Abstract
Accurate detection and quantification of mRNA isoforms from nanopore long-read sequencing remains challenged by technical noise, particularly in single cells. To address this, we introduce Isosceles, a computational toolkit that outperforms other methods in isoform detection sensitivity and quantification accuracy across single-cell, pseudo-bulk and bulk resolution levels, as demonstrated using synthetic and biologically-derived datasets. Here we show Isosceles improves the fidelity of single-cell transcriptome quantification at the isoform-level, and enables flexible downstream analysis. As a case study, we apply Isosceles, uncovering coordinated splicing within and between neuronal differentiation lineages. Isosceles is suitable to be applied in diverse biological systems, facilitating studies of cellular heterogeneity across biomedical research applications.
Subject terms: Computational biology and bioinformatics, Transcriptomics, RNA sequencing, Transcriptomics, Computational models
Analysis of nanopore long-read sequencing is challenged by technical noise, particularly in single cells. Here, authors introduce Isosceles, a toolkit for accurate isoform detection, quantification, and flexible downstream analysis of long-read data at single-cell, pseudo-bulk and bulk resolutions.
Introduction
Alternative splicing (AS) contributes to the generation of multiple isoforms from nearly all human multi-exon genes, vastly expanding transcriptome and proteome complexity across healthy and disease tissues1. However, current short-read RNA-seq technology is restricted in its ability to cover most exon-exon junctions in isoforms. Consequently, the detection and quantification of alternative isoforms is limited by expansive combinatorial possibilities inherent in short-read data2. Short read lengths can impose additional challenges at the single-cell level. For example, nearly all isoform information is lost with UMI-compatible high-throughput droplet-based protocols which utilize short-read sequencing at the 3′ or 5′ ends3. Recent advances in long-read sequencing technologies provide an opportunity to overcome these limitations and study full-length transcripts and complex splicing events at both bulk and single-cell levels, yet downstream analysis must overcome low read depth, high base-wise error, pervasive truncation rates, and frequent alignment artifacts4. To approach this task, computational tools have been developed for error prone spliced alignment5 and isoform detection/quantification6–13. However, these tools vary widely in accuracy for detection and quantification14, their applicability to bulk or single-cell resolutions, and in their capabilities for downstream analysis.
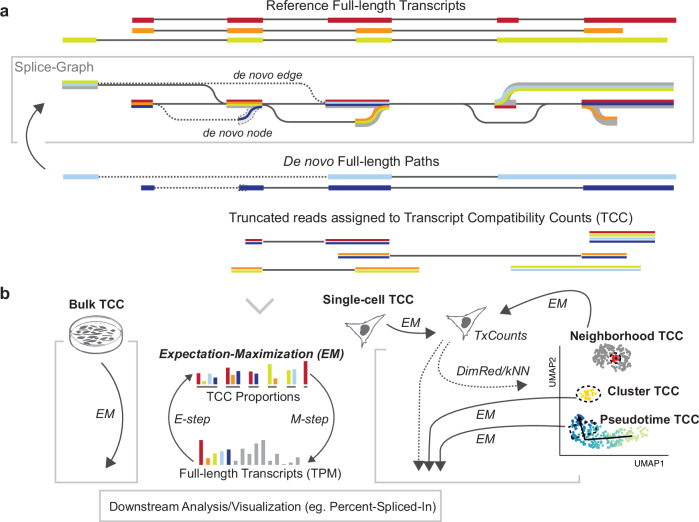
Here we present Isosceles (the Isoforms from single-cell, long-read expression suite); a computational toolkit for reference-guided de novo detection, accurate quantification, and downstream analysis of full-length isoforms at either single-cell, pseudo-bulk, or bulk resolution levels (https://github.com/Genentech/Isosceles). In order to achieve a flexible balance between identifying de novo transcripts and filtering misalignment-induced splicing artifacts, the method utilizes acyclic splice-graphs to represent gene structure15. In the graph, nodes represent exons, edges denote introns, and paths through the graph correspond to whole transcripts (Fig. 1a). The splice-graph and transcript set can be augmented from observed reads containing novel nodes and edges that surpass reproducibility thresholds through a de novo discovery mode, enhancing the adaptability of the analysis. In the process, sequencing reads are classified relative to the reference splice-graphs as either node-compatible (utilizing known splice-sites) or edge-compatible (utilizing known introns), and further categorized as truncated or full-length (Fig. 1a). Full-length reads can be directly assigned to known transcripts, meanwhile those representing novel transcript paths are assigned stable hash identifiers. These identifiers facilitate ease of matching de novo transcripts across data from the same genome build, irrespective of sequencing run, biological sample, or independent studies. In contrast, truncated reads may introduce ambiguity in terms of their transcript of origin, reflecting a challenge commonly found in short-read data analysis. To address this, we utilize a concept developed for short-read methods, Transcript Compatibility Counts (TCC)16, as the intermediate quantification of all reads. TCCs are used to obtain the maximum likelihood estimate of transcript expression through the expectation-maximization (EM) algorithm (in ref. 17,18; see Methods). This approach tackles another challenge: accurately quantifying transcripts at multiple single-cell resolution levels. First, transcripts can be quantified through EM within single-cells, which can be subsequently used to obtain a neighbor graph and low dimensional embedding (eg. with common tools like Seurat19). Second, transcripts can be quantified at the pseudo-bulk level through EM on the TCCs summed within cell groupings (Fig. 1b). This configuration enables versatility of quantification; pseudo-bulk can be defined by the user in numerous ways, such as through marker labeling, clustering, windows along pseudotime, or for each cell based on its k-nearest neighbors (kNN). Downstream statistical analysis and visualization for percent-spliced-in and alternative start and end sites is seamlessly integrated to facilitate biological interpretation of isoforms. Our performance evaluations demonstrate that these features act together to enhance the accuracy of isoform detection and quantification, particularly at lower expression levels. These findings support Isosceles as a robust and performant tool for long-read transcriptome analysis across resolution levels.
Fig. 1. Schematic overview of Isosceles design.
a Splice-graph building and path representation of transcripts (colored lines). Augmentation with de novo nodes and edges (dashed). Ambiguous reads are assigned to Transcript Compatibility Counts (TCCs) to be quantified using the expectation-maximization (EM) algorithm (bottom; panel b). b The Isosceles approach to multi-resolution quantification using the EM algorithm. Transcripts quantified from single-cell TCCs using EM (gray cell, right) can be used for dimensionality reduction (DimRed) with UMAP or to derive a k-nearest neighbors graph (kNN). The original single-cell TCCs can be grouped based on user-defined pseudo-bulk definition and transcripts re-quantified, either for clusters/markers or for each cell based on its neighborhood from kNN. Figure 1/panel b, created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Results
Isosceles is accurate for transcript discovery and quantification
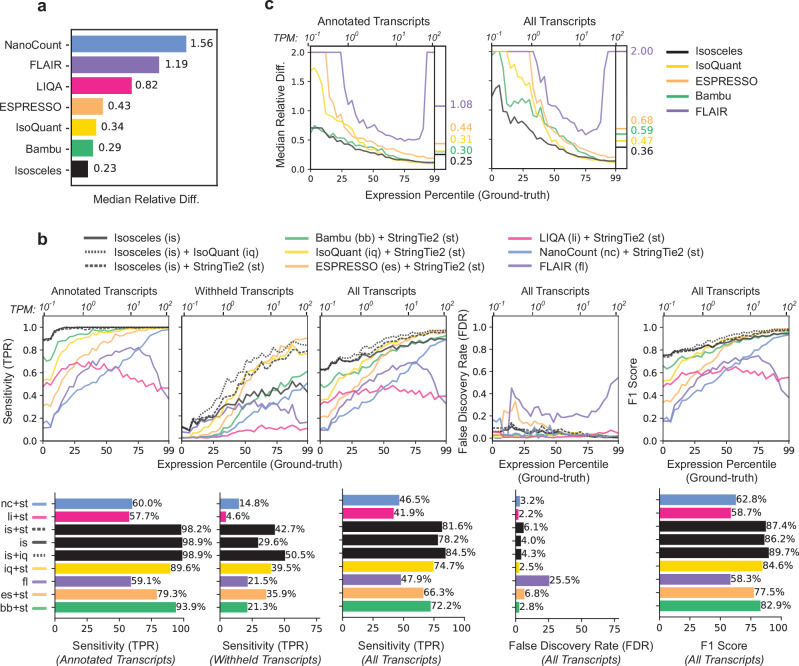
To robustly assess Isosceles performance against a wide-array of currently available software6–13, we simulated ground-truth nanopore reads from reference transcripts proportional to the bulk expression profile of an ovarian cell line, IGROV-1, using NanoSim20 (see Methods). In the evaluation of annotated transcript quantification against the ground-truth, Isosceles outperforms other programs, achieving a highly correlated Spearman coefficient of 0.96 (Supplementary Fig. 1b). Bambu was the next best method at 0.92, while both IsoQuant and ESPRESSO were lower at 0.88. Assessing quantification error through absolute relative difference, Isosceles decreases median and mean error by 21% compared to the next most accurate method, Bambu (0.23 vs. 0.29 and 0.41 vs. 0.52; Fig. 2a and Supplementary Fig. 1a). Importantly, the reduction in error over other methods is even more pronounced, demonstrating ~45% lower error than the median performer ESPRESSO, and 67-85% lower error than the worst performer NanoCount due to lack of detection of many simulated transcripts (Fig. 2a and Supplementary Fig. 1a).
