Abstract
Macrophages may acquire a reparative phenotype that supports tissue repair and remodeling in response to tissue injury. However, the metabolic requirements underpinning this process are incompletely understood. Here, we show that posttranslational modification (PTM) of PPARγ regulates lipid synthesis in response to wound microenvironmental cues and that metabolic rewiring orchestrates function of reparative macrophages. In injured tissues, repair signaling leads to decreased macrophage PPARγ threonine 166 (T166) phosphorylation, which results in a partially active PPARγ transcriptional program comprised of increased binding activity to the regulator regions of lipid synthesis-associated genes, thereby increased lipogenesis. The accumulated lipids serve as signaling molecules, triggering STAT3-mediated growth factor expression, and supporting the synthesis of phospholipids for the expansion of the endoplasmic reticulum (ER), which is required for protein secretion. Genetic or pharmacological inhibition of PPARγ T166 phosphorylation promotes the reparative function of macrophages and facilitates tissue regeneration. In summary, our work identifies PPARγ T166-regulated lipid biosynthesis as an essential pathway for meeting the anabolic demands of the activation and function of macrophages and provides a rationale for potential therapeutic targeting of tissue repair.
Subject terms: Monocytes and macrophages, Phosphorylation, Lipid signalling, Skin diseases
Macrophages with a reparative phenotype are important for tissue repair and have distinctive metabolic features. Here authors show that metabolic rewiring of macrophages during wound healing involves dephosphorylation of the transcription factor PPARγ, which results in activation of target genes that regulate lipid biosynthesis.
Introduction
The restoration of tissue integrity and homeostasis after injury is a complex biological process that proceeds in the following sequential phases: inflammation, tissue formation and maturation1. Among all cell types, macrophages play critical roles in supporting each phase of wound repair by rapidly adopting diverse activation states in response to local tissue microenvironment cues2,3. When damage occurs, macrophages polarize to a classically activated phenotype and exhibit proinflammatory functions such as phagocytosis, antigen presentation, and proinflammatory cytokine production4. When the wound-healing response is well organized and controlled, macrophages shift toward a resolution phenotype to inhibit inflammation by releasing anti-inflammatory cytokines2,5. Although early studies investigating the contribution of macrophages to wound repair focused on their role as immune cells, it is now clear that macrophages exhibit much more complex roles not only in inflammation control but also in the mechanisms of tissue regeneration6–8. Macrophages are an important source of growth factors, including platelet-derived growth factor (PDGF), transforming growth factor-β1 (TGF-β1), and vascular endothelial growth factor (VEGF), which induce re-epithelialization and neovascularization by promoting cellular proliferation and blood vessel development4,9. Macrophage depletion leads to less efficient repair and regeneration following injury10. However, the precise molecular mechanisms that drive and sustain the macrophage reparative phenotype remain poorly understood.
There is a growing understanding that cellular metabolic profiles are associated with the balance of macrophage activation and function11. Macrophages with different activation states exhibit distinct metabolic programs in response to activation to meet their bioenergetic needs11,12. In addition, metabolic intermediates can function as signaling molecules to facilitate the polarization of macrophages13,14. For instance, lipopolysaccharide (LPS) strongly induces glycolysis in classically activated macrophages to meet energy demands12,15. Moreover, this process increases succinate levels, which block hypoxia-inducible factor-1α (HIF-1α) degradation to sustain the proinflammatory phenotype16,17. However, the metabolic requirements of macrophages in the repair phase remain unclear. Studies of metabolic programs fueling macrophage activation have largely focused on catabolic pathways, such as glycolysis and oxidative phosphorylation (OXPHOS)18, while anabolic processes, including the synthesis of lipids, amino acids, and nucleotides, are also essential for proliferation and cytokine production in immune cells19–21. The production and secretion of repair factors are crucial for the function of reparative macrophages. It is imperative to further investigate the anabolic processes of macrophages.
To determine the functional interplay between cellular metabolism and specific repair effector functions in injury-associated macrophages, we conduct a comprehensive analysis of these macrophages at the transcriptional and functional levels. The single-cell transcriptome atlas reveals that fatty acid synthesis (FAS) activities are consistently enhanced following stimulation with healing signals in 9 tissue injury models. This metabolic reprogramming is precisely regulated by PPARγ T166 (corresponding to the T136 site in PPARγ1) phosphorylation. Intracellular fatty acids induce STAT3-mediated growth factor expression and support the expansion of the ER, which is required for the secretion of reparative proteins. Genetic and pharmacological inhibition of PPARγ T166 phosphorylation markedly improve tissue repair in vivo and the migration of primary dermal fibroblasts (DF) in vitro. Collectively, our results identify a function and mechanism of PPARγ in reparative macrophages and open avenues for therapeutically modulating macrophage polarization to promote wound repair.
Results
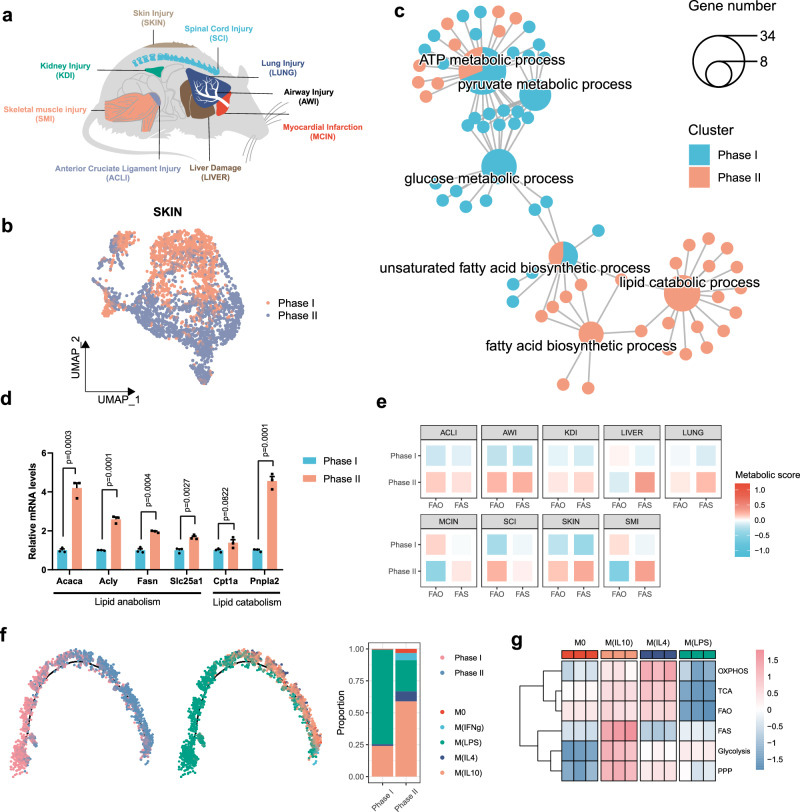
The increase in lipid anabolic metabolism is a feature of reparative macrophages during wound healing
To explore the metabolic pathways that are used by reparative macrophages, we obtained single-cell RNA sequencing (scRNA-seq) data on monocyte-macrophage lineage cells from mouse wound healing models in 9 tissue types, including airway, joint, heart, kidney, liver, lung, skeletal muscle, skin, and spinal cord (Fig. 1a). After strict quality control and filtration, we collected a total of 59332 macrophages derived from the inflammatory phase (phase I) and reparative phase (phase II) (Supplementary Fig. 1a, b). Considering the heterogeneity of macrophages and batch effects among different datasets, we analyzed each dataset independently. Using skin injury as an example, we found that inflammatory phase and reparative phase macrophages clustered predominantly in disparate populations, as shown by uniform manifold approximation and projection (UMAP) (Fig. 1b). Gene ontology (GO) enrichment analysis showed that inflammatory phase macrophages had significantly upregulated genes associated with glycolytic processes, while reparative phase macrophages highly expressed genes associated with lipid metabolic processes (Fig. 1c). To verify this metabolic difference, we sorted wound macrophages in the inflammatory and reparative phases in a full-thickness excisional skin injury model. As expected, we found significantly elevated expression of genes associated with lipid metabolism in skin macrophages in the reparative phase (Fig. 1d). Intriguingly, we observed consistent upregulation of FAS activity in reparative phase macrophages in all injury models, while the other types of metabolism exhibited various regulation patterns in phase II macrophages (Fig. 1e and Supplementary Fig. 1c). The differences in metabolic features indicate the flexibility of macrophages in their energy usage to meet the bioenergetic demands of cellular activation, but FAS is a more conserved metabolic pathway for the activation of reparative macrophages.
Fig. 1. Fatty acid synthesis is a feature of reparative macrophages.
a Tissue injury models included in the analysis. b The scRNA-seq analysis of skin wound macrophages of phase I (1 dpi) and phase II (6 dpi). UMAP highlights wound healing phases. c GO enrichment analysis showing the highly enriched metabolic pathway associated with phase I and phase II. Pie charts represent enriched biological processes and the ratio of gene number that enriched in each group, small dots represent genes associated with the processes. The size of each pie indicates the number of genes enriched in a specific term. d The mRNA expression of lipid metabolism-associated genes in sorted wound macrophages isolated at 1 and 6 dpi (n = 3 mice per group). e Heatmaps showing ssGSEA scores of phase I and phase II macrophages using signature genes associated with FAO (mmu00071) and FAS (mmu00061) pathways. f Developmental trajectory of skin wound macrophages. Sample are colored by wound healing phases or macrophage subtypes. g Heatmap showing expression patterns of glycolysis-, tricarboxylic acid cycle (TCA)-, OXPHOS-, pentose phosphate pathway (PPP)-, FAO-, and FAS-associated signature genes among macrophage subtypes from GSE131364 dataset. Unless specified otherwise, the data are presented as means ± s.e.m. (error bar) and compared using the two-tailed Student’s t test. Source data are provided as a Source Data file.
Considering the strong influences of the tissue microenvironment, we further confirmed lipid synthesis activity in reparative macrophages in vitro. Polarized macrophages in vitro are instrumental in analyzing macrophage function, and we tested polarizing conditions to generate reparative macrophages. We used a skin injury model and found a significant increase in the secretion of IL-10 during the tissue repair phase (Supplementary Fig. 2a). SingleR revealed that 58.9% of macrophages in the reparative phase exhibited an IL-10-induced phenotype (Fig. 1f). IL-10-treated bone marrow-derived macrophages (BMDM) showed upregulated expression of tissue repair-related genes (Supplementary Fig. 2b), and significantly promoted the migration of skin DFs (Supplementary Fig. 2c). These results suggest that IL-10 polarizes macrophages toward a typical reparative phenotype.
We retrieved public transcriptomes of IL-10-induced macrophages M(IL10) and showed that M(IL10) exhibited a distinct metabolic pattern compared to M0 (undifferentiated), M(LPS) (classically activated), or M(IL4) (alternatively activated) macrophages (Fig. 1g). Interestingly, M(IL10) and M(IL4) exhibited increased lipid metabolic activity, while M(IL4) showed higher expression levels of fatty acid oxidation (FAO)-related genes, and M(IL10) macrophages had strongly upregulated FAS genes (Fig. 1g). We also observed that IL-10 enhanced FAS-related gene expression in human macrophages (Supplementary Fig. 1d, e). In addition, previously reported reparative macrophage induction strategies, that used dexamethasone or TGF-β1, induced highly expressions of lipid metabolism-associated genes, especially FAS-related genes in macrophages (Supplementary Fig. 1f). Collectively, our data revealed that reparative macrophages possess a distinct metabolic program characterized by a significant increase in lipid synthesis-related genes.
Lipid synthesis is required for the activation and function of reparative macrophages
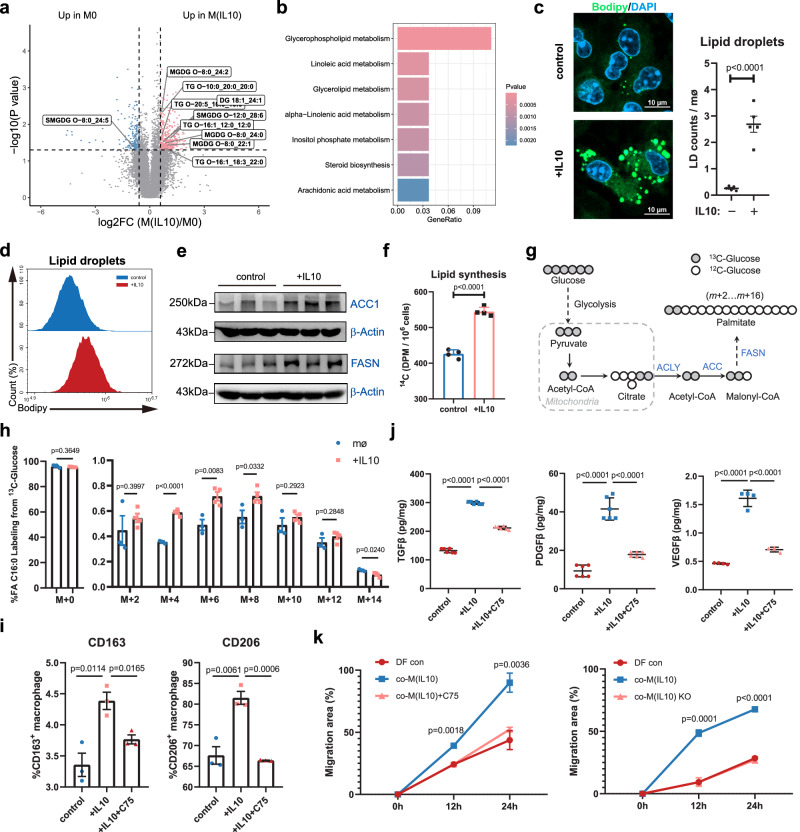
To investigate the lipid metabolic impact of reparative signaling in macrophages, we performed lipidomic analysis (Supplementary Fig. 3a). We observed that glycerolipids were highly accumulated in M(IL10) (Fig. 2a). Pathway enrichment analysis validated the alteration of glycerolipid metabolism in IL-10-induced macrophages (Fig. 2b). Consistently, M(IL10) exhibited markedly increased staining with neutral lipid dye (BODIPY 493/503), which is indicative of increased lipid accumulation (Fig. 2c, d). Similarly, reparative phase (6 days post injury, 6 dpi) wound macrophages accumulated more lipids than macrophages isolated from the early phase (1 dpi) of skin injury (Supplementary Fig. 3b). Stimulating macrophages with IL-10 increased intracellular triglycerides (TG) and nonesterified fatty acids (NEFA) (Supplementary Fig. 3c). These results indicate that reparative signaling induces lipid accumulation in macrophages.
