Abstract
Background:
High red meat and/or processed meat consumption are established colorectal cancer (CRC) risk factors. We conducted a genome-wide gene-environment (GxE) interaction analysis to identify genetic variants that may modify these associations.
Methods:
A pooled sample of 29,842 CRC cases and 39,635 controls of European ancestry from 27 studies were included. Quantiles for red meat and processed meat intake were constructed from harmonized questionnaire data. Genotyping arrays were imputed to the Haplotype Reference Consortium. Two-step EDGE and joint tests of GxE interaction were utilized in our genome-wide scan.
Results:
Meta-analyses confirmed positive associations between increased consumption of red meat and processed meat with CRC risk (per quartile red meat OR = 1.30; 95%CI = 1.21–1.41; processed meat OR = 1.40; 95%CI = 1.20–1.63). Two significant genome-wide GxE interactions for red meat consumption were found. Joint GxE tests revealed the rs4871179 SNP in chromosome 8 (downstream of HAS2); greater than median of consumption ORs = 1.38 (95%CI = 1.29–1.46), 1.20 (95%CI = 1.12 −1.27), and 1.07 (95%CI = 0.95 – 1.19) for CC, CG and GG, respectively. The two-step EDGE method identified the rs35352860 SNP in chromosome 18 (SMAD7 intron); greater than median of consumption ORs = 1.18 (95%CI = 1.11–1.24), 1.35 (95%CI = 1.26–1.44), and 1.46 (95%CI = 1.26–1.69) for CC, CT, and TT, respectively.
Conclusions:
We propose two novel biomarkers that support the role of meat consumption with an increased risk of CRC.
Impact:
The reported GxE interactions may explain the increased risk of CRC in certain population subgroups.
INTRODUCTION
Colorectal cancer (CRC) is currently the third most common cancer worldwide, and second leading cause of cancer death (1). It is estimated that at least ~50% of CRC cases and CRC deaths could be attributed to modifiable lifestyle factors (2–4). The main established modifiable CRC risk factors are high consumption of processed meat and/or red meat, consumption of alcoholic drinks, smoking, being overweight or obese, low consumption of foods containing dietary fiber, low consumption of whole grains, and low physical activity (5–7). Based on the existing literature, the World Cancer Research Fund has concluded that there is strong evidence that red meat and/or processed meat consumption increases the risk of CRC (5). Moreover, based on epidemiological, animal, and mechanistic data, the International Agency for Research on Cancer (IARC) classified consumption of processed meat as a group 1 carcinogen (i.e., an established cause of CRC), and red meat as a group 2a carcinogen (i.e., a probable cancer agent) (6,8). These classifications were in great part based on the evidence for CRC.
There are several mechanisms that have been proposed to explain the relationship between consumption of red or processed meat and CRC risk. Among them is the presence of carcinogenic N-nitroso compounds (NOC) in processed meat that can also be formed endogenously in the gut after consumption of red meat (9,10). The abundance of heme iron in red meat, combined with the presence of gut bacteria can facilitate this carcinogenic process. NOC primarily produced through bacterial decarboxylation of amino acids in the presence of a nitrosating agent may cause damage and inflammation to the gut lining. Furthermore, the existence of a persistent intestinal dysbiosis might exacerbate the carcinogenic process, as it has been linked to chronic gut inflammation (11–14). Additional carcinogens linked to red meat and/or processed meat are heterocyclic amines (HCAs) (15) and polycyclic aromatic hydrocarbons (PAHs) (16), which can be formed by different cooking methods (17,18).
In addition to modifiable risk factors, there are common genetic variants that have been linked to CRC. To date, there are over 200 genetic variants that were identified with genome-wide association studies (GWAS) that altogether explain ~20% of the heritability in CRC risk (19–21). Gene-environment (GxE) interactions refers to the phenomenon in which the effects of genetic variations at an individual level are modified by environmental factors and vice versa. In other words, it is the concept that environmental factors can impact the expression/function of genes, and that genetic factors can influence a person’s sensitivity or reaction to environmental factors. It has been speculated that these GxE interactions may explain part of the large fraction of missing heritability (22). There have been multiple studies that have explored the role of common genetic variants as potential modifiers of the effect of processed meat or red meat (23), most of them focused on candidate genes and single nucleotide polymorphisms (SNPs). There have only been two genome-wide GxE scans for red and processed meat (24,25), and only one of these reported a significant GxE interaction with processed meat (24). A more recent review and evaluation of all available data assigned this finding a moderate plausibility score and overall concluded that most studies of GxE in CRC to date have been underpowered to detect GxE interactions (23).
In this study, we applied powerful methods in a genome-wide GxE scan to identify possible interactions between common variants and red meat and/or processed meat intake and CRC risk, using a large CRC pooled dataset.
MATERIALS and METHODS
Study participants
A total of 27 studies (17 prospective cohorts; 10 case-controls) were included in this study from three CRC genetic consortia: the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO), the Colorectal Cancer Transdisciplinary Study (CORECT) and the Colon Cancer Family Registry (CCFR) (19,20,26) (Supplementary Table 1). For cohort studies, nested case-control sets were assembled via risk-set sampling, while population-based controls were used for case-control studies. Controls were mainly matched on age (study-specific range), sex, race, and enrollment date/trial group, when applicable (for the SELECT trial). Cases were defined as colorectal adenocarcinoma (the most common type of CRC) and were confirmed by medical records, pathology reports, or death certificate information. All participants gave written informed consent and studies were approved by their respective institutional review boards. Analyses were limited to individuals of European ancestry, based on self-reported race and ethnicity, and clustering of principal components with the 1000 Genomes Europeans (EUR) superpopulation. We excluded individuals based on cryptic relatedness or duplicates, genotyping/imputation errors, and age outliers. Individuals missing both red meat and processed meat intake or any of the adjustment covariates we considered were excluded. We further excluded studies of individuals diagnosed with advanced adenomas only. The final pooled sample included 29,842 CRC cases and 39,635 controls.
Environmental exposure data
Lifestyle and environmental risk factors were collected by in-person interviews, phone interviews or structured self-administered questionnaires. Harmonized quality-control checks were performed in each study following similar criteria (27). Common data elements (CDEs) were defined a priori and through an iterative process, and responses from study questionnaires and data dictionaries were mapped to these CDEs. All definitions, permissible values, and standardized coding were tracked in a database via SAS and T-SQL. Data checks were performed to identify outliers and other errors. We developed a pooled variable that tracked consumption of red meat intake (beef, pork, lamb), and another one that tracked consumption of processed meat (bacon, sausages, luncheon/deli meats, hot dogs), each expressed as servings per day. Many of the red meat variables in the pooled studies had included processed meats as part of the red meat definition. Of the total study population, 64,152 individuals had both red and processed meat consumption information; 5,325 had information only on red meat intake. We evaluated these two exposures using sex- and study-specific quartiles in which study-specific quartiles were computed and then the median of intake within each quartile was obtained. Body Mass Index (BMI) was calculated as the weight of each participant (in kg) divided by the square of height (in m2). A BMI variable was used that captured 5 kg/m2 increments. In addition, the World Health Organization (WHO) pre-defined BMI cut-points were used: normal weight (18.5–<25 kg/m2), overweight (>=25.0–<30 kg/m2), and obesity (>=30 kg/m2) (28). Total energy intake was calculated from food frequency questionnaires. For studies with partially missing data, total caloric intake was imputed using the mean value in the study. Studies with missing energy intake estimates were set to zero.