Fig. 2. Quantification and transcript detection benchmarks against ground-truth using simulated long-read data.
a Median relative difference of transcripts per million [TPM] values as defined by abs(ground_truth - predicted) / ((ground_truth + predicted)/2) for each method on reference transcripts. b Downsampling benchmarks for 30% transcripts withheld. Transcript detection defined as TPM > 0, the TPR, FDR and F1 score metrics as a function of the expression percentile (primary x-axis) and TPM values (secondary x-axis) of the simulated transcripts. For each program, the better of either single-program or pre-detection combination is plotted (see Supplementary Fig. 2 for all combinations), with TPR stratified by annotated, withheld and all transcripts alongside FDR and F1 score, with overall values plotted as bars below the graphs. c Median relative difference of annotated and withheld transcripts (30% downsampling) as a function of the simulated expression level, as defined for panel b. Source data are provided as Source Data files.
Since detection of both known and novel transcripts is a major attraction of long-read sequencing, we investigated the ability of various methods to detect 10%, 20% or 30% of transcripts when they are withheld from the annotation file (3269, 6537 and 9801 transcripts respectively; 30% in Fig. 2b, 10-30% in Supplementary Fig. 2a-c). Here, detection is defined as output of a transcript annotation with a splicing structure correctly matching a simulated transcript (irrespective of transcript start/end positions) and a quantification value greater than zero in transcripts per million (TPM > 0). We calculate the true-positive rate (TPR) as the number of correct transcripts detected from the total number with reads simulated. The false-discovery rate (FDR) is defined as the percentage of incorrect transcripts out of the total detected. The overall F1-score is computed as the harmonic mean of sensitivity (TPR) and precision (1-FDR). Notably, most methods output low TPR even for transcripts that are not withheld from the annotation file, as we illustrate by separating the TPR calculations for annotated and withheld transcripts (Fig. 2b left, Isosceles=98.9% vs. median other=69.7%). Methods such as NanoCount and LIQA do not have a de novo detection mode, so we benchmark them with a pre-detection step using StringTie221, adding this step to other tools for consistency (eg. Bambu, ESPRESSO, and also include IsoQuant alongside single-method detection for Isosceles; Fig. 2b, Supplementary Fig. 2, dashed lines). While ESPRESSO and IsoQuant alone have modestly higher single-method TPR for withheld transcripts than Isosceles (1.0 and 6.0 percentage points respectively; Supplementary Fig. 2a), Isosceles demonstrates the highest single-program TPR across all transcripts, achieving 78.2%, compared to the next best method, IsoQuant, which has a TPR of 74.2% (Supplementary Fig. 2a, middle). Moreover, combining Isosceles with pre-detection by IsoQuant outperforms all other methods and combinations for withheld and annotated transcripts, achieving an 84.5% TPR overall (Fig. 2b). Importantly, Isosceles exhibits this relative gain in sensitivity at lower expression levels than other methods (<10 TPM), and at a reasonable FDR of 4.3%, which is comparable to other programs (Fig. 2b; median FDR of 3.0%). Taken together, Isosceles presents the highest F1-score overall both independently (86.2%; Supplementary Fig. 2a) and with pre-detection using IsoQuant (89.7%; Fig. 2b). When considering the relative difference of quantification for annotated and withheld transcripts, Isosceles performs at 16.7% to 76.9% decrease in median error compared to other methods on annotated transcripts and 23.4% to 82.0% when including de novo (withheld) transcripts across the range of expression levels (Fig. 2c left & right; Supplementary Fig. 3a). Similar to detection sensitivity, the most pronounced improvement in quantification accuracy occurs for the lowest half of expressed transcripts.
In addition to read simulations, we benchmarked performance in the context of nanopore sequencing of synthetic molecule spike-ins. To investigate quantification accuracy, two mixtures containing Sequins22 were analyzed and compared to ground-truth values. Isosceles, Bambu, and IsoQuant achieved equally high Spearman correlations for both mixtures (0.97 and 0.98; Supplementary Fig. 3b), with Isosceles slightly outperforming the others with a lower mean relative difference for the first mixture (0.71 vs. 0.74). To evaluate transcript detection in a synthetic setting that does not resemble a model organism, we utilized SIRV spike-in samples23 alongside annotated and withheld transcript sets (Supplementary Fig. 3c-e). Isosceles, Bambu, and IsoQuant all showed reasonably high precision (96-99%) for read assignment to correct annotations vs. over-annotation decoys (see Methods, Supplementary Fig. 3e). Similar performance was also achieved for transcript detection with all three methods F1-scores falling into the range of 76-78% (Supplementary Fig. 3c). Despite identifying fewer withheld transcripts, Isosceles, with zero false positives, outperformed IsoQuant, which had four false positives and missed five annotated structures (Supplementary Fig. 3d). Taken together, these data suggest Isosceles is able to perform in both well-annotated and non-model organism contexts.
Isosceles also shows favorable performance with the PacBio long-read sequencing platform. We compared nanopore reads from the Nanopore WGS Consortium and PacBio reads from the ENCODE Consortium24 to short-read Illumina quantifications for the same cell line (GM12878; see Methods). We find that Isosceles, IsoQuant and Bambu all perform well in the ONT vs. Illumina comparison, however Isosceles displays slightly higher Spearman correlation for PacBio vs. Illumina (Supplementary Fig. 4a). Lastly, computational speed and RAM usage are important metrics that impact the overall usability and feasibility of large-scale analysis efforts. Benchmarking a 5 M read PromethION IGROV-1 sample, Isosceles emerged as one of the more efficient tools, finishing approximately two hours sooner than the median performer IsoQuant (93.0% of total CPU time) and outperforming the slowest software, ESPRESSO, by two days and two hours (33.4% of total CPU time; Supplementary Fig. 4b).
