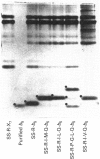
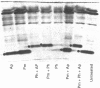
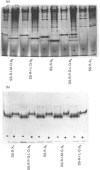
Abstract
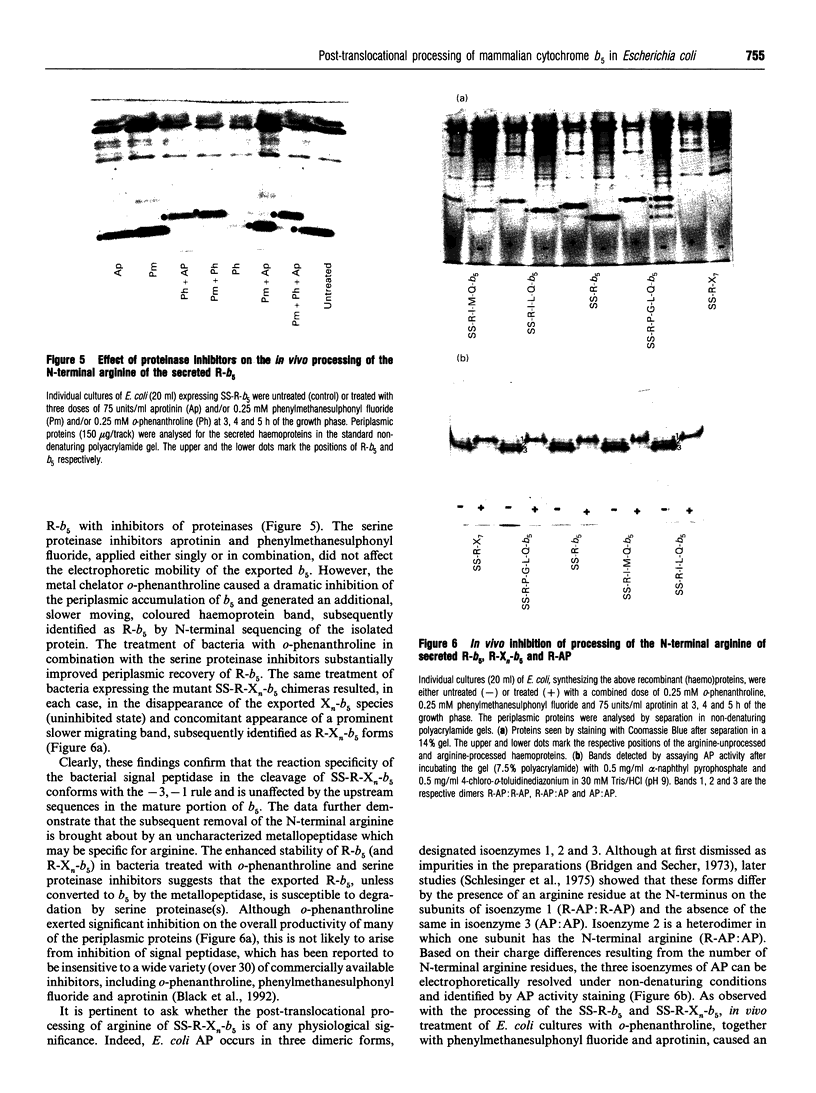
A chimeric precursor interlinked by an arginine residue between the full-length signal sequence of alkaline phosphatase and the eukaryotic cytoplasmic cytochrome b5 was constructed. Expression of the chimeric precursor protein in Escherichia coli resulted in efficient export of spectrally authentic cytochrome b5 into the periplasm [Karim, Harding, Evans, Kaderbhai and Kaderbhai (1993) Bio/Technology 11, 612-618]. On sequencing, the apparent absence of arginine at the N-terminus of the secreted cytochrome b5 implied that the chimera was either miscleaved by signal peptidase or further processed following signal excision by an uncharacterized peptidase. The influence of the N-terminal region of cytochrome b5 on the unusual processing of the chimeric precursor was investigated by engineering a number of variant forms in which the region between Arg+1 and the mature portion of cytochrome b5 was extended and varied. Observations of the in vivo processed patterns of these variant cytochrome b5 forms exported into the periplasm revealed that the absence of arginine was due to neither miscleavage of the translocated precursor by the signal peptidase nor the nature of the early region of cytochrome b5. In fact, the selective excision of the arginine residue occurred subsequent to signal sequence deletion by an aminopeptidase which was sensitive to the metal chelator o-phenanthroline. We show that this aminopeptidase also participates in the trimming of the N-terminal arginine residue of the bacterial alkaline phosphatase to generate the three isoenzymes in the periplasm.