Genotyping and imputation
Details on quality control and genotyping metrics were previously published (19,26). Several genotyping arrays were used and are summarized in Supplementary Table 1. Briefly, exclusion criteria included: single nucleotide polymorphisms (SNPs) with a missing call rate of >2-to-5%, departure from Hardy-Weinberg equilibrium (HWE) (P<1×10−4), inconsistencies between self-reported and genotyped sex, and discordant genotype calls within duplicate samples. Genotypes were imputed to the Haplotype Reference Consortium (HRC) panel (39.1 million variants) using the University of Michigan Imputation Server (29,30), and converted into a binary format for data management and analyses using the BinaryDosage R package (https://cran.r-project.org/web/packages/BinaryDosage). Imputed SNPs were restricted based on a pooled minor allele frequency (MAF) ≥1% and imputation accuracy (R2>0.8). After imputation and quality control, a total of approximately 7.2 million SNPs were selected for analyses. Principal component analysis (PCA) for population stratification assessment was performed using PLINK1.9 on 30,000 randomly sampled imputed SNPs with MAF > 5% and R2>0.99.
Statistical analyses
The associations of red meat or processed meat intake variables with CRC risk were assessed by meta-analysis of study-specific estimates, adjusted by sex, age and total energy intake. Between-sex, between-tumor site, between-study design as well as between-study heterogeneity and inconsistency were investigated using the heterogeneity Chi-squared and I2 statistic (31). Between-study heterogeneity represents the proportion of total variation in effect estimates attributable to between-study variance. Potential outlier studies were assessed by estimating the posterior probability of outliers based on mixture random effects using the “outlierProbs” function of metaplus R package (32). No outlier studies (posterior probability >0.99) were identified. We also assessed the relationship between red meat or processed meat with CRC risk stratified by sex, tumor localization (proximal colon, distal colon, or rectum) and study design (case-control or cohort study).
To identify novel GxE interactions for CRC, we performed genome-wide scans using the GxEScanR R package (https://cran.r-project.org/web/packages/GxEScanR), which implements several interaction testing methods. Imputed allelic dosages were modelled as continuous variables. In addition to the standard 1 degree-of-freedom (1-df) test of GxE interaction based on logistic regression, we utilized the more powerful two-step EDGE method (33) and the 3-degree-of-freedom (3-df) joint test (34). All models were adjusted for age, sex, study, total energy intake (kcal/day), and the first three principal components to account for ancestry. We considered a P value of <5×10−8 statistically significant. We also calculated odds ratios (OR) for red meat intake stratified by genotype and for genotype stratified by meat intake to examine patterns of sub-group-specific associations. A detailed description of the notation and methods used in the GxE analysis has been previously published (33–34).
Functional annotation plots for GxE findings and regional plots were also generated. Regional plots enable inspection of the strength and extent of association signal, linkage disequilibrium (LD), and position of findings relative to genes in the region. Regional plots were generated using the software LocusZoom v1.3.32. Measures of LD were estimated using 1000G EUR study population controls.
Data availability
The data generated in this study are available upon request from the corresponding author.
RESULTS
Red meat and processed meat consumption and CRC risk
CRC cases were slightly older, were more likely living with obesity, had a higher total energy intake (2011kca/day ± 730.8 versus 1920kcal/day ± 706.6, p<0.001), and consumed more servings of red (0.60 ± 0.46 versus 0.54 ± 0.44, p<0.001) and/or processed meat (0.36 ± 0.35 versus 0.28 ± 0.35, p<0.001) per day when compared to controls (Table 1).
Table 1.
Summary statistics for demographic and CRC related risk factors, by case/control status
| Cases | Controls | ||
|---|---|---|---|
|
|
|||
| N = 29842 | N = 39635 | P-value | |
|
|
|||
| Red meat intake (servings/day)a | |||
| Mean (SD) | 0.60 (± 0.46) | 0.54 (± 0.44) | <0.001 |
| Missing | 70 (0.2%) | 69 (0.2%) | |
| Processed meat intake (servings/day)a | |||
| Mean (SD) | 0.36 (± 0.35) | 0.28 (± 0.29) | <0.001 |
| Missing | 2930 (9.8%) | 2395 (6.0%) | |
| Ageb | |||
| Mean (SD) | 64.2 (± 10.8) | 63.5 (± 9.42) | <0.001 |
| Sex | |||
| Female | 14569 (48.88%) | 19615 (49.52%) | 0.082 |
| Male | 15273 (51.2%) | 20020 (50.48%) | |
| Height (cm) | |||
| Mean (SD) | 169 (± 9.56) | 169 (± 9.53) | 0.314 |
| Missing | 411 (1.4%) | 282 (0.7%) | |
| BMI | |||
| Mean (SD) | 27.4 (± 4.90) | 27.0 (± 4.62) | <0.001 |
| Missing | 1492 (5.0%) | 1459 (3.7%) | |
| BMI | |||
| Normal (18.5 to <25 kg/m2) | 9286 (31.1%) | 13570 (34.2%) | <0.001 |
| Overweight (>=25 to <30 kg/m2) | 11915 (39.9%) | 16263 (41%) | |
| Obese (>=30 kg/m2) | 6903 (23.1%) | 8071 (20.4%) | |
| Missing | 1738 (5.8%) | 1731 (4.4%) | |
| Total Energy Intake (kcal/day)b | |||
| Mean (SD) | 2011 (± 730.8) | 1920 (± 706.6) | <0.001 |
| Missing | 7475 (25%) | 14998 (37.8%) | |
1 serving is equivalent to 70.9 grams or 2.5 ounces.
Mean imputed for partially missing data. Studies with missing variable not used for mean estimation.