Isosceles outperforms other methods at single-cell resolution
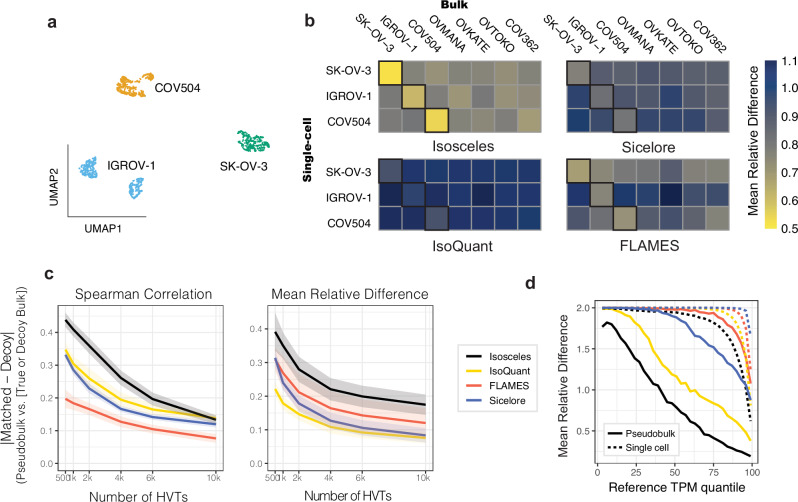
While known ground-truth values are effective for benchmarking performance, the analysis of true biological data introduces additional complexities that synthetic and simulated data may not fully capture. To address this, we benchmark each method’s fidelity of quantification for the same biological sample and ability to differentiate decoy samples across bulk and single-cell resolutions. We perform nanopore sequencing on 10X Genomics single-cell libraries from the pooling of three ovarian cancer cell lines, IGROV-1, SK-OV-3, and COV504, noting that the cells separate into three clusters by transcript expression and that each cluster corresponds to a separate genetic identity according to Souporcell25 (Fig. 3a; see Methods). Conducting bulk nanopore sequencing in parallel on MinION and PromethION platforms, we investigate the consistency of those same cell lines as well as the ability to distinguish against four additional ovarian cancer cell lines sequenced as decoys, namely COV362, OVTOKO, OVKATE, and OVMANA. We find that Isosceles consistently maintains the lowest mean relative difference (24-43% less than other methods) and the highest Spearman correlation (0.87 for Isosceles vs. 0.75 for the next highest, Sicelore) amongst methods quantified on the same cell line in bulk and pseudo-bulk (Fig. 3b and Supplementary Fig. 5a). We further find that this performance is recapitulated when comparing across technical runs, between platforms, and independent of the number of cells included or transcripts compared for IGROV-1 (Supplementary Fig. 4c-d). Isosceles’ application of the EM algorithm is designed to result in greater usage of ambiguous reads, which may ultimately provide higher apparent read depths, and influence quantification accuracy of both matched and decoy comparisons. Therefore, to ensure the observed results reflect accuracy and not merely precision, we stringently consider the consistency of difference between matched and decoy comparisons. To enhance discriminatory power we investigate highly variable transcripts (HVT) between cell lines as determined by each program26. While all methods perform better using fewer HVTs, Isosceles exhibits a 1.3- to 1.4-fold greater absolute difference in Spearman correlation than the next best method, IsoQuant, using between 500 and 4,000 HVTs (Fig. 3c). This outperformance is also observed for mean relative difference as compared to the next best method, FLAMES, and is statistically significant across HVT numbers for both (p value < 5.3×10−5 for Spearman and p value < 3.4 × 10−3 for mean relative diff. vs. next best methods; Wilcoxon paired signed-rank test, see Methods; Fig. 3c). To provide orthogonal support for this conclusion, we simulated 100 cells at approximately 50k reads per cell and 5 M bulk reads for each sample using NanoSim with single-cell and bulk error models respectively (see Methods). We repeated the same benchmark, comparing matched and decoy metrics derived either from each method’s estimates based on simulated reads or from the ground-truth expression profiles used for the simulations. Isosceles outperforms other methods by 1.4 to 2.4-fold across metrics, with the exception of IsoQuant, which is equivalent to Isosceles for Spearman correlation only (p value = 0.5; Supplementary Fig. 5c). Last, we compare the simulated single-cell and pseudo-bulk quantifications for IGROV-1 to ground-truth. While all methods show inflated error for single-cells compared to pseudo-bulk, Isosceles harbors lower average error than other methods for both, demonstrating quantification accuracy even in a data-sparse context (Fig. 3d).
Fig. 3. Quantification fidelity and cell type benchmark using single-cell nanopore data.
a 2D UMAP embedding of transcript-expression level quantifications from nanopore data of pooled IGROV-1, SK-OV-3, and COV504 ovarian cell lines, subsequently colored by genetic identity (according to Souporcell). b Mean relative difference (color scale) of each program’s quantifications across resolutions (pseudo-bulk vs. bulk data) for the top 4,000 highly variable transcripts. c Absolute difference between matched and decoy cell lines across a range of 500-10,000 highly variable transcripts (HVT) comparing mean relative difference and Spearman correlation metrics (shaded ribbons provide the upper and lower bounds of std. error). d Mean relative difference (as defined for Fig. 2a) between ground-truth and estimated TPM values from simulated reads at pseudo-bulk (solid lines) and single-cell level (dashed lines). Source data are provided as Source Data files.
Isosceles enables biological discovery with single-cell data
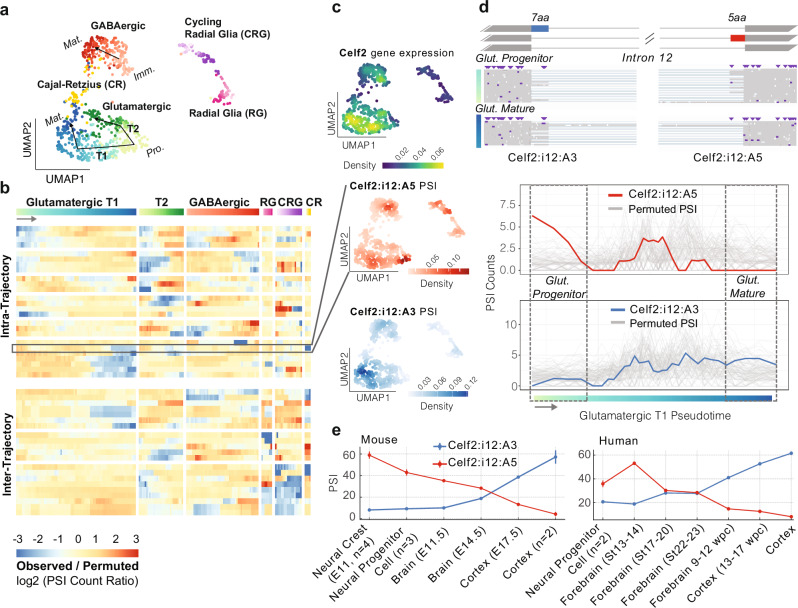
Isosceles’ capabilities for accurate and flexible quantification also enhance downstream analysis and biological discovery. To demonstrate, we reanalyzed 951 single-cell nanopore transcriptomes from a mouse E18 brain. Investigating transcriptional markers (Supplementary Fig. 6), we observe the major cell types identified in the original study using Sicelore9. Isosceles quantifications provide greater resolution however, separating differentiating glutamatergic neurons into two distinct trajectories instead of one (annotated here as T1 and T2), in addition to the single GABAergic trajectory using Slingshot27 (Fig. 4a). We also observe separation of radial glia and glutamatergic progenitor cells, which were connected in the original study. Isosceles’ versatility of pseudo-bulk quantification coupled to generalized linear models (GLM), further distinguishes downstream experimental design capabilities for biological discovery. For example, to investigate transcriptional dynamics within trajectories we apply the EM algorithm to pseudo-bulk windows, quantifying transcript expression as a function of pseudotime. To summarize individual transcript-features, Isosceles provides the inclusion levels of alternative splicing (AS) events, such as alternative exons and splice sites quantified as percent-spliced-in2,28 [PSI] or counts-spliced-in [CSI] (see Methods). In order to test for differential inclusion versus exclusion as a function of pseudotime (or any other condition), Isosceles seamlessly integrates with the DEXseq package29 to utilize GLMs in the context of splicing (see Methods). Applying the method identifies 25 AS events changing within trajectories as well as 21 changing between trajectories respectively (Supplementary Data 1). Isosceles also implements the ‘isoform switching’ approach utilized in the original study (see Methods). However, we note that applying this method only identifies transcripts changing between major clusters, and none within glutamatergic or GABAergic neurogenesis trajectories (including the exemplar genes Clta and Myl6 presented in the original study; eg. Supplementary Fig. 7a).
Fig. 4. Analysis of 951 single-cell nanopore transcriptomes from a mouse E18 brain.
a 2D UMAP embedding from PCA performed jointly on variable gene and transcript features. Gradient coloring by pseudotime according to each trajectory. Neural Progenitor Cells are abbreviated Pro., Immature neurons Imm. and Mature neurons Mat. T1 and T2 describe the two trajectories of Glutamatergic neurogenesis observed. b Heatmap of significant AS events colored by the ratio of observed CSI vs. permuted CSI for permutations within (top) or across all (bottom) trajectories. c UMAP density column from top to bottom: Celf2 gene expression, Celf2 alternative 5′ splice site (A5) in intron 12 (Celf2:i12:A5, chr2:6560659-6560670; row highlighted in panel b), and juxtaposed alternative 3′ splice site (A3) for intron 12 (Celf2:i12:A3, chr2:6553965-6553982). d AS event diagram on the top of Celf2 gene intron 12 where exons are shown as boxes and introns as lines, with the A5 event in red, and the A3 event in blue, with reads from cells in the beginning and the end of the glutamatergic T1 trajectory shown below respectively (boxed regions from the bottom panel). In the bottom panel are plots of CSI for windows along pseudotime for the observed data (A5, red) and (A3, blue) plotted over the background permutations in gray. e Mean PSI values of sample group quantifications from the human (Hsa38, left) and mouse (Mmu10, right) VastDB splicing databases30. Standard error is provided as bars (for sample groups with n > 1 samples), with source accession identifiers for each sample provided in Supplementary Data 2 and raw values in Source Data.
