Abstract
In the developing vertebrate retina, diverse neuronal subtypes originate from multipotent progenitors in a conserved order and are integrated into an intricate laminated architecture. Recent progress in mammalian photoreceptor development has identified a complex relationship between six key transcription-regulatory factors (RORβ, OTX2, NRL, CRX, NR2E3 and TRβ2) that determine rod versus M cone or S cone cell fate. We propose a step-wise ‘transcriptional dominance’ model of photoreceptor cell fate determination, with the S cone representing the default state of a generic photoreceptor precursor. Elucidation of gene-regulatory networks that dictate photoreceptor genesis and homeostasis will have wider implications for understanding the development of nervous system function and for the treatment of neurodegenerative diseases.
“Tamaso ma jyotir Gamaya ...” (lead us from darkness to light)
In Sanskrit from the discussion of the life force ‘Prana’ in Brihadaranyaka Upanishad (1.3.28) (800–2000 bc)
For thousands of years, humankind has sought light and vision to promote growth, survival and intellectual development. Darkness and the loss of sight are among our most basic fears. In humans, many higher-order neuronal functions including behaviour, learning, memory and emotions are dictated, to a considerable extent, by vision1. Importantly, almost 30% of the sensory input to the brain originates in the retina, which is referred to as the “window to the brain”1,2. Given the ease of diagnosis and quantification of visual defects, it is not surprising that as many as 30% of approximately 20,000 entries in the Online Mendelian Inheritance in Man (OMIM) database (see the online links box) reveal some association with the eye, vision and/or visual dysfunction.
The retina is an excellent model for studying complex features of the nervous system, such as cell lineage and connectivity in general and visual function in particular. The vertebrate retina consists of six major types of neurons (rod and cone photoreceptors, and horizontal, bipolar, amacrine and ganglion cells) that exhibit laminar organization and form numerous parallel microcircuits for integration and processing of visual signals2–4 (FIG. 1a). The process of vision begins at the photoreceptors, which are unique sensory neurons that are specialized to capture light quanta. The chemical output of photoreceptors is integrated and processed by interneurons (bipolar, horizontal and amacrine cells) and transmitted to visual centres in the brain by ganglion cells. Abnormalities, dysfunction and/or death of retinal photoreceptors constitute the primary cause of visual impairment or blindness in most retinal diseases, such as Leber’s congenital amaurosis (LCA), retinitis pigmentosa and macular degeneration.
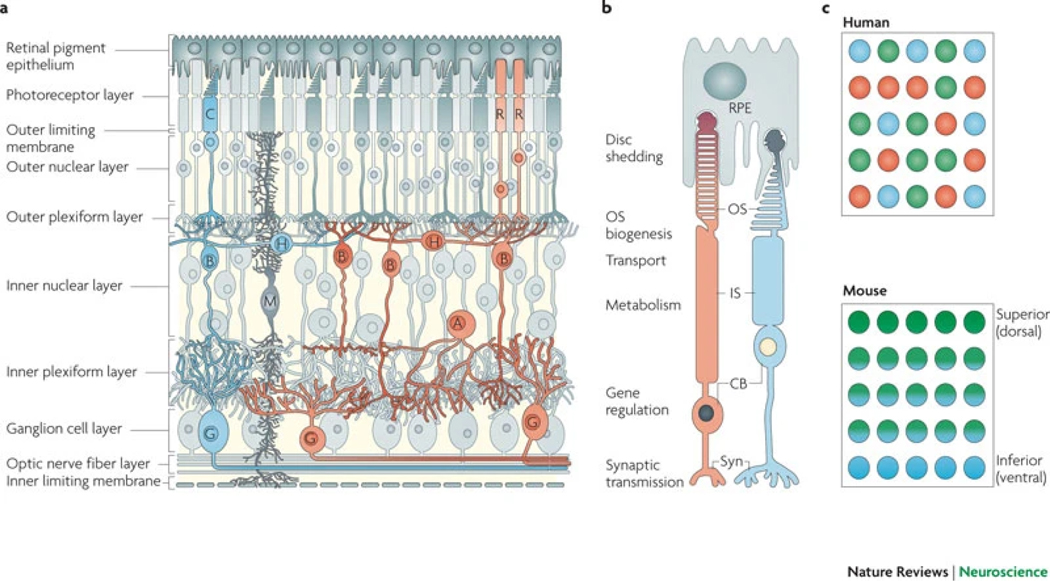
Figure 1: Functional circuitry of the retina.