Fig. 2. Limiting de novo lipid synthesis reduces reparative function of macrophages.
a Volcano plot showing changes of lipid metabolites in BMDMs with or without IL-10 treatment (n = 4 per group). b GO enrichment analysis showing the pathway alterations in BMDMs with or without IL-10 treatment. c Representative images of BODIPY staining in control or IL-10-treated BMDMs (n = 5 biological replicates). Scale bar, 10 μm. d BODIPY staining analysis by flow cytometry in control or IL-10-treated macrophages. e Protein expression of ACC1 and FASN in BMDMs in response to IL-10. β-Actin was used as loading control. f Lipid synthesis rate in control and IL-10 BMDMs. The data represent the incorporation of 14C-glucose in the lipid fraction of n = 4 biological replicates per group. g Diagram of de novo synthesis of fatty acids from [U-13C] glucose. h The percentages of isotopomers of FA 16:0 (palmitate) after trace to [U-13C] glucose in BMDMs in the presence or absence of IL-10. n = 3 (control) or n = 4 (IL-10) biological replicates. i Flow cytometry assay of CD163 and CD206 on BMDMs with IL-10 or C75 treatment (n = 3 biological replicates). j TGF-β1, PDGFβ and VEGFβ in cell culture medium is determined by ELISA upon stimulation with IL-10 or C75. n = 6 (TGF-β1 and PDGFβ) or n = 4 (VEGFβ) independent experiments. k The scratch wound closure rate of DFs co-cultured with M(IL10) or treatment with C75 or M(IL10) knockout (KO) Fsan for 24 h (n = 3 biological replicates). Data were analyzed by one-way analysis of variance (ANOVA) followed by a Dunnett’s multiple-comparisons test. Unless specified otherwise, the data are presented as means ± s.e.m. (error bar) and compared using the two-tailed Student’s t test. Source data are provided as a Source Data file.
In line with an accumulation of lipidome, the protein expression of metabolic enzymes involved in FAS (ACC1 and FASN) was also strongly induced in M(IL10) and reparative phase macrophages of skin injury (Fig. 2e and Supplementary Fig. 3d). Considering that glucose is the major substrate for FAS, we traced the fate of glucose in reparative macrophage using isotope labeling. Using C14-labeled glucose, we observed that IL-10-stimulated macrophages substantially increased C14-glucose incorporation into lipid components (Fig. 2f). We further observed that M(IL10) cells had significantly increased palmitic acid and stearic acid labeling from [U-13C] glucose tracers (Fig. 2g, h and Supplementary Fig. 3e). These data confirm that the accumulated lipids in reparative macrophages are derived from de novo lipid synthesis pathway.
To further investigate the lipid synthesis activity of reparative macrophages in wound healing processes, we generated cardiotoxin (CTX)-induced skeletal muscle injury and CCl4-induced acute liver injury models (Supplementary Fig. 4a–d). Consistent with the findings observed in skin injury models, reparative macrophages from both muscle and liver injury models exhibited enhanced lipid accumulation and upregulated expression of lipid synthesis-related genes (Supplementary Fig. 4e–h). Together, these results demonstrate that increased de novo lipid synthesis activity is a feature of reparative macrophages.
We then examined whether perturbations in the de novo lipid synthesis in macrophages impacted reparative functions. C75, a competitive fatty acid synthase (FASN) inhibitor, significantly inhibited IL-10-induced CD163 and CD206 expression (Fig. 2i), and impaired the secretion of reparative cytokines by macrophages (Fig. 2j and Supplementary Fig. 5a). Knockout of FASN encoding gene Fasn also significantly inhibited the reparative phenotype of M(IL10) (Supplementary Fig. 5b, c). Importantly, both FASN inhibition and Fasn knockout in M(IL10) decreased dermal fibroblasts migration in scratch tests (Fig. 2k and Supplementary Fig. 5d, e). Collectively, these results confirm the strong upregulation of lipid synthesis and demonstrate that de novo lipid synthesis is required for the reparative function of macrophages.
PPARγ T166 phosphorylation regulates lipid synthesis in reparative macrophages
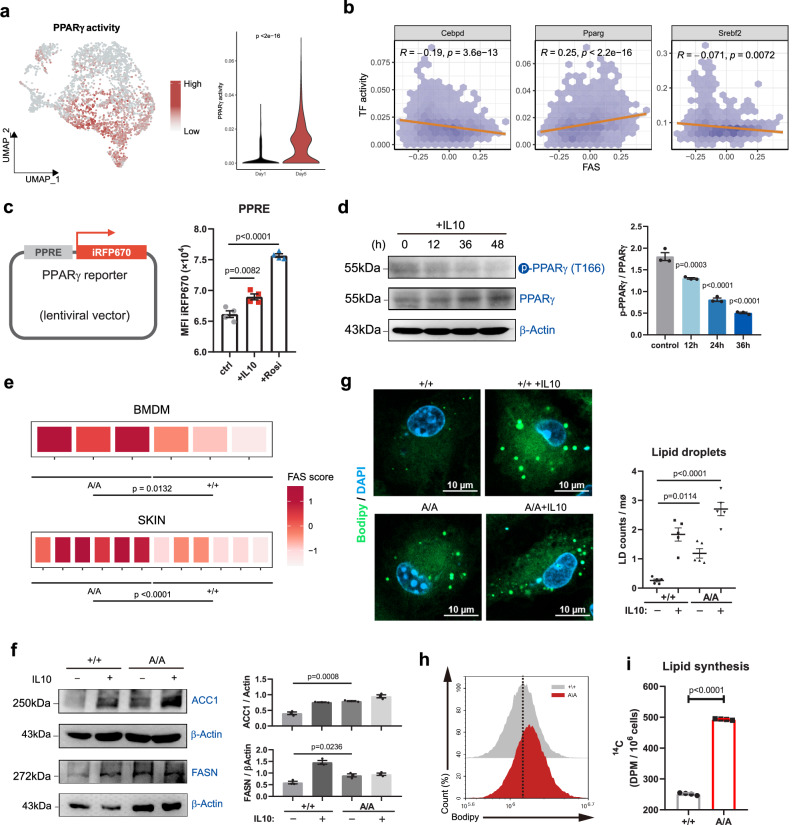
To investigate the regulator of lipid metabolism in reparative macrophages, we performed transcription factor activity analysis using SCENIC. We found that the transcriptional activity of 3 lipid metabolism-related transcription factors, including CEBPδ, PPARγ and SREBF2, increased in reparative phase macrophages (Fig. 3a and Supplementary Fig. 6a, b). However, only PPARγ transcriptional activity positively correlated with FAS in macrophages (Fig. 3b). The activation of PPARγ in reparative macrophages was validated by a fluorescence reporter assay (Fig. 3c). Using PPARγ-knockout macrophages, we confirmed that PPARγ regulated lipid synthesis activity in macrophages (Supplementary Fig. 6c, d). In addition, PPARγ deficiency downregulated the expression of reparative markers CD163 and CD206, while upregulated the expression of proinflammatory makers CD80 and MHCII in macrophages (Supplementary Fig. 6e), further indicating the strong association between lipid synthesis activity and reparative phenotype in macrophages. These results suggest that lipid synthesis in reparative macrophages is regulated by PPARγ activity. Then, we investigated the molecular mechanism through which PPARγ could regulate lipid synthesis in reparative macrophages. First, we observed no significant changes in the expression levels or cellular localization of PPARγ in macrophages after IL-10 treatment (Supplementary Fig. 6f). Considering that PPARγ activity is heavily regulated by posttranslational modification (PTM)22, we next measured three important phosphorylation sites (S112, T166 and S273) of PPARγ. We found that the levels of T166 phosphorylation gradually increased with increasing inflammatory stimulation time, and decreased in response to IL-10 stimulation (Fig. 3d and Supplementary Fig. 6g). The levels of the other two phosphorylation sites (S112 and S273) showed no significant changes in response to IL-10 (Supplementary Fig. 6h), suggesting that reparative signaling induces PPARγ activity through dephosphorylating T166 site in macrophages.
Fig. 3. PPARγ T166 dephosphorylation regulates lipid synthesis activity of skin wound macrophages.
a UMAP and violin plots showing the PPARγ activity in macrophages from skin wound healing phase I and phase II. b Correlations between FAS-related gene expression pattern and transcriptional activity of CEBPδ, PPARγ and SREBF2 in skin wound macrophages. c Schematic illustration indicating the reporter plasmid structure (left), representative flow cytometry histograms showing the reporter fluorescence levels (right) (n = 4 biological replicates). d Representative immunoblot of phospho-T166-PPARγ (pPPARγT166) and total PPARγ in BMDMs treated with IL-10. β-actin was used as a loading control (n = 3 biological replicates). Data were analyzed by a one-way ANOVA followed by a Dunnett’s multiple-comparisons test. e Heatmap showing expression patterns of FAS-associated signature genes between PPARγ+/+ and PPARγA/A BMDMs or skin wound macrophages (6 dpi). f Protein expression of ACC1 and FASN in BMDMs from PPARγ+/+ or PPARγA/A mice in response to IL-10. β-actin was used as loading control. n = 3 biological replicates. g Representative pictures of BODIPY staining in PPARγ+/+ and PPARγA/A BMDMs in response to IL-10. The lipid droplets were quantified (n = 5 biological replicates). Scale bar, 10 μm. Data were analyzed by a one-way ANOVA followed by a Tukey’s multiple comparisons test. h Lipid droplets in PPARγ+/+ and PPARγA/A BMDMs were quantified by flow cytometry. Dashed line indicates peak of the fluorescent signal. i Lipid synthesis rate in PPARγ+/+ and PPARγA/A BMDMs. The data represent the incorporation of 14C-glucose in the lipid fraction of n = 4 biological replicates per group. Unless specified otherwise, the data are presented as means ± s.e.m. (error bar) and compared using the two-tailed Student’s t test. Source data are provided as a Source Data file.
To further determine the role of PPARγ T166 phosphorylation in macrophages, we generated homozygous knock-in mice (PPARγT166A/T166A, hereafter referred to as PPARγA/A) to abrogate the phosphorylation of PPARγ at T166. We performed RNA sequencing on wild-type (PPARγ+/+) and PPARγA/A BMDMs. The results revealed distinct transcriptional features in PPARγ+/+ and PPARγA/A macrophages (Supplementary Fig. 7a), and PPARγA/A macrophages negatively enriched with pro-inflammation-related GO items (Supplementary Fig. 7b), indicating T166A induces a typical reparative phenotype in macrophages. A similar result was observed in macrophages isolated from reparative phase (6 dpi) wound (Supplementary Fig. 7c, d). Then, we found that both PPARγA/A BMDMs and skin wound macrophages exhibited significantly upregulated levels of and FAS activity (Fig. 3e and Supplementary Fig. 7e). Consistent with these findings, PPARγ T166 dephosphorylation increased the expression of ACC1 and FASN in macrophages (Fig. 3f). There were also noticeable increases in intracellular TGs and lipid droplets in PPARγA/A macrophages compared to wild-type cells (Fig. 3g, h and Supplementary Fig. 7f). Moreover, lipogenesis assays using C14-labeled glucose showed increased incorporation of labeled glucose-derived carbon into lipids in PPARγA/A macrophages compared to control cells (Fig. 3i). Collectively, we reveal that PPARγ T166 dephosphorylation is critical in promoting lipid generation in reparative macrophages.