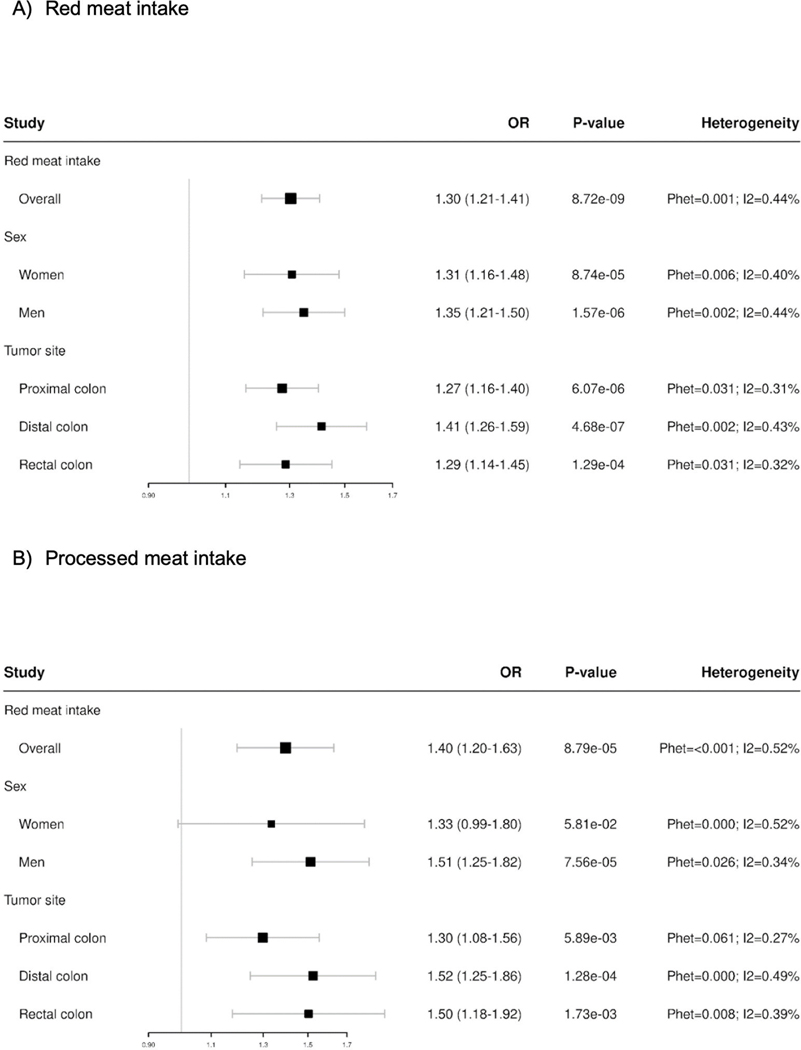
Meta-analyses of exposure main effects adjusting for age, sex, and total energy intake, showed associations between intake of red meat (per quartile increase servings/day OR=1.30; 95%CI 1.21–1.41) and intake of processed meat and CRC risk (per quartile increase OR=1.40; 95%CI 1.20–1.63) (Figure 1). We observed significant between study heterogeneity that was largely limited to case-control studies (Phet = 0.001, I2 = 65% & Phet = 0.008, I2 = 61% for red meat and processed meat, respectively) (Supplemental Figure 1). For both exposures, estimates of association were greater in magnitude in meta-analyses of case-control studies (red meat meta-OR = 1.45; 95% CI = 1.22–1.72; processed meat meta-OR = 1.56; 1.22–2.00) than cohort studies (red meat meta-OR = 1.21; 95% CI = 1.08–1.35; processed meat meta-OR = 1.16; 0.98–1.37) (Supplemental Figure 1). Analysis stratified by sex suggested a slightly higher association for consumption of processed meat and CRC risk among men (OR = 1.51; 95%CI 1.25–1.82) compared to women (OR = 1.33; 95% CI = 0.99–1.80) (Figure 1). There were no substantial sex differences for the association between CRC and red meat intake. The association between red meat consumption and CRC risk was slightly higher for distal colon compared to proximal or rectal; whereas for processed meat consumption the association was higher for both distal colon and rectal localization compared to proximal location, albeit overlap in the confidence intervals was observed and these differences are not statistically significant (Figure 1). We obtained similar estimates when performing pooled analyses of associations further adjusted by the first three ancestry principal components. Whereas there was evidence for heterogeneity across studies (Supplemental Figure 1), when considering cohort and case-control studies separately, we decided to conduct GxE testing across all studies combined given that a positive association was reported in both study types.
Figure 1.
Results from overall association between colorectal cancer and A) red meat intake and B) processed meat intake, overall and stratified by sex and tumor site. Models are adjusted for age, sex, and total energy intake. Meat intake servings per day were coded as median of sex/study specific quartiles, modeled as a continuous variable.
Genome-wide interaction scan results
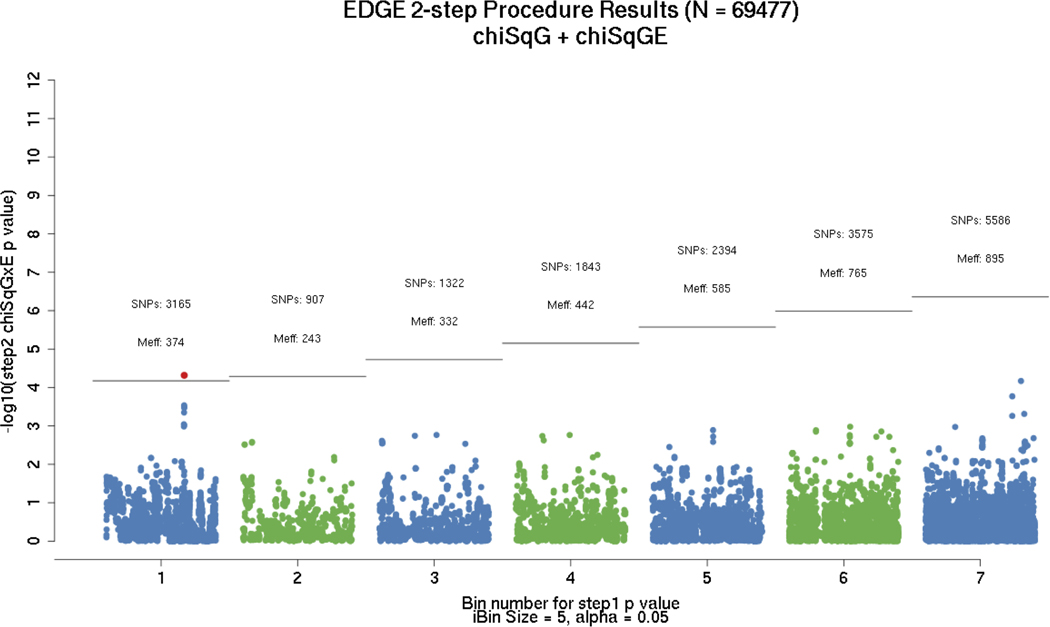
Evaluation of red meat consumption interaction using the two-step EDGE method identified a statistically significant interaction of red meat with rs35352860 (p-value = 2 × 10−8) which maps to chromosome 18 in an intronic region of the SMAD7 (SMAD family member 7) gene (Figure 2, Table 2).
Figure 2.
Results from EDGE two-step GxE testing procedure using expectation-based SNP partitioning with principal components approach for effective number of tests adjustment (59). Numbers reflect total number of SNPs assigned to each partition, with the number of effective tests listed below. Only the first 7 bins were plotted; significant hit corresponds to rs35352860 (SMAD7 region).
Table 2.
Main results from genome-wide interaction scans of red meat consumption
| Meat intake variable | SNP | Chr | BP Position | Gene | Ref | Alt | Alt Allele Freq (1000G EUR) | Method | P-value G | P-value GxE | P-value 3-df |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Red meat | rs4871179 | 8 | 122247679 | Downstream HAS2 | C | G | 0.33 | Joint test (3-df) | 3.99E-04 | 1.00E-06 | 3.05E-08 |
| Red meat | rs35352860 | 18 | 46453754 | SMAD7 | C | T | 0.26 | Two-step | 4.75E-21 | 2.0E-8 | 7.16E-23 |
SNP, single nucleotide polymorphism; Chr, chromosome; df, degrees of freedom; BP Position, base pair position based on NCBI Build37.
Notes: Imputed SNPs were coded as expected gene dosage. Multiplicative interaction terms were modelled as the product of red meat and each SNP of interest. All statistical tests were two-sided. Models were adjusted for age, sex, study, total energy intake (kcal/day), and the first three principal components to account for ancestry.