a | Organization of retinal circuits. Rod (R) and cone (C) photoreceptors have cell bodies (CBs) in the outer nuclear layer and extend inner segments (IS), which contain metabolic machinery, and outer segments (OS), which associate with retinal pigment epithelial (RPE) cells. Photoreceptor axons terminate in the outer plexiform layer and synapse with horizontal (H) and bipolar (B) cells in the inner nuclear layer, which also contains Müller glial (M) and amacrine (A) cells. Bipolar cells relay signals to amacrine and ganglion (G) cells through synapses in the inner plexiform layer. Ganglion cell axons project towards the optic nerve head and carry signals to the brain. End-feet of Müller glia form the outer and inner limiting membranes. A representative cone pathway is shown in blue and a representative rod pathway is shown in orange. b | Illustrations of rod and cone morphologies that include subcellular locations of functions. c | A surface-view representation of cone distribution across the mammalian retina. In humans, cones express S opsins (peak sensitivity to blue light), L opsins (peak sensitivity to red light) or M opsins (peak sensitivity to green light) in a mosaic-like pattern. In mice, cones express S opsins and M opsins in opposing distribution gradients along the superior (M opsin-high) to inferior (S opsin-high) axis. Syn, synaptic terminus.
In this Review, we synthesize current knowledge of photoreceptor development and propose a framework for the regulatory transcriptional networks that determine photoreceptor cell fate in the mammalian retina. We briefly describe genetic defects that result in syndromic or non-syndromic photoreceptor degeneration due to aberrant differentiation, function or homeostasis, and discuss potential treatment strategies for neurodegenera tive blinding diseases based on the current understanding of transcriptional networks.
Photoreceptors: types and organization
Mammalian photoreceptors are highly compartmentalized for specialized functions and have an intimate relationship with the retinal pigment epithelium (RPE), which supports photoreception (FIG 1a,b). Cone photoreceptors respond to bright light, mediate colour vision and permit high resolution of visual images. Rod photoreceptors function only under conditions of low light and can respond to single light quanta, being a hundred-fold more sensitive than cones5 (see the Webvision website, listed in Further information)
Across the retina, rods and various cone subtypes are arranged in an elegant laminar and non-random pattern (mosaic)6–8 (FIG. 1c) that must be sequentially generated with the right number of components, correct spatial organization and appropriate wiring to interneurons for serial and parallel processing of visual information9. In mice and humans, photoreceptors constitute over 70% of retinal cells, but rods outnumber cones by 30:1 in mice and 18–20:1 in humans6,7. A major difference between humans (and diurnal primates) and mice (and most other mammals) is the presence in humans of a thin, pit-like, cone-only region in the centre of the retina, called the fovea, which is responsible for highest visual acuity. In humans, the density of rods increases from the fovea to the periphery of the retina, with the highest rod density in the parafoveal region10.
The mammalian retina has only one type of rod visual pigment (rhodopsin, which has a peak spectral sensitivity at ~500 nm). Most mammals (including the mouse) have two types of cone opsins that confer dichromatic colour vision: S opsin (also known as blue-sensitive opsin), which has peak sensitivity in the short wavelength (ultraviolet or blue) region of the spectrum; and M opsin (also known as green-sensitive opsin), which has peak sensitivity in the medium–long wavelength (green) region of the spectrum. Duplication of an ancestral M opsin-like gene on the X chromosome has provided humans and diurnal primates with an additional opsin, L opsin (also known as red-sensitive opsin), which is sensitive to longer wavelength (red) light and, together with S opsin and M opsin, confers trichromatic colour vision11. Each human cone expresses only one of the three opsins, giving a mosaic-like pattern over the retina12. In mice and other rodents, M opsins and S opsins are expressed in opposing gradients across the retina13. Cones in mid-retinal regions express varying amounts of both M and S opsin14,15, leading to the concept of S- or M-dominant cone subpopulations in some species16.
Multipotent progenitors and neuronal diversity
The proportions and distribution of photoreceptor subtypes in the mature retina are optimized to the precise needs of a given species for capturing visual information in diverse habitats. The identification of genes and signals that direct a dividing retinal progenitor cell (RPC) to become a photoreceptor or another cell type has been the subject of intense investigations17–19. Classical birth-dating studies using 3H-thymidine labelling, lineage tracing experiments, or cell-type specific antibodies demonstrated a fixed chronological and overlapping order of birth for all retinal neurons from common pools of progenitor cells in the vertebrate retina20–24. The generation of functionally mature neurons from multipotent RPCs proceeds through a series of steps that increasingly restrict lineage choices and commit cells to a particular fate25–27 (FIG. 2). How is this process stringently controlled to be appropriate to the temporal and spatial context?