One major challenge in the interpretation of single-cell data at the transcript-level (or event-level) is that fluctuations in detection or quantification may be attributable to gene expression changes alone. To decouple splicing dynamics and visualize them independently, we utilize a permutation-based approach. We estimate a background distribution by shuffling each gene’s splicing quantification among cells expressing that gene (within and between trajectories). We then visualize log ratios of the observed CSI values versus the mean expected CSI from these permutations (Fig. 4b and Supplementary Fig. 7b; see Methods). Here, we observe AS events that exhibit precise changes within specific neuronal differentiation trajectories (such as only T1 or T2), including several RNA binding proteins (eg. Celf2, Hnrnpa2b1, Luc7l3, Ythdc1). Exemplifying a unique mode of alternative splicing in the gene Celf2, we observe a coordinated switch from one alternative donor splice site to an alternative acceptor splice site in the same intron as cells differentiate from glutamatergic progenitor to mature neurons (T1 trajectory, Fig. 4c-d). To validate the statistical significance of this event, we compare observed to permuted values using a stringent empirical test (see Methods). Here, we find the splicing-change is robustly independent of the overall changes in Celf2 expression that simultaneously occur (Fig. 4c-d and Supplementary Fig. 8c; p value < 3.8 × 10−4). Underscoring biological significance, we note the two alternative splice sites have orthologs in other mammalian species (as annotated in VastDB30) and high sequence conservation in the intronic region surrounding both splice sites (Supplementary Fig. 8a-b). We show the mutual exclusivity and switch-like splicing change are similarly conserved in human and mouse, recapitulating the longitudinal observation across embryonic brain samples from bulk short-read datasets30 (Fig. 4e), including an in vitro study of mouse neuronal differentiation31 (Supplementary Fig. 8d).
Discussion
In summary, Isosceles is a computational toolkit with favorable performance compared to other methods, as demonstrated through rigorous benchmarks on simulated, synthetic spike-in, and biological data from nanopore sequencing across ovarian cell lines. In these benchmarks, Isosceles performs transcript detection and quantification with accuracy, revealing improvements over existing methods that are most pronounced at lower expression levels. Notably, transcription factors and other regulatory proteins typically exhibit low gene expression levels, accompanied by rapid, fine-tuned regulation in mRNA and protein turnover rates32. Such regulatory genes are frequently the focus of single-cell biological investigations, underscoring the importance of precision in this range. Through multi-resolution sequencing of ovarian cancer cell lines, we benchmark fidelity of quantification, demonstrating Isosceles’ performant capacity to consistently reproduce results for the same sample, and to differentiate among related yet distinct samples. Such demultiplexing of pooled samples is both a practical task in single-cell analysis33, and analogous to the identification of distinct cell types or lineages in single-cell studies where technical noise and data sparsity are common challenges. For example, intrinsic differences between cell lines, even those of the same tissue origin, may be more substantial than many biological changes typically investigated in biomedical research.
We observe that some methods exhibit variability in performance between simulated and biological benchmarks (eg. Figure 3c vs. Supplementary Fig. 5c), likely attributable to inherent differences between real and simulated long-read data. However, Isosceles remains consistently performant, illustrating accurate quantification in multiple contexts. As a reference-guided method, it is designed to excel in the setting of well-annotated model organisms, such as human and mouse. However, we note that Isosceles also handles SIRV synthetic molecule spike-ins effectively, which feature non-canonical splice sites and artificial sequence content. These findings underscore Isosceles’ methodological robustness and support its utility in multiple settings.
We further illustrate that these performant capabilities are enabling in the context of biological discovery. In our case study, we utilize Isosceles to uncover the dynamics of alternative splicing in differentiating neurons. Here, Isosceles provides enhanced resolution and reveals numerous AS events not reported in the original study. Importantly, these results reveal fine-tuned regulation within fate-determined trajectories and not only between major clusters (eg. radial glia vs. mature neurons). Among these events are genes encoding disease relevant RNA binding proteins that are themselves implicated in the regulation of neuronal differentiation. The Celf2 gene, for instance, plays a central role in neurogenesis, as it modulates the translation of target mRNAs through its shuttling activity34. The example in Celf2 (presented in Fig. 4) highlights a switch-like splicing event that results in a conserved substitution of five to seven amino acids within the protein’s disordered region. This is akin to peptide changes introduced by microexons, which have been attributed functional roles in neurogenesis, including translational control of mRNAs through recruitment to membrane-less condensates, and dysregulation in disease35–37. These results demonstrate that Isosceles is an effective method for hypothesis generation and biological discovery, offering insight into the splicing dynamics of a key regulator of differentiation in our case study.
Taken together, Isosceles is a flexible toolkit for the analysis of long-read bulk and single-cell sequencing that outperforms existing methods in detection and quantification across biological resolution levels. Based on its accuracy and flexibility for experimental designs, Isosceles will significantly aid researchers in transcriptomic studies across diverse biological systems.
Methods
Isosceles Splice-graphs
Splice-graph compatibility is defined for reads using various stringency levels to match their concordance with existing knowledge. Reads are classified based on compatibility as Annotated Paths (AP), Path Compatible (PC), Edge Compatible (EC), Node Compatible (NC), De-novo Node (DN), Artifact Fusion (AF), Artifact Splice (AS), and Artifact Other (AX). AP refers to full-length transcript paths that perfectly match a reference transcript from the input gene annotation and are quantified by default. PC reads follow transcript paths that are a traversal of an AP, and may be truncated or full-length or with differing transcript start or end positions. EC reads traverse annotated splice-graph edges (introns) and may be truncated or full-length. NC reads are paths that traverse only annotated splice-graph nodes (splice-sites) but contain at least one novel edge. DN reads have paths that traverse a de novo node (splice-site). AF reads traverse paths connecting at least two splice-graphs for annotated genes that do not share introns with each other. AS reads are assigned to genes, but traverse an unknown and irreproducible node (splice-site), while AX reads lack compatibility due to ambiguous strand or lack of gene assignment.
Reads are also classified based on their truncation status, which includes Full-Length (FL), 5′ Truncation (5 T), 3′ Truncation (3 T), Full-Truncation (FT), and Not Applicable (NA). AP transcripts are automatically annotated as FL, and truncation status is checked only for PC, EC, NC, and DN transcripts. AF, AS, and AX transcripts are automatically labeled NA. Reference transcripts used for truncation status classification are recommended to be filtered to only the GENCODE ‘basic’ dataset (tag=’basic’), but also could be all transcripts in the provided annotations, as decided by the user. Full-length reads are those whose paths splice from a first exon (sharing a reference transcripts first 5′ splice site) and whose paths splice to a last exon (sharing a reference transcripts final 3′ splice site).
To add nodes with one or more de novo splice sites to the splice-graph, each splice-site must meet two conditions: it is observed in at least the minimum number of reads (default: 2) and it is connected to a known splice site in the splice-graph with least a minimum fraction (default: 0.1) of that known splice site’s connectivity. Additionally, annotations for known transcripts and genes are merged and extended based on specific criteria. For example, any annotated genes sharing introns with each other are merged into one gene and given a new gene_id & gene_symbol (comma-separated list of original Ensembl IDs and gene symbols). Annotated spliced (and unspliced) transcripts sharing the same intron structure, as well as transcript start and end bins (default bin size: 50 bp) are merged together and given a unique transcript identifier.
The method offers three modes of extending annotations to include de novo transcripts: strict, de_novo_strict, and de_novo_loose. In the strict mode, only AP transcripts are detected/quantified. In the de_novo_strict mode, AP transcripts and filtered FL transcripts of the EC and NC classes are included in quantification. In the de_novo_loose mode, AP transcripts and filtered FL transcripts of the EC, NC, and DN classes can be included.
For downstream analysis of individual transcript features, AS events are defined as the set of non-overlapping exonic intervals that differ between transcripts of the same gene. These are quantified as percent-spliced-in or counts-spliced-in according to the sum of the relative expression or the raw counts of the transcripts that include the exonic interval respectively. AS events are classified into different types similar to previous methods analyzing splicing from short-read data2, including core exon intervals (CE), alternative donor splice sites (A5), alternative acceptor splice sites (A3), and retained introns (RI). Isosceles can also quantify tandem untranslated regions in the first or last exons including transcription start sites (TSS) and alternative polyadenylation sites (TES).
