [DOI] [PubMed] [Google Scholar]
- 60.Shenoi S., Bell S., Wallace C.A., and Mueller B.A. (2015). Juvenile idiopathic arthritis in relation to maternal prenatal smoking. Arthritis Care Res (Hoboken) 67, 725–730. 10.1002/acr.22471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao W., Li C., and Deng J. (2023). A systematic review and meta-analysis of environmental factors associated with juvenile idiopathic arthritis. Int J Rheum Dis 26, 1235–1247. 10.1111/1756-185x.14729. [DOI] [PubMed] [Google Scholar]
- 62.França C.M.P., Sallum A.M.E., Braga A.L.F., Strufaldi F.L., Silva C.A.A., and Farhat S.C.L. (2018). Risk Factors Associated with Juvenile Idiopathic Arthritis: Exposure to Cigarette Smoke and Air Pollution from Pregnancy to Disease Diagnosis. J Rheumatol 45, 248–256. 10.3899/jrheum.161500. [DOI] [PubMed] [Google Scholar]
- 63.Pahl M.C., Sharma P., Thomas R.M., Thompson Z., Mount Z., Pippin J., Morawski P.A., Sun P., Su C., Campbell D.J., et al. (2023). Dynamic chromatin architecture identifies new autoimmune-associated enhancers for IL2 and novel genes regulating CD4+ T cell activation. bioRxiv, 2023.2004.2005.535731. 10.1101/2023.04.05.535731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su C., Johnson M.E., Torres A., Thomas R.M., Manduchi E., Sharma P., Mehra P., Le Coz C., Leonard M.E., Lu S., et al. (2020). Mapping effector genes at lupus GWAS loci using promoter Capture-C in follicular helper T cells. Nature Communications 11, 3294. 10.1038/s41467-020-17089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chesi A., Wagley Y., Johnson M.E., Manduchi E., Su C., Lu S., Leonard M.E., Hodge K.M., Pippin J.A., Hankenson K.D., et al. (2019). Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nature Communications 10, 1–11. doi: 10.1038/s41467-019-09302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pippin J.A., Chesi A., Wagley Y., Su C., Pahl M.C., Hodge K.M., Johnson M.E., Wells A.D., Hankenson K.D., and Grant S.F.A. (2021). CRISPR-Cas9-Mediated Genome Editing Confirms EPDR1 as an Effector Gene at the BMD GWAS-Implicated 'STARD3NL' Locus. JBMR Plus 5, e10531. 10.1002/jbm4.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palermo J., Chesi A., Zimmerman A., Sonti S., Pahl M.C., Lasconi C., Brown E.B., Pippin J.A., Wells A.D., Doldur-Balli F., et al. (2023). Variant-to-gene mapping followed by cross-species genetic screening identifies GPI-anchor biosynthesis as a regulator of sleep. Sci Adv 9, eabq0844. 10.1126/sciadv.abq0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia Q., Chesi A., Manduchi E., Johnston B.T., Lu S., Leonard M.E., Parlin U.W., Rappaport E.F., Huang P., Wells A.D., et al. (2016). The type 2 diabetes presumed causal variant within TCF7L2 resides in an element that controls the expression of ACSL5. Diabetologia 59, 2360–2368. 10.1007/s00125-016-4077-2. [DOI] [PubMed] [Google Scholar]
- 69.Berghoff S.A., Spieth L., Sun T., Hosang L., Depp C., Sasmita A.O., Vasileva M.H., Scholz P., Zhao Y., Krueger-Burg D., et al. (2021). Neuronal cholesterol synthesis is essential for repair of chronically demyelinated lesions in mice. Cell Rep 37, 109889. 10.1016/j.celrep.2021.109889. [DOI] [PubMed] [Google Scholar]
- 70.Itoh N., Itoh Y., Tassoni A., Ren E., Kaito M., Ohno A., Ao Y., Farkhondeh V., Johnsonbaugh H., Burda J., et al. (2018). Cell-specific and region-specific transcriptomics in the multiple sclerosis model: Focus on astrocytes. Proc Natl Acad Sci U S A 115, E302–e309. 10.1073/pnas.1716032115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schafflick D., Xu C.A., Hartlehnert M., Cole M., Schulte-Mecklenbeck A., Lautwein T., Wolbert J., Heming M., Meuth S.G., Kuhlmann T., et al. (2020). Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nature Communications 11, 247. 10.1038/s41467-019-14118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ota M., Nagafuchi Y., Hatano H., Ishigaki K., Terao C., Takeshima Y., Yanaoka H., Kobayashi S., Okubo M., Shirai H., et al. (2021). Dynamic landscape of immune cell-specific gene regulation in immune-mediated diseases. Cell 184, 3006–3021.e3017. 10.1016/j.cell.2021.03.056. [DOI] [PubMed] [Google Scholar]
- 73.Mostafavi H., Spence J.P., Naqvi S., and Pritchard J.K. (2023). Systematic differences in discovery of genetic effects on gene expression and complex traits. Nat Genet 55, 1866–1875. 10.1038/s41588-023-01529-1. [DOI] [PubMed] [Google Scholar]
- 74.Santori F.R., Huang P., van de Pavert S.A., Douglass E.F. Jr., Leaver D.J., Haubrich B.A., Keber R., Lorbek G., Konijn T., Rosales B.N., et al. (2015). Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab 21, 286–298. 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu X., Wang Y., Hao L.Y., Liu X., Lesch C.A., Sanchez B.M., Wendling J.M., Morgan R.W., Aicher T.D., Carter L.L., et al. (2015). Sterol metabolism controls T(H)17 differentiation by generating endogenous RORγ agonists. Nat Chem Biol 11, 141–147. 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- 76.Reina-Campos M., Heeg M., Kennewick K., Mathews I.T., Galletti G., Luna V., Nguyen Q., Huang H., Milner J.J., Hu K.H., et al. (2023). Metabolic programs of T cell tissue residency empower tumour immunity. Nature 621, 179–187. 10.1038/s41586-023-06483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.