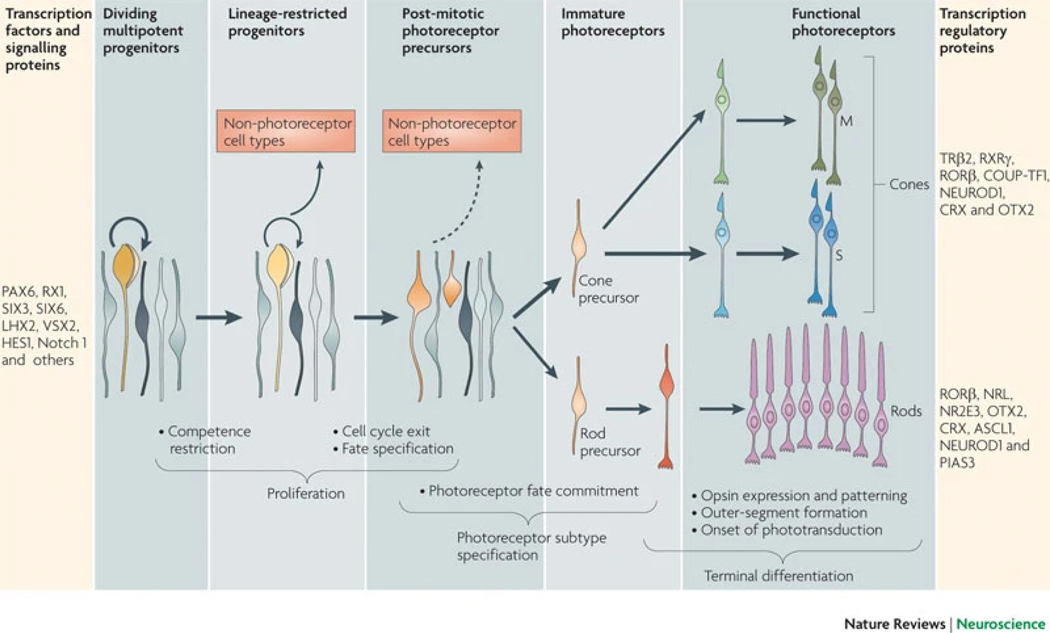
Figure 2: Stages of photoreceptor development.
Early in retinogenesis, multipotent retinal progenitor cells (RPCs) divide and produce additional multipotent progenitors (thick circular arrow) or progenitor cells that become restricted in their competence to generate various cell types (thin circular arrow). Some of these proliferating cells become restricted to a lineage that will give rise to at least one photoreceptor cell and possibly to non-photoreceptor cells. After cell cycle exit, postmitotic precursors can remain plastic. During cell type specification of photoreceptors, precursors are directed to become cones or rods that eventually express photopigments (M opsin and S opsin in cones, and rhodopsin in rods), and form outer segments and synapses. The bar on the far left lists key transcription factors and signalling proteins that maintain RPC multipotency and proliferation: paired box protein PAX6, retinal homeobox protein RX1, SIX3, SIX6, LIM–homeobox protein LHX2, visual system homeobox 2 (VSX2), HES1 and Notch 1. The bar on the far right lists key transcription regulatory proteins that are involved in cone and rod differentiation and maintenance (for cones: thyroid hormone receptor β2 (TRβ2), retinoid X receptor-γ (RXRγ), nuclear receptor RORβ, COUP transcription factor 1 (COUP-TF1; also known as NR2F1), neurogenic differentiation factor 1 (NEUROD1), cone–rod homeobox protein (CRX) and homeobox protein OTX2; for rods: RORβ, neural retina leucine zipper protein (NRL), photoreceptor-specific nuclear receptor (NR2E3), OTX2, CRX, achaete-scute homologue 1 (ASCL1; also known as MASH1), NEUROD1 and E3 SUMO-protein ligase PIAS3).