One additional SNP for red meat intake on chromosome 8 was identified based on the joint 3-df test (Table 2). This SNP, rs4871179, maps downstream of HAS2 (hyaluronan synthase 2). No statistically significant interactions were identified for processed meat. The less powerful standard 1-df GxE analysis revealed no evidence of interaction with either red meat or processed meat and genome-wide statistically significant loci (Supplemental Figure 2). No other statistically significant GxE interactions (p-value <10−8) that were also genome-wide significant GWAS loci were found.
To further explore the significant interactions, we constructed a dichotomous variable for red meat intake with median of consumption as cutoff point and evaluated the association between red meat intake and CRC risk, stratifying by genotypes of the identified loci (Table 3). For the SNP on chromosome 18 identified by the two-step method (rs4871179), red meat was associated with CRC risk within each genotype group, but the magnitude increased with every copy of the major T allele. Specifically, the red meat in relation to CRC odds ratio was OR = 1.18 (95% CI = 1.11–1.24) for those with genotype CC, OR = 1.35 (95% CI 1.26–1.44) for CT, and OR = 1.46 (1.26–1.69) for TT (Table 3; Supplemental Figure 3). For the SNP in chromosome 8, the association between red meat intake and CRC risk was significant among homozygous carriers of the more common allele (OR for CC = 1.38; 95% CI = 1.29–1.46) and heterozygous (OR for CG = 1.2; 95% CI = 1.12–1.27) with a non-statistically significant association among homozygous carriers of the minor G allele (OR for GG = 1.07; 95% CI = 0.95–1.19) (Table 3; Supplemental Figure 3).
Table 3.
Odds ratio (95% confidence interval) showing association between red meat intakea and CRC risk, stratified by genotypes of loci identified using joint testing and two-step methods
| rs4871179 (8:122247679) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CG | GG | |||||||||
| red meat intake = 0 | 1 (Reference) | 1.03 (0.98–1.09) | 1.07 (0.98–1.16) | ||||||||
| red meat intake = 1 | 1.38 (1.29–1.46) | 1.24 (1.16–1.31) | 1.14 (1.05–1.24) | ||||||||
| red meat intake by genotype | 1.38 (1.29–1.46) | 1.2 (1.12–1.27) | 1.07 (0.95–1.19) | ||||||||
|
| |||||||||||
| rs35352860 (18:46453754) |
|||||||||||
| CC | CT | TT | |||||||||
|
| |||||||||||
| red meat intake = 0 | 1.0 (Ref) | 0.83 (0.79–0.88) | 0.65 (0.58–0.73) | ||||||||
| red meat intake = 1 | 1.17 (1.11–1.24) | 1.13 (1.06–1.19) | 0.94 (0.85–1.04) | ||||||||
| red meat intake by genotype | 1.18 (1.11–1.24) | 1.35 (1.26–1.44) | 1.46 (1.26–1.69) | ||||||||
Red meat intake variable was dichotomized with median of servings/day from study specific-quartiles as cutoff point.
Notes: Models were adjusted for age, sex, study, total energy intake (kcal/day), and the first three principal components to account for ancestry.
DISCUSSION
In this large-scale genome-wide GxE analysis of over 69,000 individuals from 27 studies, we found evidence of a statistical interaction between 2 SNPs (rs35352860, rs4871179) and red meat consumption in relation to CRC risk. We did not find evidence of GxE interactions when considering consumption of processed meat. A prior publication reported a G x processed-meat interaction for the rs4143094 SNP (10p14, near GATA3). Analysis of this SNP in our current sample revealed some suggestion of an interaction (p=0.0065) but it did not achieve genome-wide significance in any of our analyses (24).
The rs35352860 SNP which maps to the SMAD7 gene was found to have the strongest evidence to support effect modification of the red meat intake and CRC risk association as reported by the p-values in the GxE scan (Table 2). The SMAD7 gene codes for an intracellular protein, traditionally considered as a negative regulator of TGF-B1 (35) by interfering with TGF-B1 signaling. Several SMAD7 polymorphisms have been previously associated with CRC risk in several GWAS studies for CRC in European and Asian individuals (36–38). Moreover, a regional plot for rs35352860 shows that the SNP aligns with multiple hits in LD previously reported by Broderick et.al (39) (Supplementary Figure 4A). Additionally, an earlier study from this consortium reported a statistically significant GxE interaction between rs4939827 and BMI (40). We conducted a sensitivity analysis after adjustment of main effects model for BMI. Results did not differ from main findings obtained in genome-wide interaction scans of red meat consumption (Supplementary Table 2).
There are no reports of a mechanism for a potential GxE interaction for the rs35352860 polymorphism and red meat. Previous genome-wide GxE interaction analyses including red meat did not report associations of genome-wide significance for SMAD7 polymorphisms (27,41). However, SMAD7 deletion has been previously associated with a protective dose-effect in overall survival and disease-free survival in tumor biopsies of CRC patients, with a greater effect on increased CRC-related death with every additional copy (42), and with overexpression in colonic adenocarcinoma cells inducing tumorigenesis (43).
Key components of red meat that have been linked to CRC risk include established carcinogens, such as heterocyclic amines and nitrosamines, as well as heme iron (6). The porphyrin structure of heme iron stimulates the production of pro-carcinogenic endogenous N-nitroso compounds as well as lipid oxidation products that can cause DNA damage (6,13,44). It is still unclear if the effects of red meat in colorectal carcinogenesis are only mediated through either of these compounds, or potential synergism between all of these. Given our observation of an effect modification of the association of red meat and CRC risk by genetic variants in SMAD7, we propose a mechanism that involves the SMAD7 protein via regulation of circulating levels of hepcidin, which is a liver-derived peptide that closely influences erythrocyte production and is the main molecule in charge of regulating systemic iron homeostasis (45). Systemically, hepcidin binds and degrades the membrane transporter ferroportin; whose function is to excrete the previously intracellular stored iron in enterocytes from a pool that contains both ferrous iron (Fe2+) from the heme pathway as well as non-heme, into the bloodstream (46). Alterations in hepcidin concentration that reduce circulating levels of hepcidin lead to increase in duodenal iron absorption and clinical iron overload, and the SMAD7 protein has been reported to function as a regulator of iron homoeostasis by reducing hepcidin expression (47). Knock-out mice of the hepcidin protein modulator were reported to have an increased risk of CRC (48). Hepcidin is canonically regulated via the bone morphogenetic protein 6 (BMP6)-SMAD1/5/8 pathway (49). However, murine models with iron overload have also proposed that the SMAD6 and SMAD7 proteins are co-regulated with hepcidin levels (50), and that suppression of SMAD7 can suppress hepcidin (50).
The SMAD7 rs35352860 SNP and several SNPs in LD align with open chromatin regions in the functional annotation plot due to the high density of mapped DNase type I cleavages (Supplemental Figure 5A). These regions correspond to DNase I hypersensitive sites, canonical epigenetic markers (H3K27ac & H3K4me) and cell lines evaluated in normal colon histological samples, tumor samples and CRC cell lines.