Blocking PPARγ T166 phosphorylation accelerates wound healing
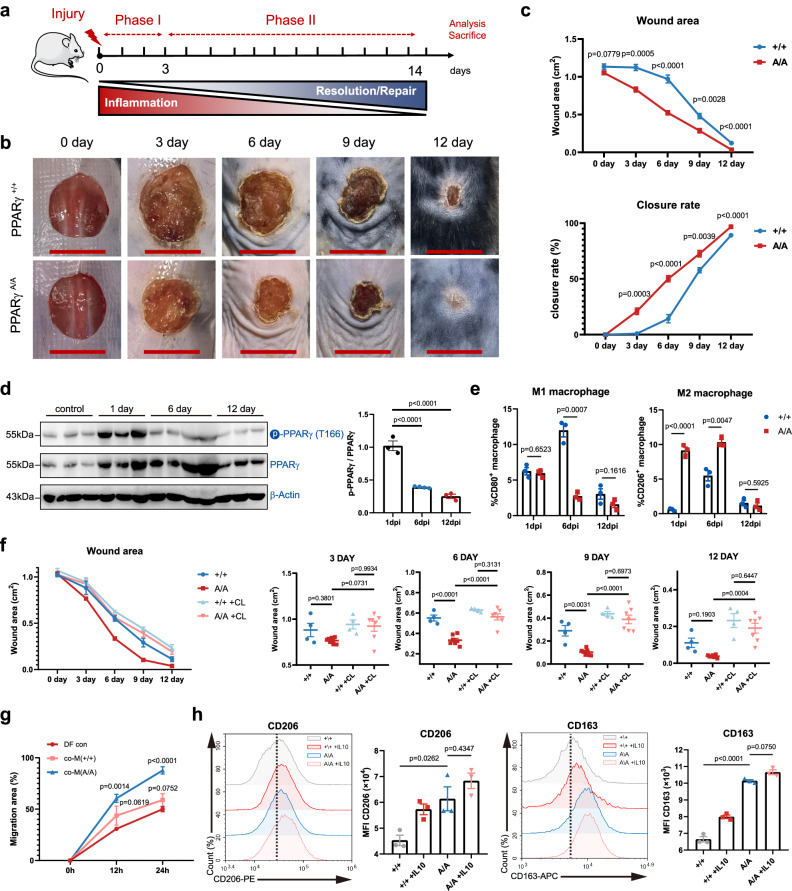
We further examined the role of PPARγ T166 dephosphorylation in the tissue repair function of macrophages. A full-thickness excisional skin injury model was created on the dorsal skin of PPARγ+/+ and PPARγA/A mice. The wound healing rates were monitored over time (Fig. 4a). Notably, no histological changes in the skin of PPARγA/A mice were observed under homeostatic conditions (Supplementary Fig. 8a), while PPARγA/A mice had smaller skin wounds than PPARγ+/+ mice from day 3 to 12 post injury (Fig. 4b, c). Histologic assessment of the wounds revealed an increase in re-epithelialization in PPARγA/A mice (Supplementary Fig. 8b), indicating an accelerated repair process in PPARγA/A mice. Importantly, skin inflammation significantly increased PPARγ T166 phosphorylation, and as the wound progressed into the repair phase, T166 phosphorylation gradually decreased (Fig. 4d). Furthermore, we tested the healing rates of PPARγA/A mice using the other injury models. In muscle injury, the cross-sectional area (CSA) of the regenerating muscle fibers was significantly larger in the PPARγA/A mice compared to PPARγ+/+ mice on 8 dpi (Supplementary Fig. 9a, b). In acute liver injury models, CCl4-induced serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activities were significantly higher in PPARγ+/+ mice on 4 dpi (Supplementary Fig. 9c, d). Besides, wound tissues from PPARγA/A mice showed a higher abundance of reparative macrophages in both muscle and liver injury models (Supplementary Fig. 9e, f). Overall, these data demonstrate the pathophysiological relevance of PPARγ T166 dephosphorylation and wound healing.
Fig. 4. Blockage of PPARγ T166 phosphorylation improves skin wound healing.
a Schematic illustration indicating the phases of skin inflammation, tissue repair after injury. b Representative time-lapse photographs of skin wounds in PPARγ+/+ and PPARγA/A mice over 12 dpi. c Wound area and closure rate in PPARγ+/+ and PPARγA/A mice over 12 dpi (n = 5 per genotype). d Protein levels of p-PPARγT166 and PPARγ were determined by western blots using the wounds from 1, 6 and 12 dpi mice. Control, 1 dpi and 12 dpi, n = 3 biologically independent mice per condition. 6 dpi, n = 4 biologically independent mice. Data were analyzed by a one-way ANOVA followed by a Dunnett’s multiple-comparisons test. e Percentage of CD80+ and CD206+ macrophages in wound tissue of 1-, 6- and 12-dpi in PPARγ+/+ and PPARγA/A mice within the CD45+ immune cell population. n = 3 biological replicates. f The quantification of wound area in PPARγ+/+ and PPARγA/A mice after treatment with CL-lipo and Lipo control. n = 4 (PPARγ+/+ and PPARγ+/+ + CL), n = 7 (PPARγA/A and PPARγA/A + CL) biologically independent samples. Data were analyzed by a one-way ANOVA followed by a Tukey’s multiple comparisons test. g The scratch wound closure rate of DFs co-cultured with PPARγ+/+ or PPARγA/A BMDMs for 24 h (n = 3 per group). Data were analyzed by a one-way ANOVA followed by a Dunnett’s multiple-comparisons test. h Representative flow cytometry histograms and quantifications of surface levels of CD163 and CD206 in PPARγ+/+ and PPARγA/A BMDMs treated with IL-10 for 48 h. n = 3 biological replicates per group. Data were analyzed by a one-way ANOVA followed by a Tukey’s multiple comparisons test. Unless specified otherwise, the data are presented as means ± s.e.m. (error bar) and compared using the two-tailed Student’s t test. Source data are provided as a Source Data file.
Next, we investigated the linkage between tissue repair and the cutaneous immune response in PPARγA/A mice. As expected, injury led to the recruitment of neutrophils, monocytes, and macrophages at the wound site, but the absolute numbers of monocytes/macrophages, T lymphocytes and granulocytes were unchanged between the genotypes after injury (Supplementary Fig. 10a). Strikingly, PPARγA/A mice showed a decrease in MHCII+ macrophage numbers but a significant increase in the number of CD206+ macrophages around the wound compared to those in PPARγ+/+ mice 6 dpi (Supplementary Fig. 10b). In addition, macrophages from wild type mice showed higher levels of the inflammatory M1 marker CD80 at 1 dpi, while macrophages from PPARγA/A mice showed significantly higher surface expression of the reparative marker CD206 at 1 and 6 dpi (Fig. 4e). Genes encoding anti-inflammatory and reparative molecules, including Arg1, Tgfb1, Col2a1 and Vegfb, were upregulated in PPARγA/A mice skin, whereas inflammatory cytokines and chemokines, such as Il6 and Ccl2, were downregulated in PPARγA/A mice skin in the reparative phase (Supplementary Fig. 10c). These data reveal that PPARγ T166 dephosphorylation promotes wounding healing by controlling macrophage reparative phenotype.
To determine the contribution of macrophages in these transgenic mice, we used a clodronate liposome-based approach to deplete macrophages from the dorsal skin of mice. As expected, intradermal injection of clodronate-encapsulated liposomes (CL-lipos) slowed the wound healing rate in wild-type mice, suggesting a crucial role for macrophages in regulating skin regeneration (Fig. 4f and Supplementary Fig. 11a). Importantly, the comparable repair rates between PPARγA/A mice and wild-type mice following macrophage depletion suggests a critical role of PPARγ T166 dephosphorylation in macrophages during tissue repair (Fig. 4f and Supplementary Fig. 11a). Moreover, compared to wild-type macrophages, PPARγA/A macrophages more rapidly promote the migration of skin fibroblasts in vitro (Fig. 4g and Supplementary Fig. 11b).
We further examined the reparative function of PPARγA/A macrophages in vitro. Intriguingly, we found that IL-10 stimulation did not further increase the levels of CD206 and CD163 in PPARγA/A macrophages, suggesting that the reparative phenotype of PPARγA/A macrophages are fully polarized (Fig. 4h). Then, we mimicked the tissue injury phase transition in vitro by shifting the stimulation from LPS + IFNγ to IL-10 (Supplementary Fig. 11c). We found that PPARγA/A BMDMs expressed significantly increased levels of reparative and anti-inflammatory genes, while the expressions of inflammatory genes were lower in PPARγA/A macrophages (Supplementary Fig. 11d), suggesting that PPARγ T166 dephosphorylation polarize macrophages toward a robust reparative phenotype.
T166 dephosphorylation promotes PPARγ binding to lipid synthesis-related gene loci
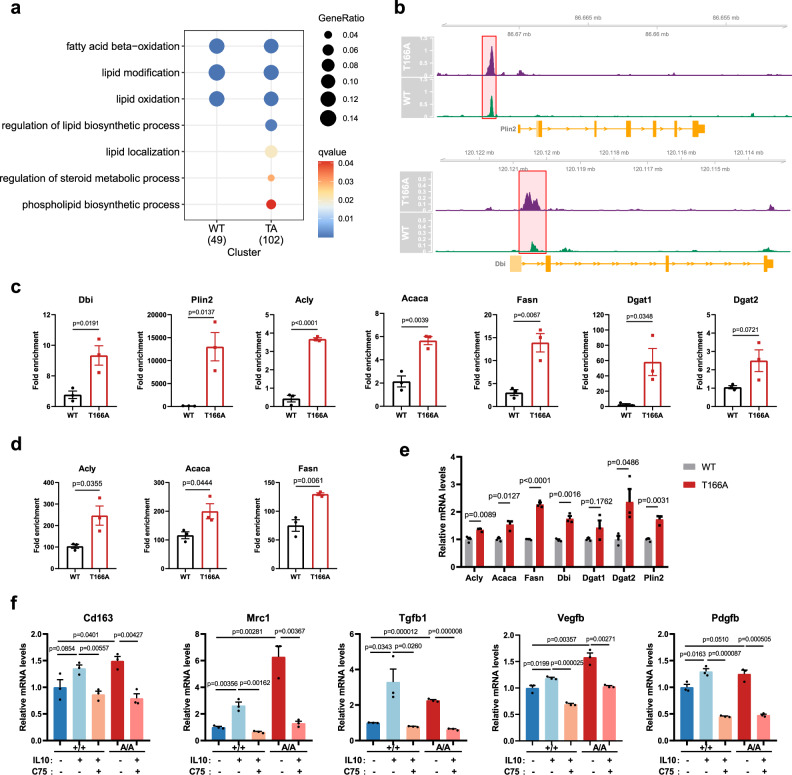
To further understand the role of PPARγ T166A-mediated macrophage reparative activity, we performed chromatin-immunoprecipitation sequencing (ChIP-seq) of PPARγ+/+ and PPARγA/A macrophages. We observed no binding of endogenous PPARγ to reparative phenotype-related gene, such as Cd163, Mrc1 and Arg1 (Supplementary Fig. 12a). Instead, the occupancy of PPARγ binding sites was highly correlated with lipid metabolism (Fig. 5a). Both wild type and T166A PPARγ binding was highly enriched at promoters of FAO-related genes, while the regulation ability of lipid synthesis and localization was significantly increased in the mutated PPARγ (Fig. 5a). Representative gene tracks of PPARγ bindings are shown in Fig. 5b. Subsequent ChIP-qPCR and CUT&RUN analysis confirmed the increases in PPARγ binding, and further revealed that the occupancy of PPARγ binding sites at Acly, Acaca, Fasn, Dgat1 and Dgat2 regulatory regions increased by T166A mutation (Fig. 5c, d). The mRNA levels of these PPARγ target genes were significantly upregulated in T166A macrophages (Fig. 5e). These data demonstrate that PPARγ T166A upregulate lipid synthesis by directly promoting lipid synthesis-related gene expression. Moreover, the expression of genes associated with phenotypic markers and reparative factors was upregulated in M(IL10) or T166A macrophages but was attenuated in macrophages exposed to the FASN inhibitor C75 (Fig. 5f). Taken together, these results provide evidence that posttranslational modification of T166 plays a key role in PPARγ-mediated transcriptional activity of lipid synthesis-related genes, leading to the acquisition of a reparative phenotype in macrophages.
Fig. 5. Blockage of T166 phosphorylation promotes PPARγ binding to lipid synthesis and accumulation-related gene loci.
a GO enrichment analysis showing the highly enriched metabolic pathway associated with WT or T166A PPARγ binding sites. b Binding peaks of WT or T166A PPARγ around indicated gene region. c ChIP-qPCR analysis of PPARγ binding to the Dbi, Plin2, Dgat1, Dgat2, Acly, Acaca and Fasn promoters in WT and T166A BMDMs. qPCR was performed with primers specific to the PPARγ-binding motifs. Data were normalized to the input. n = 3 biological replicates per group. d CUT&RUN qPCR of PPARγ binding to the indicated promoters in WT and T166A BMDMs (n = 3 biological replicates per group). Data were normalized to the spike in DNA. e The relative mRNA levels of FAS-related genes in WT or T166A BMDMs (n = 3 biological replicates per group) were assessed by RT-qPCR. f The relative mRNA levels of repair-related genes in WT or T166A BMDMs (n = 3 biological replicates per group) were assessed by RT-qPCR after treatment with IL-10 and C75 for 48 h. Unless specified otherwise, the data are presented as means ± s.e.m. (error bar) and compared using the two-tailed Student’s t test. Source data are provided as a Source Data file.