Isosceles quantification
We use the Expectation-Maximization (EM) algorithm to obtain the maximum likelihood estimate (MLE) of transcript abundances, as used previously in transcript quantification methods for short-read data such as our prior software Whippet2, or the approach’s conceptual precursors RSEM17 and/or Kallisto18. Specifically, we quantify transcript compatibility counts (TCCs) based on fully contained overlap of reads to the spliced transcript genomic intervals (including an extension [default: 100 bp] for transcript starts/ends), with strand for unspliced reads ignored by default. For computational efficiency, TCCs matching more than one gene are disallowed in the current version. The likelihood function is defined for transcript estimation as it is defined for short-read data with Whippet2, as L(α) proportional to the product, over all reads, of the sum of the probabilities α(t) of selecting a read from each compatible transcript t, divided by the effective length of t. However, due to the long length of nanopore reads, we define effective transcript length here to be the maximum of the mean read length or the transcript’s actual length, then divided by the mean read length. This directly accommodates shorter transcripts which would be fully spanned by the average read and are thus assigned an effective length of 1.0, whereas longer transcripts are represented proportionally to that value. In contrast, the user defined parameter specifying single-cell data does not use length normalization due to the anchoring of reads to the 5′ or 3′ ends of transcripts which assumes read coverage irrespective of transcript length. The EM algorithm iteratively optimizes the accuracy of transcript abundance estimates derived from TCCs, continuing until the absolute difference between transcript fractions is less than a given threshold (default:0.01) between iterations, or until the maximum number of iterations is reached (default: 250).
Simulating ONT data
In this study, the Ensembl 90 genome annotation (only transcripts with the GENCODE ‘basic’ tag) was used for all simulations, focusing specifically on spliced transcripts of protein-coding genes to exclude single-isoform non-coding genes. In order to simulate data with realistic transcriptional profiles, we quantified the expression of reference annotations in IGROV-1 cells using publicly available short-read data ([sample, project] accession IDs: [SRR8615844, PRJNA523380]; https://www.ebi.ac.uk/ena/browser/view/SRR8615844) and Whippet v1.7.3 using default settings. Only transcripts with non-zero expression in IGROV-1 were retained for simulations. For detection benchmarks, the Ensembl 90 annotation file (in Gene Transfer Format [GTF]) was randomly downsampled such that the longest transcript of each gene was always retained to ensure at least one full-length major isoform for each gene (by 10%, 20%, and 30% downsampling, where 99.8-100.0% of downsampled transcripts had unique exon-intron architectures). To assess performance in de novo transcript detection, each program was run individually (if de novo detection was supported) and in tandem with StringTie2 for a pre-detection step (also including IsoQuant for pre-detection with Isosceles). IsoQuant was executed in a similar manner to other programs but in a consecutive two-step process (where the first IsoQuant run identifies de novo transcripts which are concatenated to the original annotations in a second run) instead of a single-run due to significant improvement in performance observed (Fig. 2b-c and Supplementary Fig. 2 for IsoQuant two-step results; IsoQuant single-run results in Isosceles_Paper: reports_static/simulated_bulk_benchmarks_isoquant.ipynb).
In order to simulate Oxford Nanopore Technologies (ONT) reads using NanoSim, we trained error models on bulk nanopore RNA-Seq FASTQ files concatenated from sequencing three cell lines: SK-OV-3 (SRR26865806), COV504 (SRR26865804), and IGROV-1 (SRR26865803). Nanopore single-cell RNA-Seq (nanopore scRNA-Seq) read models were also generated from the pooled set of the aforementioned cell lines (SRR26865982). A total of 100 million reads were simulated from each error model and then the first 12 million reads deemed alignable by NanoSim were extracted.
Read model error rates:
| Bulk RNA-Seq | scRNA-Seq | |
|---|---|---|
| Mismatch rate | 0.02982687241070872 | 0.02866079657952386 |
| Insertion rate | 0.024056603631908934 | 0.024409736896819117 |
| Deletion rate | 0.04654204334915349 | 0.030249793440489687 |
| Total error rate | 0.10042551939177115 | 0.08332032691683267 |
To align the simulated reads provided in BAM format to all benchmark programs, Minimap2 was employed, using Ensembl 90 introns given in a BED file and applying a junction bonus parameter of 15 (with the exception of NanoCount, which required read alignment directly to the transcriptome). For the scRNA-Seq ONT dataset used to create the read model, various tools detected a similar number of cells (~2460), but the median number of unique molecular identifiers (UMIs) per cell differed. The Sicelore preprocessing of ONT scRNA-seq, identified between 3,000 and 6,000 UMIs per cell, which were provided in BAM format for biologically derived data benchmarks to Sicelore, IsoQuant, and Isosceles with cell barcode and UMI tags annotated (Fig. 3a-b). In contrast, FLAMES, with its own UMI detection and deduplication processes, detected around 13,500 UMIs per cell. To strike a balance between the varying results from different tools, a compromise of 10,000 reads per cell was chosen for this study.
To simulate scRNA-Seq ONT data, a BAM file containing aligned simulated reads from the scRNA-Seq read model was randomly downsampled 100 times using samtools, with a subsampling proportion of 0.000833. This resulted in approximately 10,000 reads out of the original 12 million for each BAM file. A custom Python script (see supplemental Benchmark commands) was used to assign unique cell barcode sequences and UMI sequences for each read within the 100 BAM files. These subsampled BAM files were then merged and sorted using samtools.
Synthetic molecules and platform-comparison data processing
The data used for comparative analysis of results from different sequencing platforms included FASTQ files for PacBio (ENCODE: ENCFF450VAU) and ONT (cDNA Pass basecalls from the Nanopore WGS Consortium GitHub repository: https://github.com/nanopore-wgs-consortium/NA12878/blob/master/RNA.md23), as well as Illumina short read transcript quantifications (ENCODE: ENCFF485OUK) for the GM12878 cell line. Long reads were aligned to the reference genome using Minimap2 as discussed previously for simulated data (although in the PacBio dataset, the ‘-ax splice:hq’ parameter was used instead of ‘-ax splice’). Transcripts with >1 TPM in Illumina quantifications (intersected with the Ensembl 90 transcript IDs utilized in this study to account for annotation discrepancies with Ensembl 95 annotation from ENCFF485OUK) were selected, and for those with one-to-many matches of Ensembl IDs, ground-truth values were aggregated.
The alignment file in BAM format for ‘Nanopore cDNA Pass’ reads aligned to the SIRV sequences (SIRV set 3, Lot No. 001485) was downloaded from the Nanopore WGS Consortium GitHub repository (see above). The three top performing tools Isosceles, Bambu and IsoQuant were benchmarked on both insufficient annotations (44 annotated SIRV isoforms [24 withheld], compared to 68 isoforms in the correct annotations) and over-annotations (68 annotated SIRV isoforms with an additional 32 decoy isoforms) obtained from Lexogen’s website (https://www.lexogen.com/sirvs/download). For the over-annotations, the fraction of reads assigned to correct transcripts (read assignment precision) was calculated for each tool (utilizing both SIRV transcripts and 92 unspliced ERCC sequences). In case of insufficient annotations, transcript detection (comprising both annotated and withheld) was measured with the precision, recall, and F1 score metrics on spliced (SIRV) data only, with the metrics being calculated on the level of unique transcript splicing structures.
Nanopore raw read files in FASTQ format were obtained from SRA for Sequin mix A data (SRR14286054) and mix B data (SRR14286063), then aligned using Minimap2 and processed using individual tools. Sequin reference sequences and annotations used for the analysis were downloaded from (https://github.com/XueyiDong/LongReadRNA/tree/master/sequins/annotations) as described previously22,38. Quantifications from each tool were compared to ground-truth Sequin expression values for mix A and mix B in order to calculate Spearman correlations and mean relative differences for each mix as well as for concatenated expression values from both mixes.