High accessibility to DNase Hypersensitive Sites increases the likelihood of transcriptional alterations due to cis-regulatory elements, namely: enhancers, promoters, silencers, insulators, and locus control regions. We hypothesize a mechanism in which over-expression of the SMAD7 molecule effectively inhibits hepcidin production. This in turn, may prevent hepcidin binding to ferroportin, thus preventing the internalization of the transmembrane transporter with increased output of iron through the basolateral membrane into the bloodstream. Free circulating iron can contribute to free radical formation, facilitating a pro-inflammatory, pro-carcinogenic state. Therefore, we speculate that among carriers of a SMAD7 variant that leads to overexpression of this protein, the effects of excess iron will be worsened by a diet with high consumption of red meat, due to an impaired inability to increase hepcidin production in response to heme iron intake levels.
The variant in the 8q24 region (rs4871179) resides downstream of the HAS2 gene (Supplementary Figure 4B). The functional annotation plot did not portray accessible chromatin regions (Supplementary Figure 5B). There is evidence that this gene may play a role in colorectal carcinogenesis. Specifically, the HAS2 gene is a member of the Glycosyltransferase family 2, which confers glycosylation profiles to molecules in the endoplasmic reticulum and Golgi apparatus. Alterations of these glycosylation profiles in cancer cells have been associated with carcinogenesis, and tumor progression in the past (51,52). The HAS2 gene, catalyzes the synthesis of Hyaluronan Acid (HA), one of the main extracellular matrix components, has been reported to be overexpressed in CRC (53) and has been associated with tumorigenesis and metastasis in breast cancer (54,55). Inhibition or reduced expression of HAS2 and/or HAS3 decreases metastatic colon carcinoma cell adhesion to laminin (55), and increased apoptosis and reduced metastasis in experimental models (54). Higher HA levels are associated with a worse prognosis in CRC patients, with an inverse correlation between percentage of HA-positive carcinoma cells and cancer-related survival rate and recurrence-free survival (56). There are no reports that link HAS2 to possible effects of red meat consumption, or report GxE interactions with red meat consumption and CRC risk.
Our study has several strengths, including our large sample size with uniformly harmonized data and systematic quality control across all pooled studies, which allowed for consideration of tumor anatomical localization, gender, and study design in the meta-analyses. Another key strength is the implementation of powerful approaches for GxE analyses, including the two-step methods that improve our ability to identify novel interactions that were not detected using the traditional 1-df GxE test. As well, we were able to consider previously reported confounders of both the exposure and the genotypes. Among the limitations of our study is the fact that consumption of red meat and processed meat was obtained via questionnaires, and that many of the included studies are case-control studies where the exposure was assessed after the cancer was diagnosed. However, case-control study participants were asked to report on intake typically 1 to 2 years before diagnosis/selection into the study. Therefore, we cannot exclude the possibility of misclassification and/or recall bias. For cohort studies, risk factors were assessed at the study-specific reference time, which aligns with the time of blood draw or buccal collection. A comprehensive description has been previously published (57). Moreover, four studies did not report total caloric intake which was set to zero in the analyses (i.e., ASTERISK, DACHS, PHS, UKB) (Supplementary Table 1). Nevertheless, sensitivity analyses confirmed that, since the interaction models included a fixed effect indicator for study, subjects in those four studies did not contribute information for estimating the energy effect. However, they did contribute to the estimation of all other effects including GxE interaction. Furthermore, confounding analyses revealed that GxE estimates for both our significant SNPs changed <0.04% on the Odds Ratio scale when an adjusted model was compared to an unadjusted model without total caloric intake. Additionally, study-specific quartiles were created to evaluate meat consumption which do not account for absolute differences in exposure variable. However, given differences in assessment tool, the use of study-specific quantiles is a valid commonly used approach in pooled analysis of nutritional exposures (58). As well, other behavioral patterns (e.g., exercise) were not considered as confounders in the analysis. Finally, we acknowledge that this study pooled data from most studies conducted among populations of European ancestry, and thus these findings may not apply to other racial and ethnic populations.
In summary, we report two novel GxE interactions for red meat consumption and colorectal cancer risk. Our strongest finding is in a SNP in the SMAD7 gene, which provides further supportive evidence for a role of heme iron in the carcinogenic pathway of red meat consumption and CRC development.
Supplementary Material
ACKNOWLEDGMENTS:
ASTERISK: We are very grateful to Dr. Bruno Buecher without whom this project would not have existed. We also thank all those who agreed to participate in this study, including the patients and the healthy control participants, as well as all the physicians, technicians, and students. CCFR: The Colon CFR graciously thanks the generous contributions of their study participants, dedication of study staff, and the financial support from the U.S. National Cancer Institute, without which this important registry would not exist. The authors would like to thank the study participants and staff of the Seattle Colon Cancer Family Registry and the Hormones and Colon Cancer study (CORE Studies). CLUE II: We thank the participants of Clue II and appreciate the continued efforts of the staff at the Johns Hopkins George W. Comstock Center for Public Health Research and Prevention in the conduct of the Clue II Cohort Study. Cancer data was provided by the Maryland Cancer Registry, Center for Cancer Prevention and Control, Maryland Department of Health, with funding from the State of Maryland and the Maryland Cigarette Restitution Fund. The collection and availability of cancer registry data is also supported by the Cooperative Agreement NU58DP006333, funded by the Centers for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the Department of Health and Human Services. CPS-II: The authors express sincere appreciation to all Cancer Prevention Study-II participants, and to each member of the study and biospecimen management group. The authors would like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention’s National Program of Cancer Registries and cancer registries supported by the National Cancer Institute’s Surveillance Epidemiology and End Results Program. The authors assume full responsibility for all analyses and interpretation of results. The views expressed here are those of the authors and do not necessarily represent the American Cancer Society or the American Cancer Society – Cancer Action Network. DACHS: We thank all participants and cooperating clinicians, and everyone who provided excellent technical assistance. EDRN: We acknowledge all contributors to the development of the resource at University of Pittsburgh School of Medicine, Department of Gastroenterology, Department of Pathology, Hepatology and Nutrition and Biomedical Informatics. EPIC: Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization. Harvard cohorts: The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We acknowledge Channing Division of Network Medicine, Department of Medicine, Brigham, and Women’s Hospital as home of the NHS. The authors would like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention’s National Program of Cancer Registries (NPCR) and/or the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) Program. Central registries may also be supported by state agencies, universities, and cancer centers. Participating central cancer registries include the following: Alabama, Alaska, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Hawaii, Idaho, Indiana, Iowa, Kentucky, Louisiana, Massachusetts, Maine, Maryland, Michigan, Mississippi, Montana, Nebraska, Nevada, New Hampshire, New Jersey, New Mexico, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Puerto Rico, Rhode Island, Seattle SEER Registry, South Carolina, Tennessee, Texas, Utah, Virginia, West Virginia, Wyoming. The authors assume full responsibility for analyses and interpretation of these data. Kentucky: We would like to acknowledge the staff at the Kentucky Cancer Registry.