Initially, it was proposed that the developmental potency of uncommitted progenitors changes with developmental age28. According to a more detailed model29 both intrinsic control mechanisms of the progenitor cells and the environment change over time, and changes in competence generate different neurons. The progressive-restriction and competence models for generating cellular diversity provided a valuable framework for subsequent developmental and molecular genetic studies. A complex interplay of transcription factors (including homeodomain proteins such as paired box protein PAX6, retinal homeobox protein RX1, SIX3, SIX6, visual system homeobox 2 (VSX2) and LIM–homeobox protein LHX2) is implicated in an evolutionarily conserved programme that establishes the anterior neuroectodermal region for eye formation and maintains the retinogenic potential of progenitor cells30–34. The transient and quantitatively precise expression of regulatory transcriptional proteins, which are often bHLH transcription factors and homeodomain transcription factors18,19,26, mediates the acquisition of specific cell fates and provides a starting point for the elucidation of regulatory pathways underlying neurogenesis. It has emerged that many transcription factors (including PAX6, VSX2 and neurogenic differentiation factor 1 (NEUROD1)) participate at an early point in cell fate specification as well as later in functional maturation and maintenance (homeostasis).
The timing of photoreceptor genesis
In mammals, the genesis of photoreceptors occurs over a long temporal window20,21,35,36 (FIG. 3). After exit from final mitosis, functional maturation of a committed photoreceptor precursor (rod or cone) can take weeks to months depending on the species (FIG. 3). The process of photoreceptor development can be divided into five major steps (FIG. 2): first, proliferation of multipotent RPCs; second, restriction of the competence of RPCs; third, cell fate specification and commitment to photoreceptor precursors during or after final mitosis; fourth, expression of photoreceptor genes, such as those for phototransduction and morphogenesis; and fifth, axonal growth, synapse formation and outer segment biogenesis. In humans, all photoreceptors are generated prenatally. The first cones and rods are born around foetal week (Fwk) 8 and Fwk 10, respectively37,38. S opsin mRNA is produced as early as Fwk 12, followed by rhodopsin, M opsin and L opsin transcripts at Fwk 15 (REF. 38) (FIG. 3). At birth, opsin expression patterns are reasonably well established, but photoreceptors are still immature. In the mouse, photoreceptor development is less advanced than in humans at birth, and the eyes of the newborn pup remain closed for almost 2 weeks. Cone genesis is essentially complete at birth (having started at embryonic day (E) 11), and S opsin is expressed at late embryonic stages. In mice, rods are born both pre- and postnatally. The peak of rod genesis occurs in the first few postnatal days, accompanied soon thereafter by the expression of rhodopsin20,21 (FIG. 3). Expression of M opsin by cones begins around postnatal day 6 and lags behind that of other opsins. As photoreceptors mature, the opsin levels increase substantially, outer segments elaborate towards the RPE and synapses form with horizontal and bipolar neurons.
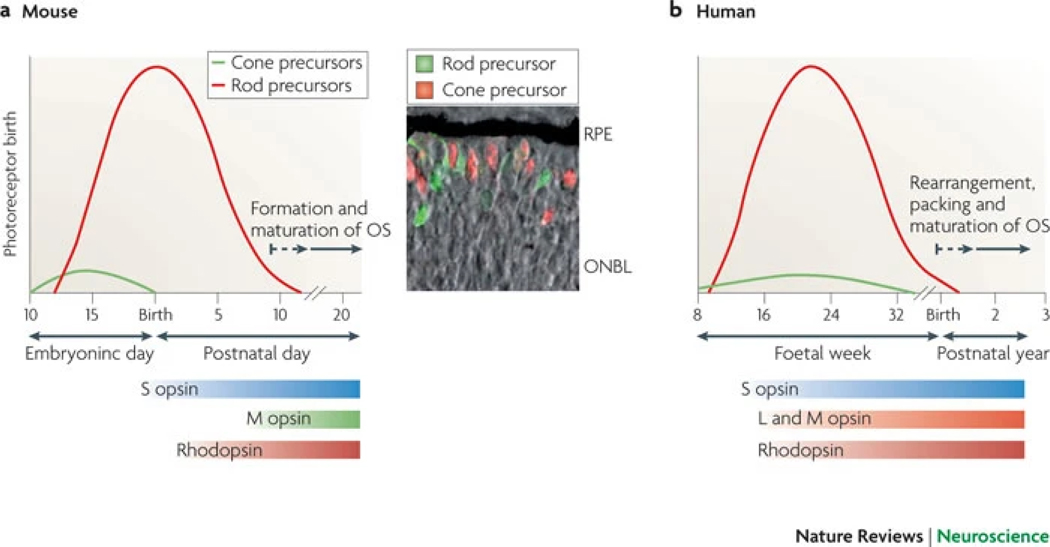
Figure 3: Photoreceptor genesis and maturation in mice and humans.