Intracellular lipids promote growth factor expression by activating STAT3 transcriptional activity
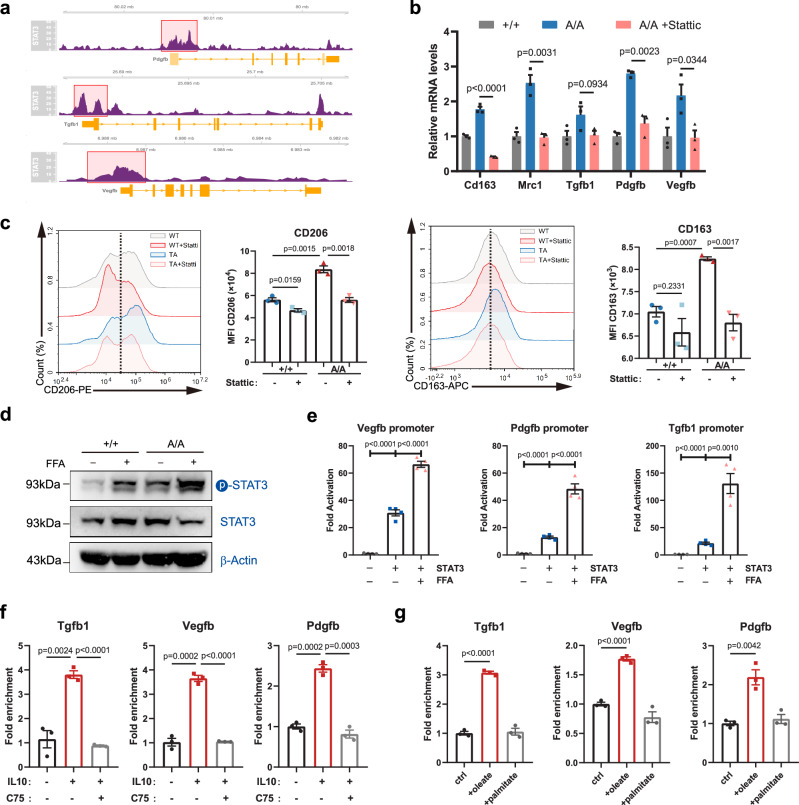
We further explored the mechanism by which intracellular lipids could affect macrophage reparative functions. Previous studies have revealed that intracellular lipids may serve as signaling molecules to induce the activation of STAT323–25, which directly regulates the expression of growth factors, impacting cellular proliferation, survival, differentiation, and other physiological processes26,27. Thus, we reasoned that the de novo-synthesized lipids in macrophages would be essential for the increased expression of growth factors mediated by STAT3 activation. We showed that STAT3 was substantially enriched in the regulator regions of Vegfb, Pdgfb, and Tgfb1 in macrophages (Fig. 6a). A STAT3 inhibitor, Stattic, substantially inhibited the increase in the expression of growth factors and concomitant reparative macrophage activation induced by PPARγ T166A mutation (Fig. 6b, c). However, JAK inhibition did not impact the reparative phenotype of T166A macrophages (Supplementary Fig. 12b). These data indicate that PPARγA/A macrophage exert reparative function through activating STAT3 in a JAK independent manner. Next, we evaluated the role of lipids in STAT3 activation in reparative macrophages. Free fatty acids (FFA) were employed to elevate intracellular lipid levels of macrophages. FFAs addition in macrophage culture significantly induced STAT3 phosphorylation, which was essential for STAT3 activation (Fig. 6d). Luciferase reporter assays showed that FFAs strongly induced STAT3-mediated Vegfb, Pdgfb, and Tgfb1 expression (Fig. 6e). CUT&RUN assays revealed that IL-10 strongly enhanced the binding of STAT3 to the regulatory regions of Vegfb, Pdgfb, and Tgfb1, and FAS inhibition using C75 significantly decreased this effect (Fig. 6f). Furthermore, STAT3 knockout strongly reduced IL-10-induced expression and secretion of reparative factors (Supplementary Fig. 12c–e), and the effects of C75 or FFAs were markedly abolished in macrophages lacking STAT3 (Supplementary Fig. 12e, f). These results suggest that intracellular lipids induce reparative factor expressions by promoting STAT3 activation. Besides, we also found that unsaturated fatty acid, such as oleic acid, induced higher levels of STAT3 activation (Fig. 6g). Collectively, our data indicate that the increased flux of de novo FAS is essential for STAT3 activation and growth factor expression.
Fig. 6. Intracellular lipids are required to support the production of growth factor by macrophages.
a Binding peaks of STAT3 around indicated gene region. b mRNA expression of the reparative markers Cd163, Mrc1, Tgfb1, Pdgfb and Vegfb in PPARγ+/+ and PPARγA/A BMDMs treated with STAT3 inhibitor Stattic (5 μM). n = 3 biological replicates per group. c Representative flow cytometry histograms and quantifications of surface levels of CD163 and CD206 in PPARγ+/+ and PPARγA/A BMDMs treated with stattic for 48 h (n = 3 biological replicates per group). d The protein expression of phosphorylated (Tyr705) STAT3 and total STAT3 in PPARγ+/+ and PPARγA/A BMDMs in response to free fatty acid (FFA). β-Actin was used as a control. e Transcriptional assays of fatty acid on STAT3-driven gene transcription in dual-luciferase reporter assay. Vegfb, Pgdfb and Tgfb1 promoter was transfected in HEK293T cells with or without STAT3 plasmid in response to FFA. n = 4 biological replicates per group. f CUT&RUN qPCR of STAT3 binding to the indicated promoters in BMDMs treated with C75 in response to IL-10 (n = 3 biological replicates per group). Data were normalized to the spike in DNA. g CUT&RUN qPCR of STAT3 binding to the indicated promoters in BMDMs treated with oleate or palmitate (n = 3 biological replicates per group). Data were normalized to the spike in DNA. Unless specified otherwise, the data are presented as means ± s.e.m. (error bar) and compared using the two-tailed Student’s t test. Source data are provided as a Source Data file.
Fatty acids support the expansion of the endoplasmic reticulum, which is required for the secretion of reparative proteins
FAS plays a pivotal role in supplying building blocks for the synthesis of phospholipids (Supplementary Fig. 13a). Phospholipids are integral components of biological membranes that support cell proliferation, protein transport and secretion28. We observed significant alterations in phospholipid metabolic pathways in reparative macrophages using lipidomic analysis (Fig. 2b). We also found a notable increase in the mRNA levels of genes related to phospholipid metabolism, such as Chka, Chkb, Cds1, Pcyt1a, Pcyt2 and Plpp2, in IL-10-treated or PPARγ T166A-mutated macrophages (Supplementary Fig. 13b). In addition, the expression of phospholipid metabolism genes was significantly attenuated in C75-treated macrophages compared with treated controls (Supplementary Fig. 13c). We further addressed the role of phospholipids in membrane system remodeling in reparative macrophage. Although macrophages are typically not proliferative in vitro, there is evidence that macrophages possess the capability of proliferating in vivo in response to tissue microenvironment cues29. However, we observe no proliferation of macrophages decreased during reparative phase (Supplementary Fig. 13d). Thus, we hypothesized that the main role of phospholipids was as essential factors for the endoplasmic reticulum (ER) and Golgi to support the synthesis, transport, and secretion of reparative proteins. Consistent with this hypothesis, we found that ER compartments were considerably expanded in macrophages after stimulation with IL-10 or in T166A macrophages (Supplementary Fig. 13e, f). Using [U-13C] glucose tracer, we observed that IL-10 treatment and T166A mutation strongly upregulated the isotope ratio of 13C/12C in ER (Supplementary Fig. 13g). Collectively, our results demonstrate that FAS supports the expansion of the ER, which is required for the secretion of reparative proteins through the generation of phospholipids.
Topical inhibition of PPARγ T166 phosphorylation accelerates wound healing
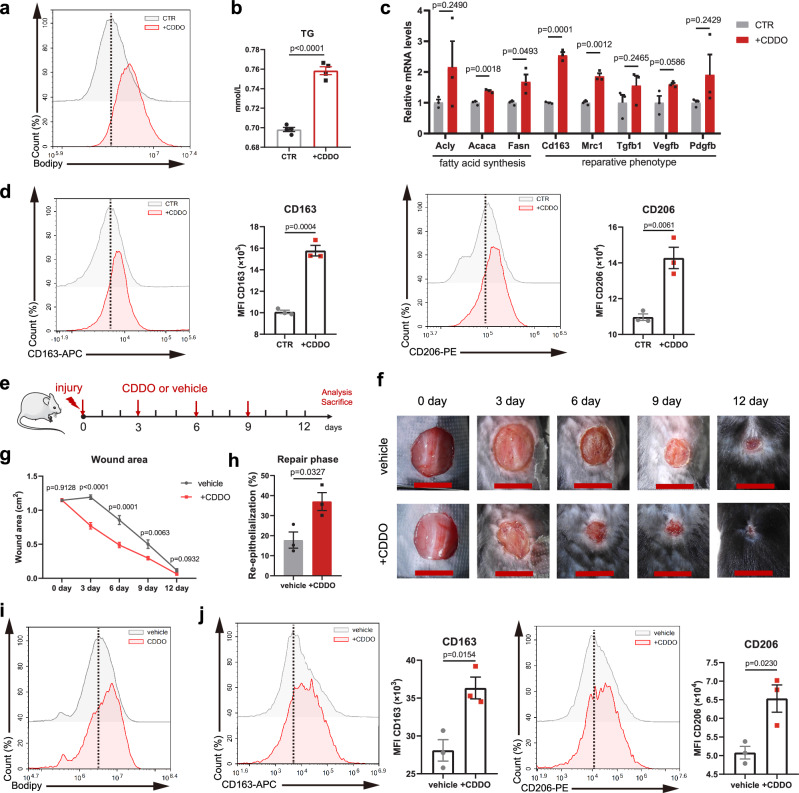
To apply these findings to clinically translatable outcomes, we tested the effects of local PPARγ T166 phosphorylation inhibition on skin wound healing. We previously found that bardoxolone (CDDO) selectively inhibited PPARγ T166 phosphorylation30. Thus, we further investigated whether CDDO could affect macrophage activation and metabolic regulation by treating BMDMs with CDDO. The results showed that CDDO-stimulated macrophages exhibited increased lipid accumulation and lipogenesis-related gene expression (Fig. 7a–c). More importantly, CDDO induced a reparative phenotype in macrophages (Fig. 7c, d). We further investigated the specificity of CDDO in macrophages. The lipid synthesis activity and reparative phenotype induced by CDDO were significantly abolished in PPARγ-knockout macrophages (Supplementary Fig. 14a–c). However, CDDO treatment did not further enhance the reparative phenotype and lipid anabolism in PPARγ T166A mutated cells (Supplementary Fig. 14d–f). Additionally, although CDDO is known to induce Nrf2 activation, the levels of Nrf2 did not affect CDDO-induced lipid synthesis and reparative phenotype (Supplementary Fig. 14g–i). These results reveal that CDDO induces macrophages reparative phenotype through PPARγ T166 dephosphorylation.
Fig. 7. CDDO accelerates healing of murine full-thickness excisional wounds.
a BODIPY staining analysis by flow cytometry in control (CTR) or CDDO-treated macrophages. Dashed line indicates peak of the fluorescent signal. b TG content of BMDMs in response to CDDO (n = 4 biological replicates). c The relative mRNA levels of FAS- and repair-related genes in control or CDDO-treated BMDMs were assessed by RT-qPCR. n = 3 biological replicates per group. d Representative flow cytometry histograms and quantifications of surface levels of CD163 and CD206 in control or CDDO-treated BMDMs (n = 3 biological replicates per group). e Schematic illustration of experimental design. f Representative time-lapse photographs of skin wounds in mice over 12 dpi, treated with vehicle or CDDO ointment. g Wound area after vehicle and CDDO treatment of C57BL/6 mice. (n = 10 per group). h Quantitative analysis of wound re-epithelialization of 6 dpi wound sections from vehicle (n = 3) and CDDO-treated (n = 3) mice. i BODIPY staining analysis by flow cytometry in MACS-sorted wound macrophages at 6 dpi from vehicle and CDDO-treated mice. Dashed line indicates peak of the fluorescent signal. j Representative flow cytometry histograms and quantifications of surface levels of CD163 and CD206 in MACS-sorted wound macrophages at 6 dpi from vehicle and CDDO-treated mice, n = 3 biological replicates per group. Unless specified otherwise, the data are presented as means ± s.e.m. (error bar) and compared using the two-tailed Student’s t test. Source data are provided as a Source Data file.