Biological data processing
The bulk RNA-Seq data (GSE248114) included Promethion data, featuring eight sequencing libraries for seven ovarian cancer cell lines (OVMANA, OVKATE, OVTOKO, SK-OV-3, COV362, COV504, and IGROV-1), as well as two technical replicates for IGROV-1. For MinION platform data, two technical replicates for IGROV-1 were sequenced. Factors such as RAM performance and program speed determined the number of reads simulated in bulk simulations and downsampled in bulk data. For example, for performing cross platform correlations, the Promethion data was downsampled to 5 million reads to make it more comparable to MinION (~6-7 million raw reads) and pseudo-bulk scRNA-Seq (3.5-4.5 million UMIs per cluster, as detected by Isosceles) in terms of total read depth. This decision was also influenced by an issue with IsoQuant (https://github.com/ablab/IsoQuant/issues/69), which limited its ability to process large read files in our hands. Notably, this issue persisted on a cluster node with 20 CPUs of 2.4 GHz and allocated 230 GB of RAM.
The scRNA-Seq data (GSE248115) consisted of a mix of three cell lines (SK-OV-3, COV504, and IGROV-1). The Illumina sequencing (SRR26865983) was preprocessed using CellRanger (Version 6.0.1). For ONT sequencing data (SRR26865982) we considered two barcode preprocessing methods (Sicelore and wf-single-cell) for cell barcode (CBC) and unique molecular identifier (UMI) detection. We observe similar average Spearman correlation (0.85 vs 0.88) and mean relative diff. (0.57 vs 0.60) between the same cell lines in pseudo-bulk and bulk between the two. However, better performance was achieved with Sicelore preprocessing for matched vs. decoy (0.26 vs 0.16 for Spearman correlation, 0.22 vs 0.14 for mean relative diff.). Therefore, we used Sicelore preprocessing to annotate the CBC and UMI tags in the ONT sequencing BAM files for Isosceles, Sicelore, and IsoQuant (Supplementary Fig. 5d; see below).
Mitochondrial transcripts common to all method’s output were removed, as they were strong outliers across methods. Additionally, three specific transcripts outliers across methods were removed: ENST00000445125 (18 S ribosomal pseudogene), ENST00000536684 (MT-RNR2 like 8), and ENST00000600213 (MT-RNR2 like 12).
Benchmarks using biological data
The correlation and relative difference analyses (Supplementary Fig. 4c) compared annotated transcripts between bulk RNA-Seq data from two Promethion and two MinION sequencing replicates of IGROV-1, both within each platform (using replicates) and between platforms (using averaged data for each platform). For each comparison, only transcripts with a mean expression of at least 1 TPM were used. In Supplementary Fig. 4d, scRNA-Seq and bulk RNA-Seq data were also compared, again considering only annotated transcripts. For each program, the IGROV-1 scRNA-Seq pseudo-bulk cluster (according to genetic identity from Souporcell) was compared with the averaged bulk RNA-Seq IGROV-1 expression values from two replicates for each platform. Analyses were also restricted to transcripts with an expression of at least 1 TPM in the single-cell RNA-Seq results. Comparisons were made for each platform using top k cells (highest UMI count) using the top 5000 transcripts (highest mean expression) to ensure a comparable number of transcripts across software package, and top N transcripts (highest mean expression) for 64 top cells (highest UMI count) (Supplementary Fig. 4d).
For Fig. 3a-c, scRNA-Seq and bulk RNA-Seq data analysis was conducted using Bioconductor packages (eg. scran, scater, etc.) on the transcript level for cells with at least 500 genes, for a range of top highly variable transcript numbers (500, 1,000, 2,000, 4,000, 6,000 and 10,000), as determined by the scran::getTopHVGs function39. Heatmaps were generated to show correlations and mean relative difference between scRNA-Seq pseudo-bulk results for three cell line clusters and Promethion bulk RNA-Seq results for seven ovarian cancer cell lines, similarly only including annotated transcripts. IGROV-1 expression was averaged from two replicates. To compare difference between matched and decoy metrics (Spearman correlation and mean relative difference), we calculated the absolute difference and computed the upper and lower bounds of the standard error using error propagation (as sqrt(se(x)^2 + se(y)^2)). To assess the overall significance of Isosceles results compared to each program in matched versus decoy metrics, we computed the differences between each matched cell line and the mean of decoys across a range of 500-10,000 HVTs. The set of differences is then compared against the matched results from Isosceles using a Wilcoxon matched-pairs signed-rank test.
For the simulated data version of Fig. 3c presented in Supplementary Fig. 5c, nanopore reads were simulated for SK-OV-3, IGROV-1, OVMANA, OVKATE, OVTOKO and COV362. These were based on short read TPM values obtained from Whippet v1.7.3 and Ensembl 90 transcripts with the GENCODE ‘basic’ tag (excluding mitochondrial transcripts) and mean expression of at least 0.1 TPM across all analyzed cell lines. 5 M reads were produced by NanoSim for both bulk RNA-Seq and scRNA-Seq read models, which were aligned to the genome using Minimap2. For the latter, cell barcodes randomly selected from 100 sequences and unique UMI sequences were added to the BAM files. Simulated bulk RNA-Seq and scRNA-Seq samples were analyzed as described for biological data presented in Fig. 3c.
We also perform this benchmark for Isosceles on the BAM files obtained from Sicelore and wf-single-cell (for the latter, Minimap2 alignment junction bonus of 15 was specified using the ‘resources_mm2_flags’ flag and the expected number of cells was set to 2,000). As wf-single-cell doesn’t produce a deduplicated BAM file, UMI deduplication using UMI-Tools was applied. Isosceles results for both BAM files were compared for the top 4,000 highly variable transcripts, defining the choice of Sicelore for single-cell barcode preprocessing used in the manuscript (see Supplementary Fig. 5d).
Case-study analysis of biological data
For the case-study in Fig. 4, the raw reads were pre-processed to identify cell barcodes (CBC) and unique molecular identifiers (UMI) according to the Sicelore workflow. The reads were subsequently aligned to the reference genome mm10/GRCm38 (with annotations derived from GENCODE M25), using Minimap2 with a junction bonus of 15, which targeted both annotated introns from Gencode M25 and those extracted from the VastDB mm10 GTF file30. The aligned reads with CBC and UMI annotations were subsequently quantified with Isosceles. The 951-cell dataset was filtered to exclude cells that expressed fewer than 100 genes. For dimensionality reduction, we combine Isosceles gene and transcript counts, culminating in the total identification of 3760 variable features (with a target of 4000), comprising 1735 genes and 2025 transcripts. We applied Principal Component Analysis (PCA), calculating 30 components using the scaled expression of the variable features. Cells were clustered using Louvain clustering (with resolution parameter of 2) on the Shared Nearest Neighbor (SNN) graph (setting a k-value of 10). The clusters’ identities were determined through gene set scores, particularly the mean TPM values of markers delineated in the original study (see Supplementary Fig. 6). Additional marker genes were identified via the scran::findMarkers function requiring the t-test FDR to be significant (q value < 0.05) in at least half of the comparisons to other clusters (selecting top 5 markers of each cluster).
Pseudotime analysis was performed using Slingshot for differentiating glutamatergic neurons (identifying two trajectories, T1 and T2), differentiating GABAergic neurons, radial glia, cycling radial glia and Cajal-Retzius cells (with one trajectory each). To implement the original ‘isoform switching’ analysis, pairs of clusters were compared, detecting marker transcripts through the specific scran::findMarkers function (Wilcoxon test). We filter for transcripts of the same gene showing statistically significant differences in opposite directions (i.e. one upregulated in one cluster, the other in another cluster). To analyze splicing changes within each trajectory, we used Isosceles to calculate aggregated TCC values for windows along pseudotime, defining the window size as 30 cells and the step size as 15 cells. AS events from variable transcripts abiding by further criteria were selected for downstream analysis. First, mean PSI values across all cells from the trajectory were between 0.025 and lower than 0.975 to exclude constitutively included/excluded events. Second, at least 30 cells must have values not equal to 0, 1, or 0.5, and 30 cells must have a value above 0.1 to select against events with only low counts. Redundant PSI events, identical in read counts profiles within a trajectory, were excluded, and those with >0.99 spearman correlation were excluded from visualization in Fig. 4b and Supplementary Fig. 7b. For comparative analysis, percent-spliced-in (PSI) count values are denoted as counts-spliced-in (CSI) and defined by PSI * gene counts. These are juxtaposed with exclusion PSI counts, calculated as [(1 - PSI value) * gene counts] and the inclusion/exclusion pair input into DEXSeq29. For each intra-trajectory comparison, our experimental design encompassed ‘~sample + exon + pseudotime:exon‘. Meanwhile, the inter-trajectory analysis included all trajectories with a design of ‘~sample + exon + pseudotime:exon + trajectory:exon‘, compared against a null model of ‘~sample + exon‘ using the LRT test.