The relative numbers of cone and rod precursor cells that are born over time are shown for mouse and human retinogenesis. a | In mice, cones are generated prenatally as early as embryonic day (E) 11. Rods, which vastly outnumber cones, are generated both before and after birth, from around E12 to postnatal day (P) 10. Expression of S opsin begins at ~E18, that of M opsin at ~P6 and that of rhodopsin at ~P2. Photopigment levels increase substantially until after weaning in mice. As photoreceptors mature, outer segments (OS) and synapses form. The inset panel shows double fluorescent detection of cone (thyroid hormone receptor β2-positive cells (red staining)) and rod precursors (neural retina leucine zipper protein (NRL) -positive cells (green staining)) in mouse retina at E18, superimposed on a phase contrast picture that reveals the outer neuroblastic layer (ONBL) (D. Sharlin, Alok Swaroop and D. Forrest, unpublished data). Newly generated photoreceptors tend to reside near the edge of the retina. b | In humans, cones and rods are generated around foetal week (Fwk) 8 and Fwk 10, respectively. Generation of cones is completed prenatally, whereas that of rods continues into the early postnatal period. Expression of S opsin is observed at ~Fwk 12, and that of L opsin, M opsin and rhodopsin at ~ Fwk 15. Functional maturation of photoreceptors continues postnatally. RPE, retinal pigment epithelium. Data are taken from Refs 14, 20, 21, 38, 80, 151, 152.
From progenitors to photoreceptor precursors
The earliest steps in photoreceptor development involve signalling through the Notch receptor39,40. High levels of Notch seem to maintain the cycling of progenitor cells, and inhibition of Notch drives progenitors to commit to the cone or rod photoreceptor fate, depending on the developmental stage, over other cell fates. Notably, signalling molecules such as sonic hedgehog can also influence progenitor commitment by converging on downstream targets of Notch41.
Notch signals through the bHLH transcription factors HES1 and HES5, which maintain the progenitor pool by downregulating the expression of proneural bHLH proteins in the developing mouse retina39,40,42. The molecular mechanisms that generate photoreceptor precursors from RPCs, including downregulation of Notch signalling, remain uncharacterized. The proneural bHLH transcription factors (such as NEUROD1, neurogenin 2, NEUROD4 (also known as MATH3), protein atonal homologue 7 (ATOH7; also known as MATH5) and achaete-scute homologue 1 (ASCL1; also known as MASH1)) have redundant and poorly defined roles in photoreceptor development42,43. Their promiscuous roles in early neuronal development make it difficult to determine specific instructive or participatory function in progenitor competence, lineage restriction and/or photoreceptor fate specification (FIG. 2). Each step in early differentiation is probably regulated by combinatorial and transient actions of several transcription factors and signalling proteins.
Transcription factors in differentiation
Although we have only a limited understanding of the signals that prompt a progenitor to become a photoreceptor precursor, we have better insights into subsequent events that direct a photoreceptor precursor to differentiate into a rod or a cone. Here, we discuss six transcription factors that dictate key steps of photoreceptor differentiation in the developing mammalian retina — namely, homeobox protein OTX2, cone–rod homeobox protein (CRX), neural retina leucine zipper protein (NRL), photoreceptor-specific nuclear receptor (NR2E3), nuclear receptor RORβ and thyroid hormone receptor β2 (TRβ2).
OTX2 and commitment to a photoreceptor fate.
The paired-type homeodomain transcription factor OTX2 is a key regulator of the photoreceptor lineage. It is expressed during final mitosis in retinal progenitors and in early precursors (T. Furukawa, personal communication) that become committed to the photoreceptor cell fate44,45. A conditional knockout of Otx2 in immature retinal progenitors leads to almost complete loss of rods and cones, as well as a loss of bipolar and horizontal cells in mice44,45. Retrovirus-mediated expression of Otx2 in RPCs of neonatal mice can drive cells towards the rod cell fate44. As OTX2 is also expressed in other cell types throughout the brain and retina, it provides a necessary, but not a sufficient, signal to induce the photoreceptor cell fate.
CRX and terminal differentiation of photoreceptors.