Given that CDDO shaped the reparative property of macrophages in vitro, we created mice with full-thickness excisional wounds and topically applied CDDO ointment or vehicle ointment (Fig. 7e). CDDO significantly accelerated wound closure, particularly at 3-6 dpi, during which time macrophages transitioned from a proinflammatory to a reparative phenotype (Fig. 7f, g). Histological examination showed that CDDO enhanced the re-epithelialization processes during the repair phase (Fig. 7h and Supplementary Fig. 15a). Furthermore, we isolated F4/80+ macrophages from wounds and found increased lipid accumulation and reparative phenotype polarization in CDDO-treated mice compared to vehicle-treated mice (Fig. 7i, j). Thus, the effects of local PPARγ T166 phosphorylation inhibition are similar to those of the PPARγ T166A mutation on improving skin wound healing. To further examine the therapeutic effect of CDDO to human wounds, we performed scratch tests using human macrophages. The results showed that CDDO treated human macrophages significantly promoted human skin fibroblasts (HSF) and human epidermal keratinocytes (HaCaT) migration (Supplementary Fig. 15b, c). Collectively, these results show that topical PPARγ T166 phosphorylation inhibition could accelerate skin wound healing.
Discussion
Macrophages are present in virtually all tissues in the body, where they act as pivotal players in signal responses and exhibit multiple biological functions, such as immune response, development, tissue remodeling and wound healing9,31–33. In the context of tissue repair, macrophages not only secrete an array of proinflammatory or anti-inflammatory cytokines to mediate immune response but also release many matrix metalloproteinases and growth factors9. These bioactive molecules remodel the extracellular matrix, foster angiogenesis, and ultimately facilitate wound healing. However, the molecular mechanisms that drive and sustain the reparative phenotype of macrophages remain unclear. In this study, we identified PPARγ T166 dephosphorylation-mediated lipid anabolic network as a major regulator of reparative function in macrophages. We detected the activation of FAS in reparative macrophages across 9 tissue injury models, as well as IL-10, TGF-β1 and dexamethasone induced in vitro models. We also revealed that this lipid metabolic reprogramming is conserved in mouse and human reparative macrophages, suggesting targeting this pathway may have therapeutic value. Moreover, intracellular lipids were necessary for sustaining the reparative function of macrophages, partly because fatty acids trigger STAT3-mediated growth factor expression and support the expansion of ER, which is required for the secretion of reparative proteins. Our results provide an additional rationale for the efficacy of FAS activation in the treatment of tissue injury, that offers significant potential in the development of innovative therapeutic strategies.
Recent advances in immunometabolism have revealed that macrophages with different activation states have distinct metabolic pathways to meet their demands for energy and the production of specific cell-associated factors18,34. For instance, M1 macrophages primarily rely on glycolysis for energy production, which enables them to rapidly generate ATP in response to inflammation35. M2 macrophages exhibit an OXPHOS metabolic phenotype. These cells rely on fatty acid oxidation and mitochondrial respiration for energy production36. Significant progress has been made in metabolic research on classical activation and alternative activation of macrophages, while there is limited understanding regarding the specific metabolic adaptations that are crucial for wound healing and tissue repair. We demonstrate that injury-associated macrophages undergo distinct metabolic adaptations to facilitate phase-specific repair functions after tissue injury. The most striking observations of this study is that reparative macrophages adjust their metabolism by activating a fatty acid synthetic program which is critical for supporting the growth factor production and secretion. Using scRNA-seq atlas, we emphasized the indispensable role of lipid anabolism in macrophage function, while the catabolic processes, such as glycolysis, OXPHOS and FAO, were strongly impacted by tissue microenvironments. This finding is consistent with previous study showing that both FAO and OXPHOS are dispensable for macrophage activation37,38.
By modulating macrophage metabolism, it is possible to manipulate macrophage phenotypes and functions, thereby impacting disease progression and treatment outcomes39,40. In cancer immunotherapy, reprogramming the metabolism of tumor-associated macrophages (TAM) can shift them toward an antitumor phenotype, thereby enhancing tumor elimination. Strategies such as inhibiting glycolysis and promoting OXPHOS in TAMs have shown promising results in preclinical models of cancer41,42. In chronic inflammatory diseases such as atherosclerosis, targeting macrophage lipid metabolism regulates inflammatory responses and cholesterol handling, reducing plaque formation and promoting stability36,43. These findings highlight the potential of manipulating macrophage metabolism to improve disease treatment outcomes. Therefore, targeting the metabolic regulation of macrophages during the repair phase will allow for more precise modulation of tissue repair and regeneration. In this study, we created an ointment to regulate FAS activity of injury-associated macrophages. This strategy was proven to be useful in promoting skin wound healing. Thus, further exploration of the intricate relationship between macrophage metabolism and tissue repair programs will pave the way for the development of therapies that exploit macrophage plasticity and metabolic rewiring.
PPARγ is a peroxisome proliferator-activated receptor (PPAR) family protein that has received particular attention as a critical regulator of a multitude of metabolic processes that maintain cellular energy homeostasis44. Notably, an extensive amount of literature has addressed the association between PTMs and PPARγ function in the regulation of cell differentiation, tissue development, and energy metabolism45–47. However, most of the previously published work was completed in the context of adipocytes48,49. Recent studies have revealed that PPARγ also plays a major role in macrophage differentiation and functions50–52. Conditional ablation of Pparg in macrophages leads to a pronounced delay in skeletal muscle regeneration53. Despite this, there remains a paucity of data on PPARγ PTM mechanisms that regulate macrophage phenotypes. It is worth noting that the biological function and regulation mechanism of PPARγ PTMs might be different between adipocytes and macrophages. Phosphorylation of PPARγ at S273 in macrophages regulates the expression of GDF354, thereby affecting macrophage reparative functions, while S273 phosphorylation primarily influences insulin sensitivity in adipocytes through the regulation of genes related to glucose and lipid metabolism55,56. These findings indicate that PPARγ PTMs have cell-specific roles. We previously identified T166 as a phosphorylation site of PPARγ in the DNA-binding domain30. Here, PPARγ T166 phosphorylation was proven to be a molecular regulator of macrophage activation and metabolism. Genetic blockade of T166 phosphorylation by an alanine mutation enhanced the binding of PPARγ to the regulator regions of lipid synthesis-related genes. The accumulation of lipids may allow reparative phase macrophages to increase anti-inflammatory cytokine and reparative growth factor production by activating STAT3 signaling and directly serving as membrane structures to facilitate protein secretion. In conclusion, this work establishes PPARγ as a required metabolic sensor and transcriptional regulator of skin reparative macrophages, but much more work needs to be done to explore the role of PPARγ in macrophages and other types of immune cells to better elucidate the molecular connection between PPARγ, tissue repair and lipid metabolism. This new insight could lead to the identification of PPARγ T166 as a new pharmaceutical target to facilitate the healthy repair of injured tissues.
Methods
Generation of PPARγT166A mice
All mouse procedures and experiments for this study were approved by the Institutional Animal Care and Use Committee at Nanjing University (IACUC-2207009). Animals were housed in a temperature-controlled specific pathogen-free facilities with a 12 h light/dark cycle with 45–65% relative humidity. Food and water were available ad libitum. All mice were more than 8 weeks old at the beginning of the experiments and included both males and females. Animal sample sizes were determined based on previous studies and literatures of the field using similar experimental paradigms.
Mice with the PPARγ mutation were generated using CRISPR-Cas9 strategy, as described previously57. Site-directed mutagenesis was used to alter the codon for T166 to alanine. Single-cell zygotes from superovulated C57BL/6J female mice mated to C57BL/6J males were injected with Cas9 mRNA, sgRNA and ssDNA, and cultured overnight. Transfer the two-cell embryos into oviducts of pseudopregnant Swiss Webster surrogate females. The resulting progeny gave rise to highly chimeric offspring. These chimeras were mated to C57BL/6J mice to obtain germline transmission of the mutant allele. These heterozygous mice (PPARγT166A/+) were mated to generate homozygous knock-in mice PPARγT166A/T166A. Mice were backcrossed onto C57BL/6J for 6 generations, after which mice heterozygous for the PPARγT166A allele (PPARγT166A/+) were bred to generate experimental cohorts. The PPARγT166A allele was detected by PCR genotyping with primers: 5’-CAAGACATGGTCTCCCTCTG-3’, 5’-AGTTGTCCTAGTGCCTACTG-3’ and 5’-CTGGCCTACACGAAGAAGCTC-3’. Wild-type littermates were used for experimental controls.
Animal model of skin injury
C57BL/6J mice (Strain No. N000013) were purchased from GemPharmatech Co., Ltd. Before surgery, all mice were anesthetized and dorsal area was shaved. The dorsal skins of mice were secured by inelastic medical adhesive tapes to prevent wound contraction. Subsequently, full-thickness skin excisional wounds (10 mm in diameter) were made using surgical scissors under sterile surgical conditions. To deplete the macrophages in vivo, 100 μl of 5 mg/ml of clodronate liposomes or PBS liposomes (YEASEN) were intradermally injected into wound of each mouse every other day. For local treatment of wounds with CDDO (HY-14909, MCE), either vehicle or CDDO ointment (3 mg/kg) was applied directly to the wound at 0, 3, 6, 9 dpi. The vehicle ointment was formulated by mixing sunflower oil (S304663, aladdin) and Vaseline (V105023, aladdin) in a 1:1 ratio (w/w). For the preparation of the ointment containing the PPARγ T166 inhibitor, CDDO was incorporated into the vehicle ointment with a weight ratio of 1:9. The epithelial space was measured on every 3 days. The area of the wound was calculated using ImageJ software. Then the percentage of wound area closed relative to the original wound was calculated using the following formula: relative closure rate = [1 − (wound size/original wound size)] × 100%. 12 days later, the mice were euthanized in a CO2 chamber adhering IACUC guidelines. For histologic analysis, wounds were excised at different times after injury.
Muscle injury
Mice were anaesthetized with isoflurane and 50 μl of 12 μM cardiotoxin (CTX) (RP17303, GenScript) was injected in the tibialis anterior muscle. Muscles were collected for flow cytometry analysis or for histology analysis at different time points after injury. Muscles were isolated and digested with RPMI 1640 medium containing 2 mg/ml Collagenase II (HY-E70005B, MCE), 0.5 mg/ml Collagenase IV (HY-E70005D, MCE) and 4 mg/ml Dispase (S25046, Yuanye Bio-Technology) for 1 h at 37 °C. Digested material was subsequently passed through a 40 μm cell strainer. The resultant cell suspension was washed with PBS and centrifuged at 500 × g for 5 min. Macrophages were isolated by F4/80-positive selection using anti-F4/80 magnetic beads (130-110-443, Miltenyi Biotech), as recommended by the manufacturer.
Acute liver injury
WT or PPARγA/A mice were intraperitoneally injected with a single dose of olive oil (vehicle) or 1% CCl4 (10 ml/kg, dissolved in olive oil) and sacrificed at 1 day or 4 days post treatment. Livers were perfused with HBSS solution and digested with 0.1 mg/ml DNase I (Roche) and 0.5 mg/ml collagenase IV (HY-E70005D, MCE). The cell suspension was filtered through a 70 μm cell strainer and centrifuged at 30 × g for 3 min to eliminate hepatocytes. The non-parenchymal cell fraction was washed once with PBS at 500 × g for 5 min at 4 °C. Kupffer cells were isolated using anti-F4/80 magnetic beads (130-110-443, Miltenyi Biotech), as recommended by the manufacturer. Blood from WT and PPARγA/A mice treated with oil (vehicle) or CCl4 was collected, and sera were extracted from the blood. The levels of serum ALT and AST were measured with the Alanine aminotransferase Assay Kit (C009-2-1) and Aspartate aminotransferase Assay Kit (C010-2-1) from Jiancheng Bioengineering Institute.
Cell culture experiments
HEK293T (#GNHu17), RAW 264.7 (#TCM13) were purchased from the cell bank of the Committee on Type Culture Collection of the Chinese Academy of Sciences. HSF and HaCaT cells were generous gifts from Prof. JinHui Wu’s Lab at Nanjing University. DFs were isolated from mouse skin. BMDMs were isolated from mouse bone marrow. Human macrophages were isolated from peripheral blood mononuclear cells. HEK293T, RAW 264.7, DF, HSF and HaCaT cells were cultured in DMEM. BMDMs were maintained in RPMI 1640 medium. Human macrophages were maintained in X-VIVO 15 medium (Lonza). Cells were maintained in culture supplemented with 10% FBS (GIBCO) and 1% penicillin/streptomycin at 37 °C with 5% CO2 in a humidified incubator.
BMDMs generation and in vitro treatment
BMDMs were isolated from the femurs and tibiase of C57BL/6J or PPARγA/A mice. Femurs and tibiase were placed in PBS. Using surgical scissor cuts off the ends of the bones. Rinse repeatedly with a 1 ml sterile syringe, until most of the cells in the bone marrow blow away. Total bone marrow cells were passed into a 40-μm cell strainer and centrifuged (800 × g for 5 min). Bone marrow cell pellets were resuspended in red blood cell lysis buffer for 5 min and washed in PBS. Next, bone marrow cells were differentiated for 7 days in the presence of M-CSF (50 ng/ml, Z03275, GenScript) in RPMI-1640 medium containing 10% FBS and 100 U/ml penicillin-streptomycin. Total bone marrow cells were seeded in 10-cm culture plates at a density of 5 × 106 cells per plate and cultured at 37 °C in a 5% CO2 incubator. On day 7, macrophage purity of the culture was routinely tested by the expression of F4/80 and CD11b by flow cytometry. Around 95–99% of the cells expressed high levels of F4/80 and CD11b after differentiation.