To determine ratios of observed vs. expected CSI, we shuffle TCCs across cells with non-zero counts and apply the EM algorithm, calculating PSI for each window. To obtain expected CSI we multiply the shuffled PSI values * observed gene counts. The permutations are conducted for each AS event across 100 bootstraps. For empirical statistical validation of changes between the first and last windows of a trajectory (eg. for Celf2), we fit a negative binomial distribution to each window using maximum likelihood estimation (‘fitdistrplus‘ package) on the permuted CSI, and calculate high and low one-tailed p values for the observed CSI. Combining the high and low, and low and high p values of the first and last windows respectively using Fisher’s method, we defined an overall p-value as two times the minimum combined p value. Specifically for heatmap visualization, a broad window size of 100 cells for glutamatergic & GABAergic neurons, and 50 cells for glia and CR cells, with a consistent step size of 3 cells for smoothing was utilized. The heatmap values were given as the log2 ratio of observed to expected, with a pseudocount of 0.1, defining the ratio between PSI counts and the average of the corresponding permuted PSI counts.
Benchmark command summary:
https://github.com/Genentech/Isosceles_Paper/blob/devel/Benchmark_commands.md
Software versions:
| Software | Version |
|---|---|
| Isosceles | v0.2.0 |
| Flair | v1.7.0 |
| StringTie2 | v2.2.1 |
| IsoQuant | v3.0.3 |
| NanoCount | v1.0.0.post6 |
| Sicelore | v2.0 |
| Bambu | v3.2.5 (R 4.3.0, Bioconductor 3.17) |
| FLAMES | v0.1 |
| ESPRESSO | beta1.3.0 |
| NanoSim | v3.1.0 |
| Minimap2 | v2.24-r1122 |
| wf-single-cell | v1.1.0 |
| UMI-tools | v1.1.5 |
Cell culture
All cell lines used in this study were validated by STR analysis and verified mycoplasma negative by PCR. No commonly misidentified cell lines were used in this study. IGROV1, SK-OV-3, OVTOKO, OVKATE and OVMANA cell lines were cultured in RPMI-1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 2mM L-Glutamine. COV362 and COV504 cells were cultured in DMEM supplemented with 10% FBS and 2mM L-Glutamine. Cells were cultured in 37 °C and 5% CO2 in a humidified incubator. Cell line source and catalog numbers are provided in the table below. Cells were cultured in 10cm2 plates until they reached ~60-80% confluency. For bulk analysis, RNA was purified using Qiagen’s RNeasy Plus Mini kit (Cat. #74134) according to the manufacturer’s instructions. For single-cell analysis, IGROV1, SK-OV-3 and COV504 cells were trypsinized and pooled together at a 1:1:1 ratio at a concentration of 1000 cells / μl and submitted for single cell long read sequencing.
| Cell line | Provider | Catalog number |
|---|---|---|
| IGROV-1 | NCI DCTD | |
| SK-OV-3 | ATCC | HTB-77 |
| OVTOKO | JCRB Cell Bank | JCRB1048 |
| OVKATE | JCRB Cell Bank | JCRB1044 |
| OVMANA | JCRB Cell Bank | JCRB1045 |
| COV362 | ECACC | 07071910 Lot# 07G029 |
| COV504 | ECACC | 07071902 Lot# 07I007 |
Reference. 40:
Single-cell, long-read library preparation and nanopore sequencing
Approximately 10 ng of cDNA generated from the Next GEM Single Cell 3′ Gene expression kit (10X Genomics, Cat # PN-100268) was amplified using 10uM of the biotinylated version of the forward primer and a reverse primer from the single cell 3′ transcriptomics protocol (ONT, SQK-LSK114), [Btn]_Fwd_3580_partial_read1_defined_for_3′_cDNA, 5′-/Biosg/CAGCACTTGCCTGTCGCTCTATCTTC CTACACGACGCTCTTCCGATCT-3′ and Rev_PR2_partial_TSO_defined_for_3′_cDNA, 5′-CAGCTTTCTGTTGGTGCTGATATTGCAAGCAGTGGTA TCAACGCAGAG-3′. To ensure enough cDNA was generated for the pull-down reaction (200 ng), two PCR reactions were carried out using 2X LongAmp Taq (NEB, Cat # M0287S) with the following PCR parameters 94°C for 3 minutes, with 5 cycles of 94°C 30 secs, 60°C 15 secs, and 65°C for 3 mins, with a final extension of 65°C for 5 minutes. The cDNA was pooled and cleaned up with 0.8X SPRI bead ratio and eluted in 40 μL RNAse free H20. Concentration was evaluated using the QuBit HS dsDNA assay (Thermofisher, Cat No. Q32851). The amplified cDNA was then captured using 15 μL M270 streptavidin beads (Thermofisher, Cat # 65305). Beads were washed three times with 1 mL of the 1X SSPE buffer (150 mM NaCl, 10 mM NaH2PO4, and 1 mM EDTA). Beads were then resuspended in 10 μL of 5X SSPE buffer (750 mM NaCl, 50 mM NaH2PO4, and 5 mM EDTA). Approximately 200 ng of the cDNA in 40 μL were added together with the 10 μL M270 beads and incubated at room temperature for 15 minutes. After incubation, the sample and beads were washed twice with 1 mL of 1X SSPE. A final wash was performed with 200 uL of 10 mM Tris-HCl (pH 8.0) and the beads bound to the sample were resuspended 10 μL of RNAse free H2O. PCR was performed on-bead using the unbiotinylated version of the primers from the ONT single 3’ transcriptomics protocol discussed earlier for 5 cycles according to the same PCR program shown above. A 0.8X SPRI was performed. The cDNA was eluted in 50 μL in RNAse free H2O and the concentration was evaluated with QuBit HS dsDNA assay and Tapestation D5000 DNA kit (Agilent Technologies, Cat # 5067-5589).
Library preparation for nanopore sequencing was performed according to the SQK-LSK110 protocol with the exception of the end-repair step time which was increased to 30 min. 125 fmol of final library was loaded on the PromethION using the FLO-PRO002 flow cells, R9.4.1 chemistry and sequenced for 72 h. Reads were basecalled using Guppy v5.0.11.
Statistics and reproducibility
No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank Bo Li, Hector Corrada-Bravo, William Forrest, John Marioni, Luca Gerosa, Marc Hafner, and Robert Piskol for helpful suggestions and feedback.
Author contributions
T.S.W. and M.K. conceived of and designed the software methodology and computational experiments with contributions from the other authors. M.K. implemented the Isosceles package and both M.K. and A.R. performed benchmarking analyses. M.K. and T.S.W. designed and performed the case study. K.S. performed the cell culture and A.B. and W.S. performed the sequencing protocols with preliminary analyses from D.L. T.S.W. wrote the manuscript with contributions from M.K. and all other authors.
Peer review
Peer review information
Nature Communications thanks Andrey Prjibelski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All biological sequencing data generated in the manuscript is deposited in the NCBI Gene Expression Omnibus (GEO) under GSE248118. Mouse E18 brain long-read single-cell sequencing data is available at GSE130708. Sequin spike-in ONT data is available at GSE172421. SIRV spike-in ONT data and GM12878 ONT data available from GitHub at nanopore-wgs-consortium/NA12878 [https://github.com/nanopore-wgs-consortium/NA12878]. PacBio data for GM12878 is available from the ENCODE Consortium at ENCFF450VAU and the transcript quantification file from ENCFF485OUK. Accession identifiers for source data in Fig. 4e and Supplementary Fig. 8d are listed in Supplementary Data 2. Source data are provided with this paper.
Code availability
Isosceles R package code, documentation, and vignettes are released on GitHub (https://github.com/Genentech/Isosceles)41 under an open source GPL-3 license. All benchmarking code, virtual environments, and quantification data necessary to reproduce the figures/analyses in the manuscript are similarly released (analysis code: https://github.com/Genentech/Isosceles_paper42, singularity containers: 10.5281/zenodo.8180648, benchmark quantifications: 10.5281/zenodo.8180604, raw simulated data: 10.5281/zenodo.8180695, simulated ovarian cell line bulk RNA-Seq data: 10.5281/zenodo.10895721, simulated ovarian cell line scRNA-Seq data: 10.5281/zenodo.10895894, mouse E18 brain scRNA-Seq data: 10.5281/zenodo.10028908).