The transcriptional regulator CRX was initially identified as a photoreceptor-specific retinopathy gene, but is now known to also be expressed in other mature retinal neurons46–50. In Crx-deficient mice, photoreceptors are generated but fail to express many phototransduction genes, resulting in a lack of outer segments and eventually retinal degeneration51. CRX is expressed early in postmitotic photoreceptor precursors and acts downstream of OTX2 (REFs 44,45). Its exact role at this stage remains undefined. CRX enhances the expression of photoreceptor-specific genes52,53 and is important for terminal differentiation of rods and cones; however, CRX alone does not determine specific photoreceptor cell fate51.
NRL and the specification of rod photoreceptor fate.
The decision to be or not to be a rod is largely determined by NRL, a basic motif–leucine zipper transcription factor of the Maf subfamily54. In mammals, NRL interacts with CRX and many other transcription factors and induces the expression of rod-specific genes52,55,56. Loss of NRL in mice leads to a retina with no rods and an excess of S opsin-expressing cones (S cones) because the precursors that were committed to become rods are converted to cones57. Ectopic expression of NRL in immature photoreceptor precursors results in loss of cone gene expression and likely transformation to rods58. These studies establish an essential and instructive role of NRL in determining rod cell fate.
NR2E3, a suppressor of cone genes.
The orphan nuclear receptor NR2E3 (REF. 59) is a direct transcriptional target of NRL60 and is expressed in postmitotic photoreceptors61. NR2E3 primarily suppresses the expression of cone genes, thereby irreversibly committing NRL-expressing precursors to a rod cell fate60,62–64. Mice carrying an L1 retrotransposon insertion65 in the Nr2e3 gene66 (rd7 mice) have rod-like photoreceptors that express many cone genes63,64,67. NR2E3, together with NRL and CRX, also serves as a co-activator of rod genes, but cannot substitute for NRL in rod gene activation63,68.
RORβ, a dual regulator in both rod and cone differentiation.
RORβ is an orphan nuclear receptor that is expressed in all cell layers of the neural retina and in the pineal gland69,70. The retina of Rorb−/− mice lacks rods and features an excess of S cone-like photoreceptors, therefore resembling that of Nrl−/− mice71. However, the excess cones are non-functional and more primitive than those in Nrl−/− mice. Nrl is not expressed in Rorb−/− mice, indicating that Rorb lies upstream of Nrl in the photoreceptor differentiation pathway. RORβ, like CRX, is also necessary for the formation of photoreceptor outer segments and contributes to the development of the inner nuclear and synaptic layers of the retina71. RORβ acts synergistically with CRX to initiate S opsin transcription in cones72.
TRβ2, a mediator of M cone differentiation.
The nuclear receptor TRβ2 is a ligand-regulated transcription factor73 expressed in new cones in the mammalian foetal retina74,75. In TRβ2-deficient mice, cones lack M opsin and express only S opsin74, indicating a crucial role for thyroid hormone signalling in opsin patterning and maturation of the colour visual system. TRβ2 coordinates cone opsin patterning in response to the increasing thyroid hormone levels that accompany development76.
The preceding discussion of six key transcription factors suggests an intricate regulatory network that guides the determination of photoreceptor cell fate.
The ‘transcriptional dominance’ hypothesis
Although a unified pathway cannot be put forward for all vertebrates17, we have synthesized the evidence from genetic studies of key transcription factors as well as earlier proposals that cones may differentiate into a common default form before diversifying into distinct cone subtypes13,14. We propose a ‘transcriptional dominance’ model of photoreceptor cell fate determination (FIG. 4) that includes three fundamental attributes: first, that all terminally differentiated photoreceptors originate from a common postmitotic photoreceptor precursor that has the potential to form rods or any of the cone subtypes; second, that a photoreceptor precursor cell follows a default pathway to differentiate into an S cone unless additional regulatory signals direct the precursor to acquire a rod or M cone identity; and third, that the acquisition of a specific fate by a photoreceptor precursor is established by a particular set of transcription-regulatory factors gaining dominance at a given developmental stage.
Figure 4: Transcriptional dominance model of photoreceptor cell fate determination.