LCCS: We acknowledge the contributions of Jennifer Barrett, Robin Waxman, Gillian Smith and Emma Northwood in conducting this study. NCCCS I & II: We would like to thank the study participants, and the NC Colorectal Cancer Study staff. PLCO: The authors thank the PLCO Cancer Screening Trial screening center investigators and the staff from Information Management Services Inc and Westat Inc. Most importantly, we thank the study participants for their contributions that made this study possible. Cancer incidence data have been provided by the District of Columbia Cancer Registry, Georgia Cancer Registry, Hawaii Cancer Registry, Minnesota Cancer Surveillance System, Missouri Cancer Registry, Nevada Central Cancer Registry, Pennsylvania Cancer Registry, Texas Cancer Registry, Virginia Cancer Registry, and Wisconsin Cancer Reporting System. All are supported in part by funds from the Center for Disease Control and Prevention, National Program for Central Registries, local states or by the National Cancer Institute, Surveillance, Epidemiology, and End Results program. The results reported here, and the conclusions derived are the sole responsibility of the authors. SELECT: We thank the research and clinical staff at the sites that participated on SELECT study, without whom the trial would not have been successful. We are also grateful to the 35,533 dedicated men who participated in SELECT. WHI: The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf
FUNDING:
MCS and JSM received support from awards U54CA233465 and U2CCA252971 from the National Cancer Institute. Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO): National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088, R01 CA059045, U01 CA164930, R01 CA201407). This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA015704 and P30CA014089. Scientific Computing Infrastructure at Fred Hutch funded by ORIP grant S10OD028685. ASTERISK: a Hospital Clinical Research Program (PHRC-BRD09/C) from the University Hospital Center of Nantes (CHU de Nantes) and supported by the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC), the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer (LRCC). The ATBC Study is supported by the Intramural Research Program of the U.S. National Cancer Institute, National Institutes of Health, Department of Health and Human Services. CLUE II funding was from the National Cancer Institute (U01 CA086308, Early Detection Research Network; P30 CA006973), National Institute on Aging (U01 AG018033), and the American Institute for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government. Maryland Cancer Registry (MCR) Cancer data was provided by the Maryland Cancer Registry, Center for Cancer Prevention and Control, Maryland Department of Health, with funding from the State of Maryland and the Maryland Cigarette Restitution Fund. The collection and availability of cancer registry data is also supported by the Cooperative Agreement NU58DP006333, funded by the Centers for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the Department of Health and Human Services. The Colon Cancer Family Registry (CCFR, www.coloncfr.org) is supported in part by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (award U01 CA167551). Support for case ascertainment was provided in part from the Surveillance, Epidemiology, and End Results (SEER) Program and the following U.S. state cancer registries: AZ, CO, MN, NC, NH; and by the Victoria Cancer Registry (Australia) and Ontario Cancer Registry (Canada). The CCFR Set-1 (Illumina 1M/1M-Duo) and Set-2 (Illumina Omni1-Quad) scans were supported by NIH awards U01 CA122839 and R01 CA143237 (to GC). The CCFR Set-3 (Affymetrix Axiom CORECT Set array) was supported by NIH award U19 CA148107 and R01 CA81488 (to SBG). The CCFR Set-4 (Illumina OncoArray 600K SNP array) was supported by NIH award U19 CA148107 (to SBG) and by the Center for Inherited Disease Research (CIDR), which is funded by the NIH to the Johns Hopkins University, contract number HHSN268201200008I. The content of this manuscript does not necessarily reflect the views or policies of the NCI, NIH or any of the collaborating centers in the Colon Cancer Family Registry (CCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, any cancer registry, or the CCFR. COLO2&3: National Institutes of Health (R01 CA060987). CPS-II: The American Cancer Society funds the creation, maintenance, and updating of the Cancer Prevention Study-II (CPS-II) cohort. The study protocol was approved by the institutional review boards of Emory University, and those of participating registries as required. CRCGEN: Colorectal Cancer Genetics & Genomics, Spanish study was supported by Instituto de Salud Carlos III, co-funded by FEDER funds –a way to build Europe– (grants PI14-613 and PI09-1286), Agency for Management of University and Research Grants (AGAUR) of the Catalan Government (grant 2017SGR723), Junta de Castilla y León (grant LE22A10-2), the Spanish Association Against Cancer (AECC) Scientific Foundation grant GCTRA18022MORE and the Consortium for Biomedical Research in Epidemiology and Public Health (CIBERESP), action Genrisk. Sample collection of this work was supported by the Xarxa de Bancs de Tumors de Catalunya sponsored by Pla Director d’Oncología de Catalunya (XBTC), Plataforma Biobancos PT13/0010/0013 and ICOBIOBANC, sponsored by the Catalan Institute of Oncology. We thank CERCA Programme, Generalitat de Catalunya for institutional support. DACHS: This work was supported by the German Research Council (BR 1704/6-1, BR 1704/6-3, BR 1704/6-4, CH 117/1-1, HO 5117/2-1, HE 5998/2-1, KL 2354/3-1, RO 2270/8-1 and BR 1704/17-1), the Interdisciplinary Research Program of the National Center for Tumor Diseases (NCT), Germany, and the German Federal Ministry of Education and Research (01KH0404, 01ER0814, 01ER0815, 01ER1505A and 01ER1505B). DALS: National Institutes of Health (R01 CA048998 to M. L. Slattery). EPIC: The coordination of EPIC is financially supported by International Agency for Research on Cancer (IARC) and also by the Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London which has additional infrastructure support provided by the NIHR Imperial Biomedical Research Centre (BRC). The national cohorts are supported by: Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), German Institute of Human Nutrition Potsdam- Rehbruecke (DIfE), Federal Ministry of Education and Research (BMBF) (Germany); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy, Compagnia di SanPaolo and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS) - Instituto de Salud Carlos III (ISCIII), Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, and the Catalan Institute of Oncology - ICO (Spain); Swedish Cancer Society, Swedish Research Council and and Region Skåne and Region Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C8221/A29017 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk; MR/M012190/1 to EPIC-Oxford). (United Kingdom). Harvard cohorts: HPFS is supported by the National Institutes of Health (P01 CA055075, UM1 CA167552, U01 CA167552, R01 CA137178, R01 CA151993, and R35 CA197735), NHS by the National Institutes of Health (P01 CA087969, UM1 CA186107, R01 CA137178, R01 CA151993, and R35 CA197735), and PHS by the National Institutes of Health (R01 CA042182).
Kentucky: This work was supported by the following grant support: Clinical Investigator Award from Damon Runyon Cancer Research Foundation (CI-8); NCI R01CA136726. LCCS: The Leeds Colorectal Cancer Study was funded by the Food Standards Agency and Cancer Research UK Programme Award (C588/A19167). MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 509348, 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database. BMLynch was supported by MCRF18005 from the Victorian Cancer Agency. MEC: National Institutes of Health (R37 CA054281, P01 CA033619, and R01 CA063464). MECC: This work was supported by the National Institutes of Health, U.S. Department of Health and Human Services (R01 CA081488, R01 CA197350, U19 CA148107, R01 CA242218, and a generous gift from Daniel and Maryann Fong. NCCCS I & II: We acknowledge funding support for this project from the National Institutes of Health, R01 CA066635 and P30 DK034987. PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS. Funding was provided by National Institutes of Health (NIH), Genes, Environment and Health Initiative (GEI) Z01 CP 010200, NIH U01 HG004446, and NIH GEI U01 HG 004438. SELECT: Research reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under Award Numbers U10 CA037429 (CD Blanke), and UM1 CA182883 (CM Tangen/IM Thompson). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Swedish Mammography Cohort and Cohort of Swedish Men: This work is supported by the Swedish Research Council /Infrastructure grant, the Swedish Cancer Foundation, and the Karolinska Institutés Distinguished Professor Award to Alicja Wolk. VITAL: National Institutes of Health (K05 CA154337). WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: https://www-whi-org.s3.us-west-2.amazonaws.com/wp-content/uploads/WHI-Investigator-Long-List.pdf
Footnotes
Conflicts of Interest:
C.M.U. has as cancer center director oversight over research funded by several pharmaceutical companies but has not received funding directly herself.