Full text
PDF

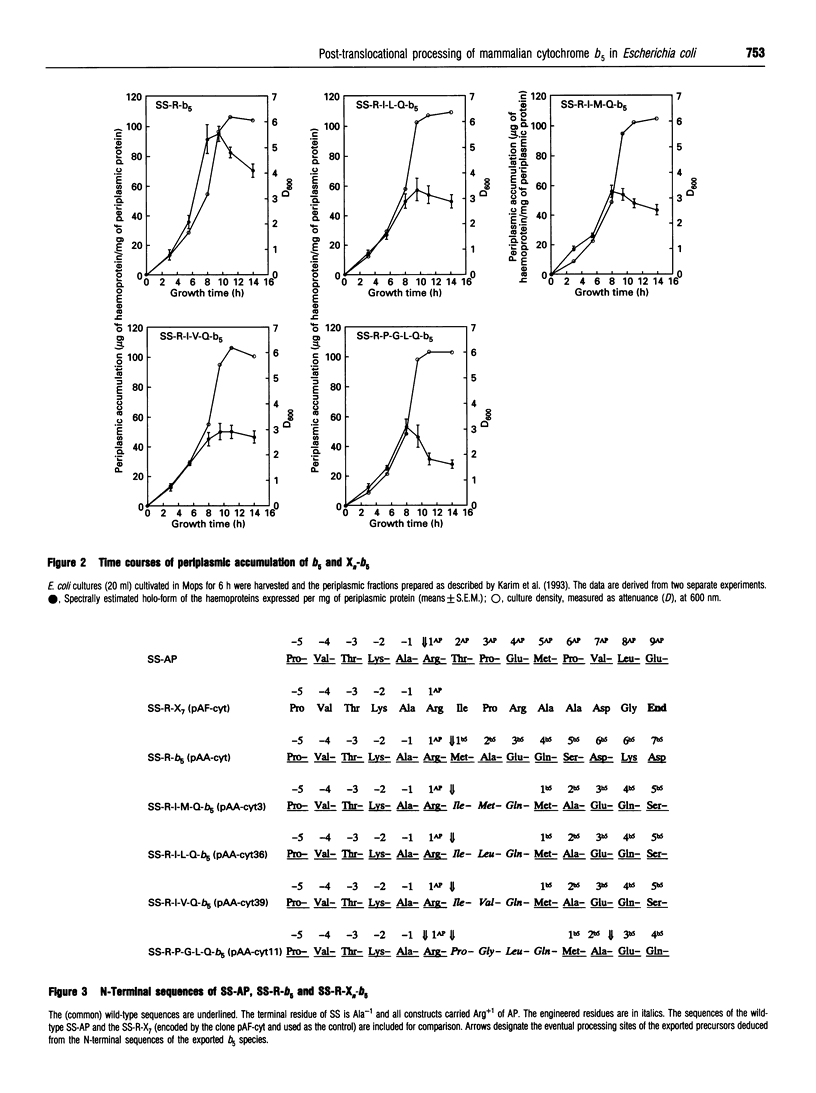


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkocy-Gallagher G. A., Bassford P. J., Jr Synthesis of precursor maltose-binding protein with proline in the +1 position of the cleavage site interferes with the activity of Escherichia coli signal peptidase I in vivo. J Biol Chem. 1992 Jan 15;267(2):1231–1238. [PubMed] [Google Scholar]
- Black M. T., Munn J. G., Allsop A. E. On the catalytic mechanism of prokaryotic leader peptidase 1. Biochem J. 1992 Mar 1;282(Pt 2):539–543. doi: 10.1042/bj2820539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bridgen J., Secher D. S. Molecular heterogeneity of alkaline phosphatase. FEBS Lett. 1973 Jan 1;29(1):55–57. doi: 10.1016/0014-5793(73)80014-6. [DOI] [PubMed] [Google Scholar]
- Chang C. N., Kuang W. J., Chen E. Y. Nucleotide sequence of the alkaline phosphatase gene of Escherichia coli. Gene. 1986;44(1):121–125. doi: 10.1016/0378-1119(86)90050-8. [DOI] [PubMed] [Google Scholar]
- Derman A. I., Beckwith J. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J Bacteriol. 1991 Dec;173(23):7719–7722. doi: 10.1128/jb.173.23.7719-7722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev I. K., Ray P. H. Signal peptidases and signal peptide hydrolases. J Bioenerg Biomembr. 1990 Jun;22(3):271–290. doi: 10.1007/BF00763168. [DOI] [PubMed] [Google Scholar]
- Fikes J. D., Barkocy-Gallagher G. A., Klapper D. G., Bassford P. J., Jr Maturation of Escherichia coli maltose-binding protein by signal peptidase I in vivo. Sequence requirements for efficient processing and demonstration of an alternate cleavage site. J Biol Chem. 1990 Feb 25;265(6):3417–3423. [PubMed] [Google Scholar]
- Gallagher J., Kaderbhai N., Kaderbhai M. A. Gene-dose-dependent expression of soluble mammalian cytochrome b5 in Escherichia coli. Appl Microbiol Biotechnol. 1992 Oct;38(1):77–83. doi: 10.1007/BF00169423. [DOI] [PubMed] [Google Scholar]