Competing interests
The authors are employees and shareholders of Genentech/Roche.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51584-3.
References
- 1.Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet40, 1413–1415 (2008). 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- 2.Sterne-Weiler, T., Weatheritt, R. J., Best, A. J., Ha, K. C. H. & Blencowe, B. J. Efficient and Accurate Quantitative Profiling of Alternative Splicing Patterns of Any Complexity on a Laptop. Mol. Cell72, 187–200.e6 (2018). 10.1016/j.molcel.2018.08.018 [DOI] [PubMed] [Google Scholar]
- 3.Ziegenhain, C. et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell65, 631–643.e4 (2017). 10.1016/j.molcel.2017.01.023 [DOI] [PubMed] [Google Scholar]
- 4.Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol.39, 1348–1365 (2021). 10.1038/s41587-021-01108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics37, 4572–4574 (2021). 10.1093/bioinformatics/btab705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, Y. et al. ESPRESSO: Robust discovery and quantification of transcript isoforms from error-prone long-read RNA-seq data. Sci. Adv.9, eabq5072 (2023). 10.1126/sciadv.abq5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang, A. D. et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun.11, 1438 (2020). 10.1038/s41467-020-15171-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian, L. et al. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol.22, 310 (2021). 10.1186/s13059-021-02525-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebrigand, K., Magnone, V., Barbry, P. & Waldmann, R. High throughput error corrected Nanopore single cell transcriptome sequencing. Nat. Commun.11, 4025 (2020). 10.1038/s41467-020-17800-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prjibelski, A. D. et al. Accurate isoform discovery with IsoQuant using long reads. Nat. Biotechnol. 41, 915–918 (2023). [DOI] [PMC free article] [PubMed]
- 11.Hu, Y. et al. LIQA: long-read isoform quantification and analysis. Genome Biol.22, 182 (2021). 10.1186/s13059-021-02399-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gleeson, J. et al. Accurate expression quantification from nanopore direct RNA sequencing with NanoCount. Nucleic Acids Res50, e19–e19 (2021). 10.1093/nar/gkab1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Y. et al. Context-Aware Transcript Quantification from Long Read RNA-Seq data with Bambu. Nat Methods20, 1187–1195 (2023). [DOI] [PMC free article] [PubMed]
- 14.Pardo-Palacios, F. J. et al. Systematic assessment of long-read RNA-seq methods for transcript identification and quantification. Nat. Methods21, 1349–1363 (2024). [DOI] [PubMed]
- 15.Heber, S., Alekseyev, M., Sze, S.-H., Tang, H. & Pevzner, P. A. Splicing graphs and EST assembly problem. Bioinformatics18, S181–S188 (2002). 10.1093/bioinformatics/18.suppl_1.S181 [DOI] [PubMed] [Google Scholar]
- 16.Ntranos, V., Kamath, G. M., Zhang, J. M., Pachter, L. & Tse, D. N. Fast and accurate single-cell RNA-seq analysis by clustering of transcript-compatibility counts. Genome Biol.17, 112 (2016). 10.1186/s13059-016-0970-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. Bmc Bioinforma.12, 323–323 (2011). 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol.34, 525–527 (2016). 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 19.Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression. Nat. Biotechnol.33, 495–502 (2015). 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang, C., Chu, J., Warren, R. L. & Birol, I. NanoSim: nanopore sequence read simulator based on statistical characterization. Gigascience6, gix010 (2017). 10.1093/gigascience/gix010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol.20, 278 (2019). 10.1186/s13059-019-1910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong, X. et al. Benchmarking long-read RNA-sequencing analysis tools using in silico mixtures. Nat. Methods20, 1810–1821 (2023). 10.1038/s41592-023-02026-3 [DOI] [PubMed] [Google Scholar]
- 23.Workman, R. E. et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods16, 1297–1305 (2019). 10.1038/s41592-019-0617-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods17, 615–620 (2020). 10.1038/s41592-020-0820-1 [DOI] [PubMed] [Google Scholar]
- 26.Luecken, M. D. & Theis, F. J. Current best practices in single‐cell RNA‐seq analysis: a tutorial. Mol. Syst. Biol.15, e8746 (2019). 10.15252/msb.20188746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom.19, 477 (2018). 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz, Y., Wang, E. T., Airoldi, E. M. & Burge, C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods7, 1009–1015 (2010). 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders, S., Reyes, A. & Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res.22, 2008–2017 (2012). 10.1101/gr.133744.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tapial, J. et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res.27, 1759–1768 (2017). 10.1101/gr.220962.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard, K. S., Gut, I. M., Lyman, M. E. & McNutt, P. M. Longitudinal RNA sequencing of the deep transcriptome during neurogenesis of cortical glutamatergic neurons from murine ESCs. F1000Research2, 35 (2013). 10.12688/f1000research.2-35.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buccitelli, C. & Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet.21, 630–644 (2020). 10.1038/s41576-020-0258-4 [DOI] [PubMed] [Google Scholar]
- 33.McFarland, J. M. et al. Multiplexed single-cell transcriptional response profiling to define cancer vulnerabilities and therapeutic mechanism of action. Nat. Commun.11, 4296 (2020). 10.1038/s41467-020-17440-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacPherson, M. J. et al. Nucleocytoplasmic transport of the RNA-binding protein CELF2 regulates neural stem cell fates. Cell Rep.35, 109226 (2021). 10.1016/j.celrep.2021.109226 [DOI] [PubMed] [Google Scholar]
- 35.Irimia, M. et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell159, 1511–1523 (2014). 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Cabau, C. et al. Kinetic stabilization of translation-repression condensates by a neuron-specific microexon. bioRxiv 2023.03.19.532587 10.1101/2023.03.19.532587 (2023).
- 37.Gonatopoulos-Pournatzis, T. et al. Autism-Misregulated eIF4G Microexons Control Synaptic Translation and Higher Order Cognitive Functions. Mol. Cell77, 1176–1192.e16 (2020). 10.1016/j.molcel.2020.01.006 [DOI] [PubMed] [Google Scholar]
- 38.Dong, X. et al. The long and the short of it: unlocking nanopore long-read RNA sequencing data with short-read differential expression analysis tools. NAR Genom. Bioinform.3, lqab028 (2021). 10.1093/nargab/lqab028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lun, A. T. L., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, M. et al. A resource for cell line authentication, annotation and quality control. Nature520, 307–311 (2015). 10.1038/nature14397 [DOI] [PubMed] [Google Scholar]
- 41.Kabza, M. & Sterne-Weiler, T. Accurate long-read transcript discovery and quantification at single-cell, pseudo-bulk and bulk resolution with Isosceles, http://github.com/Genentech/Isosceles10.5281/zenodo.12702401 (2024).
- 42.Kabza, M., Ritter, A. & Sterne-Weiler, T. Accurate long-read transcript discovery and quantification at single-cell, pseudo-bulk and bulk resolution with Isosceles, http://github.com/Genentech/Isosceles_Paper, 10.5281/zenodo.12702743 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All biological sequencing data generated in the manuscript is deposited in the NCBI Gene Expression Omnibus (GEO) under GSE248118. Mouse E18 brain long-read single-cell sequencing data is available at GSE130708. Sequin spike-in ONT data is available at GSE172421. SIRV spike-in ONT data and GM12878 ONT data available from GitHub at nanopore-wgs-consortium/NA12878 [https://github.com/nanopore-wgs-consortium/NA12878]. PacBio data for GM12878 is available from the ENCODE Consortium at ENCFF450VAU and the transcript quantification file from ENCFF485OUK. Accession identifiers for source data in Fig. 4e and Supplementary Fig. 8d are listed in Supplementary Data 2. Source data are provided with this paper.
Isosceles R package code, documentation, and vignettes are released on GitHub (https://github.com/Genentech/Isosceles)41 under an open source GPL-3 license. All benchmarking code, virtual environments, and quantification data necessary to reproduce the figures/analyses in the manuscript are similarly released (analysis code: https://github.com/Genentech/Isosceles_paper42, singularity containers: 10.5281/zenodo.8180648, benchmark quantifications: 10.5281/zenodo.8180604, raw simulated data: 10.5281/zenodo.8180695, simulated ovarian cell line bulk RNA-Seq data: 10.5281/zenodo.10895721, simulated ovarian cell line scRNA-Seq data: 10.5281/zenodo.10895894, mouse E18 brain scRNA-Seq data: 10.5281/zenodo.10028908).