BMDMs were cultured at a density of 5 × 105 cells per 12-well in 1 ml complete medium, and stimulated for 48 h with IL-10 (20 ng/ml, Z03161, GenScript), IFNγ (25 ng/ml, Z02916, GenScript), LPS (100 ng/ml, Sigma), IL-4 (20 ng/ml, Z02996, GenScript), TGF-β1 (20 ng/ml, Z03431, GenScript) or dexamethasone (100 nM, Sigma) to achieve an M(IL10), M(LPS + IFNγ),Μ(IL4), Μ(TGF-β1) or M(Dex) polarization state, respectively. In some experiments, BMDMs were treated for 48 h (unless stated otherwise) with C75 (20 μM, HY-12364, MCE), Stattic (10 μM, HY-13818, MCE), rosiglitazone (1 μM, HY-17386, MCE), sodium oleate (100 μM, O7501, Sigma-Aldrich), sodium palmitate (100 μM, P9767, Sigma-Aldrich), FFAs (100 μM, the ratio of sodium oleate and sodium palmitate concentrations at 2:1).
Skin-cell isolation
Mice were anesthetized with ketamine and killed. The dorsal skin was removed and cut into tiny pieces with scissors, followed by incubation with complete medium (RPMI 1640 and 10% FBS) containing 0.1 mg/ml DNase I (Roche), 0.5 mg/ml hyaluronidase (HY-107910, MCE) and 2 mg/ml collagenase IV (HY-E70005D, MCE) in a shaking water bath (37 °C) for 2 h. The cell suspension was filtered with a 40-μm cell nylon mesh to obtain a single-cell suspension. The remaining red blood cells were lysed using RBC lysis buffer. The acquired single-cell suspensions were processed for flow cytometry or MACS.
Magnetic-activated cell sorting
The skin digest was resuspended in MACS buffer (PBS, 2 mM EDTA, 0.5% BSA) and incubated with anti-F4/80 microbeads (130-110-443, Miltenyi Biotech) for 15 min at 4 °C. Then, the F4/80+ fraction was isolated using MACS MS columns according to the manufacturer’s instructions (Miltenyi Biotech). Samples were further processed for immunoblotting, flow cytometry analysis or gene expression analysis as described below.
Flow cytometry staining and analysis
After collection, skin digest or BMDMs were kept in FACS buffer on ice. Non-specific binding was blocked with FcR blocking reagent (130-092-575, Miltenyi Biotech) for 20 min at 4 °C. Antibodies used for flow cytometry analysis were as follows: anti-CD45 (1:100, 157214, Biolegend), anti-F4/80 (1:100, 17-4801-82, Invitrogen), anti-CD11b (1:100, 12-0112-81, Invitrogen), anti-Gr1(1:100, 552093, BD Biosciences), anti-CD3e (1:100, 12-0031-83, Invitrogen), anti-CD206 (1:100, 141705, Biolegend), anti-CD163 (1:100, 17-1631-82, Invitrogen), anti-CD80 (1:100, 12-0801-81, Invitrogen), anti-CD86 (1:100, 553692, BD Biosciences), anti-MHC class II (1:100, 12-5321-82, Invitrogen). For intracellular staining, cells were fixed and permeabilized using the Intracellular Fixation & Permeabilization Buffer Set (#88-8824-00, eBioscience) and intracellularly stained for 1 h at 4 °C with anti-CD206 or anti-CD163 antibody. The lipid content was assessed with 2 μM BODIPY 493/503 (D3922, Invitrogen), following 20 min of incubation at 37 °C. Flow cytometric analysis was performed on a NovoCyte Flow Cytometer (ACEA Biosciences Inc). The gating strategies for Fig. 4e and Supplementary Fig. 10a are presented in Supplementary Fig. 16.
Cell fractionation and immunoblot analysis
The nuclear and cytoplasmic proteins were extracted according to the manufacturer’s instructions of the kit (P0027, Beyotime). After collection, cells were washed 3 times with PBS, and lysed in RIPA buffer supplemented with protease and phosphatase inhibitor cocktail. Protein concentration was quantified by BCA reagent. Next, 50 μg of protein was then separated by electrophoresis using SDS-PAGE gels and then transferred to PVDF membranes. Membranes were blocked for 1 h in 5% BSA at room temperature. Membranes were incubated overnight with the following primary antibodies: mouse anti-PPARγ (1:200, sc-7273, Santa Cruz), rabbit anti-PPARγ (1:1000, #2443S, Cell Signalling Technology), anti-PPARγ p-Ser273 (1:1000, bs-4888R, Bioss), anti-PPARγ p-Ser112 (1:500, ab195925, Abcam), p-STAT3 (Y705) (1:2000, #9145, Cell Signalling Technology), anti-STAT3 (1:1000, BS1336, Bioworld), anti-ACC1 (1:4000, 21923-1-AP, proteintech), anti-FASN (1:5000, 10624-2-AP, proteintech), Lamin A/C (1:1000, BS1446, Bioworld), anti-β-actin (1:10,000, 700068, Zen-Bioscience) and GAPDH (1:5000, 390035, Zen-Bioscience). Mouse monoclonal antibody against p-T166 (1:500) was generated as described previously30. Signals were detected by a chemiluminescence System (Tanon-5200S) using Enhanced ECL Chemiluminescent Substrate (36222, YEASEN). The intensities of the bands were quantified by ImageJ software. The expression of proteins was normalized to a housekeeping protein (β-actin), and the phosphorylation status was determined by normalizing to a respective total protein.
Quantitative real-time PCR
Total RNA was isolated from cells or skin tissue samples with TRIzol reagent (R401-01, Vazyme). Reverse transcription was performed with HiScript III RT SuperMix (R323-01, Vazyme) kit. Quantitative RT-PCR assays were performed on a real-time PCR System (C1000, Bio-Rad) using qPCR SYBR Green Master Mix (Q111-02, Vazyme). The expression of genes of interest was normalized to housekeeping gene Rplp0, and data were normalized to the average of the control group. Primer sequences are described in Supplementary Data 1.
Single-cell transcriptomic atlas of wound macrophages
We obtained scRNA-seq datasets containing wound macrophages from 9 mouse wound healing models, including an anterior cruciate ligament injury (ACLI) model using a noninvasive single dynamic tibial compressive overload method, a polidocanol-mediated airway injury (AWI) model, a kidney injury (KDI) model by short ischemic reperfusion, an acetaminophen-induced acute liver damage (LIVER) model, a bleomycin-mediated acute lung injury (LUNG) model, a myocardial infarction (MCIN) model by left anterior descending coronary artery ligation, a spinal cord injury (SCI) model through laminectomy, a full-thickness excisional skin injury (SKIN) model, and a BaCl2-induced skeletal muscle injury (SMI) model (Supplementary Data 2). Samples were categorized into inflammatory phase (phase I) and reparative phase (phase II) according to the description provided in the source articles related to these datasets. The scRNA-seq data analysis was performed using Seurat (v4.3.0) R package58. In brief, quality control was applied to cells based on the total UMI counts and number of detected genes. Cells with less than 200 detected genes were filtered. To remove potential doublets, cells with UMI count above 40,000 and detected genes above 6000 are filtered out. The gene counts were normalized using SCTransform. For clustering, top 2000 variable genes were used, and ribosomal and mitochondrial genes were removed from the variable gene set before running PCA and identifying clusters using a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm. An annotation tool, SingleR (v2.2.0), was employed to assist cluster identification59. The gene list provided by the source article were also considered as marker genes in macrophage identification (Supplementary Data 2). Subclusters which were identified by macrophages were isolated from the original datasets and re-analyzed using above method. The ALRA method60 was used for imputing dropout genes when necessary.
Trajectory analysis
Trajectory analysis was performed by monocle (v2.30.1) R package. Monocle introduced the strategy of ordering RNA-sequencing samples in pseudotime, placing them along a trajectory according to their transcriptional similarity. To visualize the macrophage phenotype transition along the cell trajectory, dataset (GSE131364) containing M0, M(IFNγ), M(LPS), M(IL4) and M(IL10) samples was employed as the reference dataset. To train the SingleR classifier, simulated samples were synthesized from the reference dataset by randomly integrating a certain percentage of samples from the same group. The final dataset comprised of 20 samples within each group. Then, the reference data were wrapped into a SummarizedExperiment object for SingleR classification.
SCENIC analysis
Activated regulons in SKIN macrophages were analyzed using SCENIC (v1.3.1) R package61. Raw count matrix containing 2953 cells and 15,563 genes was used as input. In brief, the co-expression network was calculated by GRNBoost2 and the regulons were identified by RcisTarget. Next, the regulon activity for each cell was scored by AUCell. The BH method was used to correct multiple hypothesis.
RNA sequencing and data analysis
Full-thickness excisional skin wounds were generated on the PPARγ+/+ and PPARγA/A mice. The mice were sacrificed and wound tissues were digested into single-cell suspensions at 6 dpi. Then, skin macrophages were isolated using MACS. BMDMs were isolated from PPARγ+/+ and PPARγA/A mice and cultured at a density of 5 × 106 cells/10-cm dish. Total RNA was extracted from BMDMs and skin macrophages using TRIzol reagent according to the manufacturer’s protocol. Next generation sequencing library preparations were conducted according to the manufacturer’s protocol. The libraries were sequenced on an Illumina Novaseq 6000 Sequencer. Read quality was assessed with FastQC (v0.12.1) and adapters trimmed using Cutadapt (v4.4)62. Reads were then aligned to the mm10 genome (UCSC) using HISAT2 (v2.2.1)63 and reads in exons were counted using featureCounts function in Subread (v2.0.3) package64. Analysis of differentially expressed genes was conducted using DESeq2 (v1.40.1) R package65. Count data was transformed to log2 scale and normalized with respect to library size using rlog transformation produces. Absolute log2 fold change (log2FC) > 1.5 and p value < 0.05 was considered statistically significant.
Enrichment analysis
Gene set enrichment analysis (GSEA) was conducted and visualized using fgsea (v1.22.0) R package66. Single sample gene set enrichment analysis (ssGSEA) was performed using GSVA (v1.44.2) R package67 with custom gene lists. GO categories (biological process) enrichment analysis was performed and visualized using clusterProfiler (v4.8.3) R package68. The p values were adjusted using the FDR by BH method. Gene list of six metabolic pathway was obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG) database, including glycolysis (mmu00010), TCA (mmu00020), OXPHOS (mmu00030), PPP (mmu00061), FAO (mmu00071), and FAS (mmu00061).
Chromatin immunoprecipitation (ChIP) assays
ChIP experiments were carried out using the Enzymatic Chromatin IP Kit (#9003, Cell Signalling Technology) according to the manufacturer’s instructions. In brief, BMDMs were isolated from PPARγ+/+ and PPARγA/A mice and cultured at a density of 5 × 106 cells per 10-cm culture dish as described above. 1 × 107 cells were fixed with 1% formaldehyde in culture medium at room temperature for 10 min. Subsequently, the crosslinked cells were collected, lysed and digested with MNase. The sheared chromatin was subjected to immunoprecipitation with a rabbit anti-PPARγ antibody (1:100, #2443S, Cell Signalling Technology) or rabbit IgG control overnight at 4 °C with constant rotation. The isolated complexes were collected with Protein G magnetic beads and eluted by incubating at 65 °C in a buffer with high salt concentration. The qPCR following ChIP was performed with the primer sequences listed in Supplementary Data 1.
For ChIP-seq, libraries were prepared from purified immunoprecipitation DNA by blunting, A-tailing, adapter ligation. Library quality was assessed on the Agilent Bioanalyzer 2100 system. The ChIP-seq libraries were sequenced on an Illumina Novaseq 6000 Sequencer. Then, sequenced reads were aligned to reference mouse genome (mm10 assembly, UCSC) using Bowtie2 (v2.5.1)69, and clonal reads were removed from further analysis. After mapping reads to the reference genome, we used the MACS2 (v2.1.0)70 peak calling software to identify regions of IP enrichment over background. p value threshold of 0.0005 was used for all datasets. ChIPseeker (v1.36.0) R package71 was used to retrieve the nearest genes around the peak and annotate genomic region of the peak. Peak-related genes were used to perform enrichment analysis to identify the function enrichment results. The binding peaks were visualized using Gviz (v1.46.1) R package.
CUT&RUN assays
CUT&RUN assays were carried out using the Hyperactive pG-MNase CUT&RUN Assay Kit (HD101-01, Vazyme) according to the manufacturer’s instructions. 500,000 BMDMs from C57BL/6J mice were harvested by centrifugation and washed in wash buffer. Concanavalin A-coated beads are activated and stabilized by Ca2+ and Mn2+ cations. Cells were bound to activated Concanavalin A-coated magnetic beads, then permeabilized with Wash buffer containing 0.05% Digitonin. The bead-cell slurry was incubated with antibody buffer in a 100 μl volume at 4 °C overnight on a rotator. anti-PPARγ (1:100, #2443S, Cell Signalling Technology) or anti-STAT3 (1:100, BS1336, Bioworld) antibodies were used to IP PPARγ or STAT3 in BMDMs. After 2–3 washes in 1 ml Dig-wash, beads were resuspended in 100 μl pG/MNase and incubated for 1 h at 4 °C. After two washes in Dig-wash, beads were resuspended in ice-cold calcium incubation buffer. Tubes were replaced on ice for 1.5 h, followed by immediate addition of EGTA-STOP buffer. Beads were incubated at 37 °C for 30 min, replaced on a magnet stand and the liquid was removed to a fresh tube and DNA was extracted. The qPCR following CUT&RUN was performed with the primer sequences listed in Supplementary Data 1.