A generic photoreceptor is formed under the control of homeobox protein OTX2 and other undetermined signals. This precursor is programmed to possess a ‘default’ S cone state under the control of OTX2 (and/or cone–rod homeobox protein (CRX)) and nuclear receptor RORβ unless diverted into a rod or M cone state by additional signals. The ‘NRL control box’ determines whether a precursor becomes a rod or a cone, and the ‘thyroid hormone receptor β2 (TRβ2) control box’ prompts a cone to acquire an M opsin or an S opsin identity. Each control box is subject to modifying regulators. Induction of neural retina leucine zipper protein (NRL) and its target, photoreceptor-specific nuclear receptor (NR2E3), induces a rod state and suppresses cone genes, which consolidates the rod fate. Other factors involved in rod development include E3 SUMO-protein ligase PIAS3, neurogenic differentiation factor 1 (NEUROD1), achaete-scute homologue 1 (ASCL1; also known as MASH1), myocyte enhancer factor 2C and retinoblastoma-associated protein, probably at both early and later stages of differentiation. If NRL and NR2E3 fail to act, photoreceptor precursors follow the ‘default’ pathway to S cones unless TRβ2 and its ligand triiodothyronine (T3) induce M opsin and suppress S opsin expression in spatially restricted patterns in cone subpopulations over the retina. Other factors involved in M opsin and S opsin patterning include retinoid X receptor-γ (RXRγ), RORβ, RORα, COUP transcription factors (COUP-TFs) and CRX. NEUROD1 and bone morphogenetic protein receptor type 1A–1B contribute to inducing the expression of TRβ2 and COUP-TF, respectively.
The model predicts that the transcriptional control signals exert a profound influence at two key points during fate specification: first, the decision to form either a rod or a cone; and second, the decision for a cone to differentiate with an S cone or M cone identity (FIG. 4). It is likely that the commitment to produce a functional rod or cone is dictated by context- and time-dependent combinations of cell-specific and widely expressed transcription factors. Modest changes in the expression or activity of one or more transcriptional regulators in response to extrinsic or cell–cell communication signals might alter the overall outcome of differentiation. The model also predicts that, once the requisite numbers of committed cones are produced, by as yet unknown counting mechanisms, the rod-inducing signals will overwhelmingly direct the differentiation of precursors away from the default S cone state to the rod state.
The coordinated and precisely balanced action of the six key transcription factors (discussed above) seems to be crucial as RPCs commit to a rod or cone photoreceptor cell fate. We propose that several of these factors compete for dominance in a newborn photoreceptor precursor. These sometimes conflicting regulators would determine the particular differentiation outcome of a given precursor cell at a specific stage of development. Several (if not all) of the six factors (OTX2, CRX, RORβ, NRL, NR2E3 and TRβ2) may be expressed at varying levels in a precursor cell (FIG. 4) and engage in a transcriptional contest for dominance (‘tug of war’) that will ultimately determine the cell fate. In the default scenario, OTX2 biases the progenitor to a photoreceptor precursor fate, and CRX and RORβ initially induce the expression of S opsin. Other transcription factors including PR domain zinc finger protein 1 (also known as BLIMP1) are likely to help stabilize the commitment of OTX2-expressing precursors to a photoreceptor fate77,78. In the photoreceptor precursor, RORβ, presumably in cooperation with other specific factors, also induces NRL expression. When a certain threshold of NRL expression and/or activity is achieved, NRL may recruit CRX and activate NR2E3 expression. An NRL–NR2E3 complex then suppresses S opsin and other cone genes, while NRL–CRX-containing complexes initiate the rod differentiation programme. Below a ‘decisive threshold’ of expression and/or activity of NRL–NR2E3, the precursor differentiates by default as a cone. In these non-rod precursors, TRβ2 is crucial for the induction of M opsin in cones and differential patterning of both M opsins and S opsins.
Several lines of evidence suggest the existence of a common photoreceptor precursor. First, in the developing retina of humans and rodents, S opsin is expressed before other cone opsins (FIG. 3). Such observations in the rat led to the proposal of a default pathway in which cones initially express S opsin before a subpopulation subsequently switches to an M opsin identity79. In the human foetus, S cones cover almost 90% of the retina at Fwk 19, and this number decreases dramatically later in development, potentially reflecting a switch in opsin expression from a default S cone state to an M or L cone identity8. Second, in TRβ2-deficient mice, cones lack M opsin and instead express S opsin74, consistent with a key role for TRβ2 in M cone versus S cone patterning. Third, Nrl−/− mice possess S cones instead of rods, which supports the concept of the default S cone state57,80. Furthermore, NR2E3 and RORβ were found to act on the same transcriptional pathway as NRL in directing precursors to a rod state60,63,71. Fourth, NRL-positive precursors selected from the retina of wild-type mice at E16 express S opsin transcripts58,80, which is consistent with the existence of an early photoreceptor precursor that has a propensity to follow a default S cone pathway. For a precursor to enter into rod differentiation, the S cone state must be actively turned off by the co-expression of NRL and NR2E3. Some of the precursors begin to express NRL together with S opsin because they are not yet fully committed to a rod fate. There is likely to be an ‘interim’ state as the cone versus rod fate becomes fixed. At postnatal stages when cone genesis is complete and only rods are produced, the default S cone state of the photoreceptor precursors is likely to be transient under the influence of strong NRL activity. Lastly, the model of a default S cone state is consistent with studies on the ontogeny and evolution of photoreceptors, which suggest that the S cone represents an evolutionarily older form of photoreceptor14,17,81.