- Hultquist D. E. Methemoglobin reduction system of erythrocytes. Methods Enzymol. 1978;52:463–473. doi: 10.1016/s0076-6879(78)52051-x. [DOI] [PubMed] [Google Scholar]
- Karim A., Kaderbhai N., Evans A., Harding V., Kaderbhai M. A. Efficient bacterial export of a eukaryotic cytoplasmic cytochrome. Biotechnology (N Y) 1993 May;11(5):612–618. doi: 10.1038/nbt0593-612. [DOI] [PubMed] [Google Scholar]
- Li P., Beckwith J., Inouye H. Alteration of the amino terminus of the mature sequence of a periplasmic protein can severely affect protein export in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7685–7689. doi: 10.1073/pnas.85.20.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. Proteolysis in protein import and export: signal peptide processing in eu- and prokaryotes. Experientia. 1992 Feb 15;48(2):118–129. doi: 10.1007/BF01923506. [DOI] [PubMed] [Google Scholar]
- Nothwehr S. F., Gordon J. I. Eukaryotic signal peptide structure/function relationships. Identification of conformational features which influence the site and efficiency of co-translational proteolytic processing by site-directed mutagenesis of human pre(delta pro)apolipoprotein A-II. J Biol Chem. 1989 Mar 5;264(7):3979–3987. [PubMed] [Google Scholar]
- Oroszlan S., Luftig R. B. Retroviral proteinases. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger M. J., Andersen L. Multiple molecular forms of the alkaline phosphatase of Escherichia coli. Ann N Y Acad Sci. 1968 Jun 14;151(1):159–170. doi: 10.1111/j.1749-6632.1968.tb11886.x. [DOI] [PubMed] [Google Scholar]
- Wickner W., Driessen A. J., Hartl F. U. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem. 1991;60:101–124. doi: 10.1146/annurev.bi.60.070191.000533. [DOI] [PubMed] [Google Scholar]
- von Heijne G. The signal peptide. J Membr Biol. 1990 May;115(3):195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]