For CUT&RUN-seq, libraries were prepared from purified immunoprecipitation DNA by blunting, A-tailing, adapter ligation. Library quality was assessed on the Agilent Bioanalyzer 2100 system. The libraries were sequenced on an Illumina Novaseq 6000 Sequencer. Short reads (<20 bp) and adapter sequences were removed using Cutadapt (v4.4). Trimmed reads were aligned to the reference mouse genome (mm10 assembly, UCSC) using Bowtie2 (v2.5.1) with the “--very-sensitive --no-mixed --no-discordant” parameters. Then, the reads were filtered, retaining only the mapped read pairs that are located on the same chromosome and have a fragment length of less than 1000 bp. Subsequently, the data were converted into bedgraph file format using Bedtools (v2.31.0)72. The bedgraph files were visualized using Gviz (v1.46.1) R package.
Luciferase reporter assay
Genes encoding mouse PPARγ1 (WT or T166A) were cloned separately into the pcDNA3.1(-) vector. PPRE X3-TK-luc was a gift from Bruce Spiegelman (Addgene plasmid # 1015). HEK293T cells were transfected with PPARγ (WT or T166A), PPRE-Luc, and control Renilla plasmid pRL at a molar ratio of 50:50:1. Following an 8 h transfection period, the culture medium was replaced with fresh culture medium supplemented with 10% FBS. Subsequently, small molecules were added to the culture wells and incubated for a duration of 24 h. Cells were harvested and lysed in luciferase lysis buffer, and the luciferase activity of the cells was carried out following the dual-luciferase reporter assay system (Promega, E1910). Firefly luciferase activity obtained from each sample was normalized to the Renilla luciferase activity from the same sample.
For determination of STAT3 binding region on Vegfb, Pdgfb and Tgfb1 promoters, there promoters were amplified and cloned into pGL6 reporter plasmid (D2105, Beyotime). The constructed pGL6 vector was transfected into HEK293T cells with/without STAT3 expression plasmid and luciferase activity was measured and analyzed as above.
Tracing with [U-13C] glucose
BMDMs were left unstimulated or stimulated for 48 h with IL-10. Medium was changed to RPMI 1640 containing [U-13C] glucose (2 g/l) or 12C-glucose (2 g/l), and 48 h later cell culture plates were washed with PBS and snap-frozen in liquid nitrogen and subjected to fatty acid flux analysis (LipidALL Technologies). Lipids were extracted from BMDMs using 1 ml of ice-cold methanol. Samples were incubated for 30 min at 4 °C and centrifuged for 10 min at 14,000 × g. Supernatant was dried in a SpeedVac under OH mode. Total protein content was determined from the dried pellet using the BCA protein assay. The dried extract was reconstituted in LCMS grade methanol for fatty acid flux analysis on a system comprising an Agilent 1290 II UPLC coupled to Sciex 5600+ quadrupole-TOF MS. Fatty acids were separated on a Waters ACQUITY HSS-T3 column (3.0 × 100 mm, 1.8 μm). Information-dependent acquisition mode was used for MS/MS analyses of the metabolites. Collision energy was set at −30 ± 15 eV. Data acquisition and processing were performed using Analyst TF 1.7.1 Software (AB Sciex). All detected ions were extracted using MarkerView 1.3 (AB Sciex). PeakView 2.2 (AB Sciex) was applied to extract MS/MS data and perform comparisons with the Metabolites database (AB Sciex), HMDB and standard references to annotate ion identities.
ER expansion was assessed by culture of BMDMs in RPMI 1640 medium containing 2 g/l [U-13C] glucose and 48 h later ER were isolated with ER isolation kit (Solarbio, EX2690) according to the manufacturer’s protocol. Subsequently, the 13C/12C isotope ratio is measured by δ13C High-Precision Isotopic CO2 CRDS Analyzer (G2131-i, Picarro) and CNHS-O Elemental Combustion Analyzer (Costech ECS 4024, NC Technologies).
Lipidomic analysis
BMDMs were isolated from C57BL/6J mice and cultured at a density of 5 × 106 cells per 10-cm culture dish as described above. Subsequently, the BMDMs were either left unstimulated (M0) or stimulated with 20 ng/ml IL-10 (M(IL10)) for 48 h. Metabolite extraction from M0 and M(IL10) BMDMs was performed as follows: briefly, cells were rinsed with PBS and instantly frozen in liquid nitrogen. The collected samples were thawed on ice. 20 μl of sample was extracted with 120 μl of precooled 50% methanol, vortexed for 1 min, and incubated at room temperature for 10 min; the extraction mixture was then stored overnight at −20 °C. After centrifugation at 4000 × g for 20 min, the supernatants were transferred into new 96-well plates. The samples were stored at −80 °C prior to the LC-MS analysis. In addition, pooled QC samples were also prepared by combining 10 μl of each extraction mixture. All samples were acquired by the LC-MS system. All chromatographic separations were performed using a Thermo Scientific UltiMate 3000 HPLC. An ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.8 μm, Waters, UK) was used for the reversed phase separation. The column oven was maintained at 35 °C. The flow rate was 0.4 ml/min and the mobile phase consisted of solvent A (water, 0.1% formic acid) and solvent B (acetonitrile, 0.1% formic acid). Gradient elution conditions were set as follows: 0–0.5 min, 5% B; 0.5–7 min, 5% to 100% B; 7–8 min, 100% B; 8–8.1 min, 100% to 5% B; 8.1–10 min, 5% B. The injection volume for each sample was 4 μl. A high-resolution tandem mass spectrometer TripleTOF6600plus (SCIEX, Framingham, MA, USA) was used to detect metabolites eluted form the column. The Q-TOF was operated in both positive and negative ion modes. The acquired MS data pretreatments including peak picking, peak grouping, retention time correction, second peak grouping, and annotation of isotopes and adducts was performed using XCMS software. LC–MS raw data files were converted into mzXML format and then processed by the XCMS, CAMERA and metaX toolbox implemented with the R software. Differentially expressed lipid metabolites are list in Supplementary Data 3.
De novo lipid synthesis assay
Glucose-derived lipid biosynthesis was assessed by culture of BMDMs (2 × 106) in complete medium containing 2 μM U-14C-glucose (250 mCi/mmol, PerkinElmer), and stimulated as described in the figure legends. After 24 h incubation time in a cell incubator, the buffer was aspirated, and cells were washed with cold PBS. The lipid fraction was extracted using Folch’s extraction method. The extracted lipids were carefully transferred to scintillation vials and mixed with Opti-Fluor scintillation liquid, and incorporated radioactivity was measured with a MicroBeta Liquid Scintillation Counter (PerkinElmer).
H&E staining and immunofluorescence
Skins, muscles and livers were excised and fixed overnight in 4% formaldehyde. Subsequently, paraffin-embedded samples were segmented and stained with hematoxylin and eosin (H&E). The tissue sections (4 μm) were deparaffinized and rehydrated through xylene and graded alcohols, and washed in running water for a minimum of 5 min. Tissue sections were boiled in citrate buffer for antigen retrieval, and blocked with 1% serum goat for 60 min. The slides were stained with the primary antibody anti-F4/80 (1:100, sc-59171, Santa Cruz) at 4 °C overnight and the secondary Alexa Fluor 488-labeled goat anti-rat IgG antibodies (1:200, bs-0293G, Bioss) at room temperature for 2 h. The slides were stained with the antibody anti-CD206 (1:100, 141705, Biolegend) or anti-MHC class II (1:100, 12-5321-82, Invitrogen) at 4 °C overnight. Nuclei were counterstained with DAPI. Images were acquired using a confocal microscope (LSM980, Zeiss) and analyzed using ZEN 2.3 software.
Metabolite profiling and cytokine measurement
Triglycerides and non-esterified fatty acids were quantified using the Triglyceride assay kit (A110-1-1) and Nonesterified Free fatty acids assay kit (A042-2-1) from Jiancheng Bioengineering Institute. The levels of TGF-β1 (88-8350-22, Invitrogen), PDGFβ (RK00134, ABclonal) and VEGFβ (RK03275, ABclonal) in the supernatant of macrophages were measured by ELISA kits. The levels of IL-10 in the skin wound lysates of mice were determined by an ELISA kit (88-7105-88, Invitrogen).
Lentiviral plasmids production and cell infection
The cDNA encoding STAT3, PPARγ (WT) and PPARγ (T166A) was cloned, and inserted into the pLenti6/v5 lentiviral vector. The lentiviral GV248 plasmid encoding shRNA against Nrf2 were constructed by GeneChem Co., Ltd. The short hairpins against control (shCTR) and Nrf2 (shNrf2) targeting sequences were listed in Supplementary Data 1. The lentiviral particles were produced in HEK-293T cells by co-transfecting with psPAX2 (Addgene #12260) and pMD2.G (Addgene # 12259) using Lipofectamine 2000 (11668500, Invitrogen). 48 h after transfection, conditioned media containing lentiviruses was filtered through 0.45 μm non-pyrogenic filters. Medium containing concentrated lentivirus (by using Lenti-Concentin Virus Precipitation Solution) was used to infect target cells in the presence of 8 μg/ml polybrene. After transduction, cells were treated with blasticidin (10 μg/ml) or puromycin (2 μg/ml) for another 2 days.
CRISPR/Cas9 gene targeting in cells
Lentiviral CRISPR plasmids targeting Pparg, Fasn, Stat3 and Nrf2 were constructed by cloning gRNAs (Supplementary Data 1) into the lentiCRISPR v2 (Addgene #52961) plasmid backbone. For lentivirus production, 4 μg lentiviral CRISPR plasmids were transfected using Lipofectamine 2000 into 293T packaging cells along with 3 μg psPAX2 and 1 μg pMD2.G. RAW 264.7 cell line was transduced with lentiviral particles in the presence of 8 μg/ml polybrene. Virally transduced cells underwent puromycin selection (2 μg/ml).
Human macrophage differentiation from peripheral blood mononuclear cells
De-identified healthy donor blood samples were provided by the SAILYBIO Co., Ltd. Sample acquisitions were approved and regulated by the Shanghai Liquan Hospital Institutional Review Board (XF-WBC-220809). Peripheral blood mononuclear cells (PBMC) from healthy donors were separated by density gradient centrifugation with Lymphocyte separation medium 1.084 (40504ES, YEASEN). Monocyte-enriched fractions were purified from PBMCs immediately after isolation by positive selection with anti-CD14 magnetic beads (130-097-052, Miltenyi Biotech). For macrophage differentiation, selected CD14+ monocytes were seeded on plates in X-VIVO 15 medium (04-418Q, Lonza) supplemented with 10% FBS and 50 ng/ml GM-CSF (91102ES, YEASEN) and cultured for a further 7 days at 37 °C with 5% CO2.
In vitro wound-healing assays
The DFs (1 × 105 cells), HSFs (1 × 105 cells) or HaCaT (1 × 105 cells) were added to the bottom chamber of 12-well. BMDMs (3 × 105 cells) or human PBMC derived macrophages (3 × 105 cells) were added to the upper chamber of 12-well Transwell plates (polycarbonate filter with a 0.45-μm pore, Costar) and cultured for 24 h under standard culture conditions, respectively. On the day of the assay, the DF, HSF or HaCaT cells were wounded with a sterile 200-μl pipette tip. Cells were washed with PBS to remove cell debris before co-cultured with macrophages. The wounded areas at 0, 12 and 24 h were captured using a microscope. The area of the wound was quantified using Image J software. The results are expressed as efficiency of wound closure (1 − [(area of last frame/area of first frame) × 100%]).
Statistics and reproducibility
Statistical analysis was performed using GraphPad Prism software. One-way ANOVA-post-hoc pairwise comparison analysis was used to compare the means of more than two groups. Student’s t test (unpaired, two-tailed) was used to compare the mean value of two groups. Error bars indicated the Standard Error of Mean (s.e.m.). Differences were analyzed by unpaired Student’s t test or one-way ANOVA depending on experimental conditions. Unless otherwise indicated, the experiments were performed independently in triplicate, and n is indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
Thanks to members of Shen’s laboratory for helpful discussions, technical assistance, and critical reading of the manuscript. We thank L. Zhang and R. Ji from the State Key Laboratory of Pollution Control & Resource Reuse for their help in stable isotope tracer experimental design. This work was supported by grants from the Natural Science Foundation of Jiangsu Province (BK20222009 to P.S.), the Guangdong Basic and Applied Basic Research Foundation (2021B1515120016 to P.S.), the National Natural Science Foundation of China (82304492 to N.Y.) and the National Key Research and Development Program of China (2017YFA0506000 to P.S.). Figures 4a and 7e were modified from Servier Medical Art (http://smart.servier.com/), licensed under a Creative Commons Attribution 4.0 Unported License (https://creativecommons.org/licenses/by/4.0/).