REFERENCES
- 1.Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, et al. 2020. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Accessed 2022. [Google Scholar]
- 2.Erdrich J, Zhang X, Giovannucci E, Willett W. Proportion of colon cancer attributable to lifestyle in a cohort of US women. Cancer Causes & Control 2015;26(9):1271–9 doi 10.1007/s10552-015-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Platz EA, Willett WC, Colditz GA, Rimm EB, Spiegelman D, Giovannucci E. Proportion of colon cancer risk that might be preventable in a cohort of middle-aged US men. Cancer Causes Control 2000;11(7):579–88. [DOI] [PubMed] [Google Scholar]
- 4.Islami F, Goding-Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA: A Cancer Journal for Clinicians 2018;68(1):31–54 doi 10.3322/caac.21440. [DOI] [PubMed] [Google Scholar]
- 5.Clinton SK, Giovannucci EL, Hursting SD. The World Cancer Research Fund/American Institute for Cancer Research Third Expert Report on Diet, Nutrition, Physical Activity, and Cancer: Impact and Future Directions. J Nutr 2020;150(4):663–71 doi 10.1093/jn/nxz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvard V, Loomis D, Guyton KZ, Grosse Y, Ghissassi FE, Benbrahim-Tallaa L, et al. Carcinogenicity of consumption of red and processed meat. The Lancet Oncology 2015;16(16):1599–600 doi 10.1016/s1470-2045(15)00444-1. [DOI] [PubMed] [Google Scholar]
- 7.Petimar J, Smith-Warner SA, Rosner B, Chan AT, Giovannucci EL, Tabung FK. Adherence to the World Cancer Research Fund/American Institute for Cancer Research 2018 Recommendations for Cancer Prevention and Risk of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev 2019;28(9):1469–79 doi 10.1158/1055-9965.EPI-19-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Red Meat and Processed Meat. Lyon (FR): International Agency for Research on Cancer; 2018. (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, No. 114.) 2. CANCER IN HUMANS. Available from: https://www.ncbi.nlm.nih.gov/books/NBK507972/. 2018. [Google Scholar]
- 9.Bingham SA, Pignatelli B, Pollock JR, Ellul A, Malaveille C, Gross G, et al. Does increased endogenous formation of N-nitroso compounds in the human colon explain the association between red meat and colon cancer? Carcinogenesis 1996;17(3):515–23. [DOI] [PubMed] [Google Scholar]
- 10.Gurjao C, Zhong R, Haruki K, Li YY, Spurr LF, Lee-Six H, et al. Discovery and Features of an Alkylating Signature in Colorectal Cancer. Cancer Discov 2021;11(10):2446–55 doi 10.1158/2159-8290.CD-20-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tappel A Heme of consumed red meat can act as a catalyst of oxidative damage and could initiate colon, breast and prostate cancers, heart disease and other diseases. Medical hypotheses 2007;68(3):562–4. [DOI] [PubMed] [Google Scholar]
- 12.Cross AJ, Pollock JR, Bingham SA. Haem, not protein or inorganic iron, is responsible for endogenous intestinal N-nitrosation arising from red meat. Cancer Res 2003;63(10):2358–60. [PubMed] [Google Scholar]
- 13.Bastide NM, Pierre FH, Corpet DE. Heme iron from meat and risk of colorectal cancer: a meta-analysis and a review of the mechanisms involved. Cancer Prev Res (Phila) 2011;4(2):177–84 doi 10.1158/1940-6207.CAPR-10-0113. [DOI] [PubMed] [Google Scholar]
- 14.Seiwert N, Adam J, Steinberg P, Wirtz S, Schwerdtle T, Adams-Quack P, et al. Chronic intestinal inflammation drives colorectal tumor formation triggered by dietary heme iron in vivo. Arch Toxicol 2021;95(7):2507–22 doi 10.1007/s00204-021-03064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugimura T Carcinogenicity of mutagenic heterocyclic amines formed during the cooking process. Mutat Res 1985;150(1–2):33–41. [DOI] [PubMed] [Google Scholar]
- 16.Larsson BK. Formation of polycyclic aromatic hydrocarbons during the smoking and grilling of food. Progress in clinical and biological research 1986;206:169–80. [PubMed] [Google Scholar]
- 17.Sinha R, Rothman N, Brown ED, Salmon CP, Knize MG, Swanson CA, et al. High concentrations of the carcinogen 2-amino-1-methyl-6-phenylimidazo- [4,5-b]pyridine (PhIP) occur in chicken but are dependent on the cooking method. Cancer research 1995;55(20):4516–9. [PubMed] [Google Scholar]
- 18.Sinha R, Rothman N. Exposure assessment of heterocyclic amines (HCAs) in epidemiologic studies. Mutat Res 1997;376(1–2):195–202. [DOI] [PubMed] [Google Scholar]
- 19.Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet 2019;51(1):76–87 doi 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmit SL, Edlund CK, Schumacher FR, Gong J, Harrison TA, Huyghe JR, et al. Novel Common Genetic Susceptibility Loci for Colorectal Cancer. J Natl Cancer Inst 2018. doi 10.1093/jnci/djy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Rozadilla C, Timofeeva M, Chen Z, Law P, Thomas M, Schmit S, et al. Deciphering colorectal cancer genetics through multi-omic analysis of 100,204 cases and 154,587 controls of European and east Asian ancestries. Nat Genet 2022. doi 10.1038/s41588-022-01222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genin E Missing heritability of complex diseases: case solved? Hum Genet 2020;139(1):103–13 doi 10.1007/s00439-019-02034-4. [DOI] [PubMed] [Google Scholar]
- 23.Yang T, Li X, Montazeri Z, Little J, Farrington SM, Ioannidis JPA, et al. Gene-environment interactions and colorectal cancer risk: An umbrella review of systematic reviews and meta-analyses of observational studies. Int J Cancer 2019;145(9):2315–29 doi 10.1002/ijc.32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueiredo JC, Hsu L, Hutter CM, Lin Y, Campbell PT, Baron JA, et al. Genome-wide diet-gene interaction analyses for risk of colorectal cancer. PLoS genetics 2014;10(4):e1004228 doi 10.1371/journal.pgen.1004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueiredo JC, Lewinger JP, Song C, Campbell PT, Conti DV, Edlund CK, et al. Genotype-Environment Interactions in Microsatellite Stable/Microsatellite Instability-Low Colorectal Cancer: Results from a Genome-Wide Association Study. Cancer Epidemiology Biomarkers & Prevention 2011;20(5):758–66 doi 10.1158/1055-9965.EPI-10-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 2013;144(4):799–807 e24 doi 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutter CM, Chang-Claude J, Slattery ML, Pflugeisen BM, Lin Y, Duggan D, et al. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer research 2012. doi 10.1158/0008-5472.CAN-11-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doak CM, Wijnhoven TM, Schokker DF, Visscher TL, Seidell JC. Age standardization in mapping adult overweight and obesity trends in the WHO European Region. Obes Rev 2012;13(2):174–91 doi 10.1111/j.1467-789X.2011.00943.x. [DOI] [PubMed] [Google Scholar]