Author contributions
S. Zuo, Y. Wang, H. Bao and A. Jian performed experiments and data analysis. Y. Wang and Z. Zhang designed and conducted the bioinformatics experiments. R. Ji and L. Zhang gave technical assistance with stable isotope tracer and lipidomic experiments. N. Yang, Y. Lu, Y. Huang, M. Jia, and Q. Zhang provided scientific suggestions and contributed to the manuscript revision. P. Shen supervised the project and wrote the manuscript. The author(s) read and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks Raffaele Calogero and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
RNA-seq, ChIP-seq, CUT&RUN-seq data have been deposited in the Gene Expression Omnibus under accession code GSE269834. Publicly available RNA-seq and scRNA-seq datasets used in this study were listed in Supplementary Data 2. Source data are provided with this paper.
Code availability
The codes generated or used during the study are available at the GitHub repository (https://github.com/XiaoXxin/Macrophage_Reparative). Other data that support the findings of this study are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shiman Zuo, Yuxin Wang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51736-5.
References
- 1.Gurtner, G. C. et al. Wound repair and regeneration. Nature453, 314–321 (2008). 10.1038/nature07039 [DOI] [PubMed] [Google Scholar]
- 2.Mahdavian Delavary, B. et al. Macrophages in skin injury and repair. Immunobiology216, 753–762 (2011). 10.1016/j.imbio.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 3.Eming, S. A., Wynn, T. A. & Martin, P. Inflammation and metabolism in tissue repair and regeneration. Science356, 1026–1030 (2017). 10.1126/science.aam7928 [DOI] [PubMed] [Google Scholar]
- 4.Lucas, T. et al. Differential roles of macrophages in diverse phases of skin repair. J. Immunol.184, 3964–3977 (2010). 10.4049/jimmunol.0903356 [DOI] [PubMed] [Google Scholar]
- 5.Bosurgi, L. et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science356, 1072–1076 (2017). 10.1126/science.aai8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, S. et al. Efferocytosis fuels requirements of fatty acid oxidation and the electron transport chain to polarize macrophages for tissue repair. Cell Metab.29, 443–456.e5 (2019). 10.1016/j.cmet.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eming, S. A., Murray, P. J. & Pearce, E. J. Metabolic orchestration of the wound healing response. Cell Metab.33, 1726–1743 (2021). 10.1016/j.cmet.2021.07.017 [DOI] [PubMed] [Google Scholar]
- 8.Knipper, J. A. et al. Interleukin-4 receptor alpha signaling in myeloid cells controls collagen fibril assembly in skin repair. Immunity43, 803–816 (2015). 10.1016/j.immuni.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vannella, K. M. & Wynn, T. A. Mechanisms of organ injury and repair by macrophages. Annu. Rev. Physiol.79, 593–617 (2017). 10.1146/annurev-physiol-022516-034356 [DOI] [PubMed] [Google Scholar]
- 10.Gurevich, D. B. et al. Live imaging of wound angiogenesis reveals macrophage orchestrated vessel sprouting and regression. EMBO J.37, e97786 (2018). [DOI] [PMC free article] [PubMed]
- 11.O’Neill, L. A. & Pearce, E. J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med.213, 15–23 (2016). 10.1084/jem.20151570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van den Bossche, J., O’Neill, L. A. & Menon, D. Macrophage immunometabolism: where are we (going)? Trends Immunol.38, 395–406 (2017). 10.1016/j.it.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 13.Faas, M. et al. IL-33-induced metabolic reprogramming controls the differentiation of alternatively activated macrophages and the resolution of inflammation. Immunity54, 2531–2546.e5 (2021). 10.1016/j.immuni.2021.09.010 [DOI] [PubMed] [Google Scholar]
- 14.Dang, E. V. et al. Oxysterol restraint of cholesterol synthesis prevents AIM2 inflammasome activation. Cell171, 1057–1071.e11 (2017). 10.1016/j.cell.2017.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauterbach, M. A. et al. Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP-citrate lyase. Immunity51, 997–1011.e7 (2019). 10.1016/j.immuni.2019.11.009 [DOI] [PubMed] [Google Scholar]
- 16.Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature496, 238–242 (2013). 10.1038/nature11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills, E. L. et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell167, 457–470.e13 (2016). 10.1016/j.cell.2016.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha, S., Shalova, I. N. & Biswas, S. K. Metabolic regulation of macrophage phenotype and function. Immunol. Rev.280, 102–111 (2017). 10.1111/imr.12603 [DOI] [PubMed] [Google Scholar]
- 19.Yan, J. & Horng, T. Lipid metabolism in regulation of macrophage functions. Trends Cell Biol.30, 979–989 (2020). 10.1016/j.tcb.2020.09.006 [DOI] [PubMed] [Google Scholar]
- 20.Kelly, B. & Pearce, E. L. Amino assets: how amino acids support immunity. Cell Metab.32, 154–175 (2020). 10.1016/j.cmet.2020.06.010 [DOI] [PubMed] [Google Scholar]
- 21.Lane, A. N. & Fan, T. W. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res.43, 2466–2485 (2015). 10.1093/nar/gkv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunmeir, R. & Xu, F. Functional regulation of PPARs through post-translational modifications. Int. J. Mol. Sci.19, 1738 (2018). [DOI] [PMC free article] [PubMed]
- 23.Di Conza, G. et al. Tumor-induced reshuffling of lipid composition on the endoplasmic reticulum membrane sustains macrophage survival and pro-tumorigenic activity. Nat. Immunol.22, 1403–1415 (2021). 10.1038/s41590-021-01047-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, M. et al. A STAT3 palmitoylation cycle promotes TH17 differentiation and colitis. Nature586, 434–439 (2020). 10.1038/s41586-020-2799-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi, Y. et al. Loss of fatty acid degradation by astrocytic mitochondria triggers neuroinflammation and neurodegeneration. Nat. Metab.5, 445–465 (2023). 10.1038/s42255-023-00756-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, M. J. et al. SH2 domains serve as lipid-binding modules for pTyr-signaling proteins. Mol Cell62, 7–20 (2016). 10.1016/j.molcel.2016.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, C. et al. Noncanonical STAT3 activation regulates excess TGF-beta1 and collagen I expression in muscle of stricturing Crohn’s disease. J. Immunol.194, 3422–3431 (2015). 10.4049/jimmunol.1401779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagone, P. & Jackowski, S. Membrane phospholipid synthesis and endoplasmic reticulum function. J. Lipid Res.50, S311–S316 (2009). [DOI] [PMC free article] [PubMed]
- 29.Ruckerl, D. et al. Induction of IL-4Ralpha-dependent microRNAs identifies PI3K/Akt signaling as essential for IL-4-driven murine macrophage proliferation in vivo. Blood120, 2307–2316 (2012). 10.1182/blood-2012-02-408252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, N. et al. Blockage of PPARgamma T166 phosphorylation enhances the inducibility of beige adipocytes and improves metabolic dysfunctions. Cell Death Differ.30, 766–778 (2023). 10.1038/s41418-022-01077-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wynn, T. A. & Vannella, K. M. Macrophages in tissue repair, regeneration, and fibrosis. Immunity44, 450–462 (2016). 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wynn, T. A., Chawla, A. & Pollard, J. W. Macrophage biology in development, homeostasis and disease. Nature496, 445–455 (2013). 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosser, D. M., Hamidzadeh, K. & Goncalves, R. Macrophages and the maintenance of homeostasis. Cell Mol. Immunol.18, 579–587 (2021). 10.1038/s41423-020-00541-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wculek, S. K. et al. Oxidative phosphorylation selectively orchestrates tissue macrophage homeostasis. Immunity56, 516–530.e9 (2023). 10.1016/j.immuni.2023.01.011 [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg, G. et al. Immunometabolic crosstalk during bacterial infection. Nat. Microbiol.7, 497–507 (2022). 10.1038/s41564-022-01080-5 [DOI] [PubMed] [Google Scholar]
- 36.Wculek, S. K. et al. Metabolism of tissue macrophages in homeostasis and pathology. Cell Mol. Immunol.19, 384–408 (2022). 10.1038/s41423-021-00791-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van den Bossche, J. & van der Windt, G. J. W. Fatty acid oxidation in macrophages and T cells: time for reassessment? Cell Metab.28, 538–540 (2018). 10.1016/j.cmet.2018.09.018 [DOI] [PubMed] [Google Scholar]
- 38.Wang, F. et al. Glycolytic stimulation is not a requirement for M2 macrophage differentiation. Cell Metab.28, 463–475.e4 (2018). 10.1016/j.cmet.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gauthier, T. & Chen, W. Modulation of macrophage immunometabolism: a new approach to fight infections. Front. Immunol.13, 780839 (2022). 10.3389/fimmu.2022.780839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castegna, A. et al. Pharmacological targets of metabolism in disease: opportunities from macrophages. Pharmacol. Ther.210, 107521 (2020). 10.1016/j.pharmthera.2020.107521 [DOI] [PubMed] [Google Scholar]
- 41.Li, M. et al. Metabolism, metabolites, and macrophages in cancer. J. Hematol. Oncol.16, 80 (2023). 10.1186/s13045-023-01478-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolliniati, O. et al. Metabolic regulation of macrophage activation. J. Innate Immun.14, 51–68 (2022). 10.1159/000516780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koelwyn, G. J. et al. Regulation of macrophage immunometabolism in atherosclerosis. Nat. Immunol.19, 526–537 (2018). 10.1038/s41590-018-0113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmadian, M. et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nat. Med.19, 557–566 (2013). 10.1038/nm.3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stechschulte, L. A. et al. PPARG post-translational modifications regulate bone formation and bone resorption. EBioMedicine10, 174–184 (2016). 10.1016/j.ebiom.2016.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao, M. et al. Pathologic HIF1alpha signaling drives adipose progenitor dysfunction in obesity. Cell Stem Cell28, 685–701.e7 (2021). 10.1016/j.stem.2020.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El Ouarrat, D. et al. TAZ is a negative regulator of PPARgamma activity in adipocytes and TAZ deletion improves insulin sensitivity and glucose tolerance. Cell Metab.31, 162–173.e5 (2020). 10.1016/j.cmet.2019.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu, E. et al. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARγ. Science274, 2100–2103 (1996). 10.1126/science.274.5295.2100 [DOI] [PubMed] [Google Scholar]
- 49.Choi, J. H. et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature466, 451–456 (2010). 10.1038/nature09291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szanto, A. et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity33, 699–712 (2010). 10.1016/j.immuni.2010.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, S. et al. S100A4 enhances protumor macrophage polarization by control of PPAR-gamma-dependent induction of fatty acid oxidation. J. Immunother. Cancer9, e002548 (2021). [DOI] [PMC free article] [PubMed]
- 52.Okreglicka, K. et al. PPARgamma is essential for the development of bone marrow erythroblastic island macrophages and splenic red pulp macrophages. J. Exp. Med.218, e20191314 (2021). [DOI] [PMC free article] [PubMed]
- 53.Varga, T. et al. Macrophage PPARgamma, a lipid activated transcription factor controls the growth factor GDF3 and skeletal muscle regeneration. Immunity45, 1038–1051 (2016). 10.1016/j.immuni.2016.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall, J. A. et al. Obesity-linked PPARgamma S273 phosphorylation promotes insulin resistance through growth differentiation factor 3. Cell Metab.32, 665–675.e6 (2020). 10.1016/j.cmet.2020.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banks, A. S. et al. An ERK/Cdk5 axis controls the diabetogenic actions of PPARgamma. Nature517, 391–395 (2015). 10.1038/nature13887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li, P. et al. Adipocyte NCoR knockout decreases PPARgamma phosphorylation and enhances PPARgamma activity and insulin sensitivity. Cell147, 815–826 (2011). 10.1016/j.cell.2011.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, H., Wang, H. & Jaenisch, R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc.9, 1956–1968 (2014). 10.1038/nprot.2014.134 [DOI] [PubMed] [Google Scholar]
- 58.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e29 (2021). 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol.20, 163–172 (2019). 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linderman, G. C. et al. Zero-preserving imputation of single-cell RNA-seq data. Nat. Commun.13, 192 (2022). 10.1038/s41467-021-27729-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–1086 (2017). 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J.17, 10–12 (2011).
- 63.Kim, D. et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915 (2019). 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 65.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gennady, K., Vladimir, S. & Alexey, S. Fast gene set enrichment analysis. bioRxiv10.1101/060012 (2019).
- 67.Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics14, 7 (2013). 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol.9, R137 (2008). 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics31, 2382–2383 (2015). 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- 72.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
RNA-seq, ChIP-seq, CUT&RUN-seq data have been deposited in the Gene Expression Omnibus under accession code GSE269834. Publicly available RNA-seq and scRNA-seq datasets used in this study were listed in Supplementary Data 2. Source data are provided with this paper.
The codes generated or used during the study are available at the GitHub repository (https://github.com/XiaoXxin/Macrophage_Reparative). Other data that support the findings of this study are available from the corresponding author upon request.