- 29.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48(10):1279–83 doi 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48(10):1284–7 doi 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327(7414):557–60 doi 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beath K Metaplus: An R package for the analysis of robust meta-analysis and meta-regression. . R J 2016;8:5 doi 10.32614/rj-2016-001. [DOI] [Google Scholar]
- 33.Gauderman WJ, Zhang P, Morrison JL, Lewinger JP. Finding novel genes by testing G x E interactions in a genome-wide association study. Genetic epidemiology 2013;37(6):603–13 doi 10.1002/gepi.21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gauderman WJ, Kim A, Conti DV, Morrison J, Thomas DC, Vora H, et al. A Unified Model for the Analysis of Gene-Environment Interaction. Am J Epidemiol 2019;188(4):760–7 doi 10.1093/aje/kwy278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Troncone E, Marafini I, Stolfi C, Monteleone G. Involvement of Smad7 in Inflammatory Diseases of the Gut and Colon Cancer. International Journal of Molecular Sciences 2021;22(8):3922 doi 10.3390/ijms22083922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y, Wu W, Nie M, Li C, Wang L. SMAD7 polymorphisms and colorectal cancer risk: a meta-analysis of case-control studies. Oncotarget 2016;7(46):75561–70 doi 10.18632/oncotarget.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez-Rozadilla C, Timofeeva M, Chen Z, Law P, Thomas M, Schmit S, et al. Deciphering colorectal cancer genetics through multi-omic analysis of 100,204 cases and 154,587 controls of European and east Asian ancestries. Nat Genet 2023;55(1):89–99 doi 10.1038/s41588-022-01222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao Q, Chen J, Zhu J, Zeng S, Cai H, Zhu G. Association of several loci of SMAD7 with colorectal cancer: A meta-analysis based on case-control studies. Medicine (Baltimore) 2023;102(1):e32631 doi 10.1097/MD.0000000000032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet 2007;39(11):1315–7 doi 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 40.Campbell PT, Lin Y, Bien SA, Figueiredo JC, Harrison TA, Guinter MA, et al. Association of Body Mass Index With Colorectal Cancer Risk by Genome-Wide Variants. J Natl Cancer Inst 2021;113(1):38–47 doi 10.1093/jnci/djaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figueiredo JC, Lewinger JP, Song C, Campbell PT, Conti DV, Edlund CK, et al. Genotype-environment interactions in microsatellite stable/microsatellite instability-low colorectal cancer: results from a genome-wide association study. Cancer Epidemiol Biomarkers Prev 2011;20(5):758–66 doi 10.1158/1055-9965.EPI-10-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulay JL, Mild G, Lowy A, Reuter J, Lagrange M, Terracciano L, et al. SMAD7 is a prognostic marker in patients with colorectal cancer. Int J Cancer 2003;104(4):446–9 doi 10.1002/ijc.10908. [DOI] [PubMed] [Google Scholar]
- 43.Hu Y, He J, He L, Xu B, Wang Q. Expression and function of Smad7 in autoimmune and inflammatory diseases. J Mol Med (Berl) 2021;99(9):1209–20 doi 10.1007/s00109-021-02083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chua AC, Klopcic B, Lawrance IC, Olynyk JK, Trinder D. Iron: an emerging factor in colorectal carcinogenesis. World J Gastroenterol 2010;16(6):663–72 doi 10.3748/wjg.v16.i6.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagani A, Nai A, Silvestri L, Camaschella C. Hepcidin and Anemia: A Tight Relationship. Front Physiol 2019;10:1294 doi 10.3389/fphys.2019.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp P, Srai SK. Molecular mechanisms involved in intestinal iron absorption. World J Gastroenterol 2007;13(35):4716–24 doi 10.3748/wjg.v13.i35.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An P, Wang H, Wu Q, Wang J, Xia Z, He X, et al. Smad7 deficiency decreases iron and haemoglobin through hepcidin up-regulation by multilayer compensatory mechanisms. J Cell Mol Med 2018;22(6):3035–44 doi 10.1111/jcmm.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sivaprakasam S, Ristic B, Mudaliar N, Hamood AN, Colmer-Hamood J, Wachtel MS, et al. Hereditary hemochromatosis promotes colitis and colon cancer and causes bacterial dysbiosis in mice. Biochem J 2020;477(19):3867–83 doi 10.1042/BCJ20200392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang CY, Babitt JL. Hepcidin regulation in the anemia of inflammation. Curr Opin Hematol 2016;23(3):189–97 doi 10.1097/MOH.0000000000000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vujic Spasic M, Sparla R, Mleczko-Sanecka K, Migas MC, Breitkopf-Heinlein K, Dooley S, et al. Smad6 and Smad7 are co-regulated with hepcidin in mouse models of iron overload. Biochim Biophys Acta 2013;1832(1):76–84 doi 10.1016/j.bbadis.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015;15(9):540–55 doi 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X Alterations of Golgi Structural Proteins and Glycosylation Defects in Cancer. Front Cell Dev Biol 2021;9:665289 doi 10.3389/fcell.2021.665289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim YS, Ahn YH, Song KJ, Kang JG, Lee JH, Jeon SK, et al. Overexpression and beta-1,6-N-acetylglucosaminylation-initiated aberrant glycosylation of TIMP-1: a “double whammy” strategy in colon cancer progression. J Biol Chem 2012;287(39):32467–78 doi 10.1074/jbc.M112.370064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YH, Lee SB, Shim S, Kim A, Park JH, Jang WS, et al. Hyaluronic acid synthase 2 promotes malignant phenotypes of colorectal cancer cells through transforming growth factor beta signaling. Cancer Sci 2019;110(7):2226–36 doi 10.1111/cas.14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lien HC, Lee YH, Jeng YM, Lin CH, Lu YS, Yao YT. Differential expression of hyaluronan synthase 2 in breast carcinoma and its biological significance. Histopathology 2014;65(3):328–39 doi 10.1111/his.12390. [DOI] [PubMed] [Google Scholar]
- 56.Ropponen K, Tammi M, Parkkinen J, Eskelinen M, Tammi R, Lipponen P, et al. Tumor cell-associated hyaluronan as an unfavorable prognostic factor in colorectal cancer. Cancer Res 1998;58(2):342–7. [PubMed] [Google Scholar]
- 57.Gong J, Hutter CM, Newcomb PA, Ulrich CM, Bien SA, Campbell PT, et al. Genome-Wide Interaction Analyses between Genetic Variants and Alcohol Consumption and Smoking for Risk of Colorectal Cancer. PLoS genetics 2016;12(10):e1006296 doi 10.1371/journal.pgen.1006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim DH, Smith-Warner SA, Spiegelman D, Yaun SS, Colditz GA, Freudenheim JL, et al. Pooled analyses of 13 prospective cohort studies on folate intake and colon cancer. Cancer Causes Control 2010;21(11):1919–30 doi 10.1007/s10552-010-9620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawaguchi ES, Kim AE, Lewinger JP, Gauderman WJ. Improved two-step testing of genome-wide gene-environment interactions. Genetic epidemiology 2022. doi 10.1002/gepi.22509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.