The transient nature of plastic photoreceptor precursors at different developmental stages may make it difficult to demonstrate their existence in vivo. However, the model contains a testable prediction in that some (or all) early photoreceptor precursors co-express some or many of the key transcription factors before their final commitment to form a rod or a cone. Other bHLH transcription factors, such as ATOH7, ASCL1 and NEUROD1, are at least transiently expressed in some photoreceptor precursors and may contribute to the counting mechanism or the balance between the cone- and the rod-inducing programmes.
Cone opsin distribution patterns
For cone-committed precursors that do not receive signals to form rods, the next decision concerns the expression of S opsin versus M opsin (in most mammals) or S opsin versus either M opsin or L opsin (in humans, as discussed below)82,83. The key questions are: how is an opsin gene selected for expression, and how does this exclude the expression of other opsins in that cone photoreceptor?
Selective expression of S opsin versus M opsin.
The S opsin (Opn1sw) and M-opsin (Opn1mw) genes are located on different chromosomes (chromosome 6 and the X chromosome, respectively, in mice), and each gene is subject to independent transcriptional regulation in cone subpopulations. The mechanism of mutual exclusion of S opsin and M opsin expression has not been elucidated, but a combination of intrinsic transcriptional controls, external signals including thyroid hormone and signals that relate to spatial location on the retinal surface seem to have key roles.
TRβ2 is a major regulator of M opsin and S opsin patterning in mice but is expressed in cones in all retinal regions, suggesting that other factors contribute to the regional variations in M opsin and S opsin distribution. One such factor is the nuclear receptor retinoid X receptor-γ (RXRγ), which can heterodimerize with TRβ2 and suppress S opsin expression in regions of the superior retina in mice84. Other possible patterning factors are COUP transcription factor (COUP-TF) orphan nuclear receptors85, which help to ensure a normal distribution of S cones over the retina, and bone morphogenic protein receptor, which may control COUP-TF expression85. Factors that augment M and S opsin expression include RORα86 and, indirectly, NEUROD1, which enhances the expression of TRβ2 and M opsin in cones87,88.
The action of TRβ2 requires the thyroid hormone triiodothyronine (T3) at appropriate developmental stages76,89,90. In mice, the concentration of thyroid hormone in the circulation rises progressively during postnatal development and provides a temporal signal that induces the expression of M opsin in the second postnatal week (FIG. 3). In mice with congenital hypothyroidism, induction of M opsin expression is impaired76, whereas treatment with excessive T3 suppresses the induction of S opsin expression at early stages89. At inappropriate neonatal stages, excessive T3 promotes cone-specific cell death.
Selective expression of L opsin versus M opsin genes in humans.
A specialized mechanism of opsin patterning underlies human trichromacy and involves selective expression of L opsin or M opsin genes (OPN1LW or OPN1MW, respectively), which reside in a tandem cluster on the X chromosome82. In a given cone, expression of an L opsin or M opsin gene is selected by an upstream locus control region (LCR), thereby ensuring their mutually exclusive expression91,92. Studies in mice carrying a miniature human L opsin–M opsin gene cluster indicate that the pairing of the LCR with either the L opsin or M opsin gene promoter is determined by stochastic events and not by individual sets of opsin-specific transcription factors93. Transcription factor complexes involved in LCR-mediated activation of the L opsin or M opsin genes have not been elucidated. In human retinoblastoma cell lines that have some cone properties, assembly of regulatory complexes at the LCR determines L opsin versus M opsin expression during final mitosis94. LCR-mediated selection of opsin gene expression has also been observed in zebrafish, which differentially express a tandem array of four green-sensitive RH2 opsin genes95. TRβ2 and RXRγ in human foetal cones96 might participate in the activation of L opsin and M opsin genes, analogously to their role in M opsin induction in mice.
Other processes contribute to opsin patterning in the context of the growing eye. The retinal surface varies not only in terms of cone opsin patterning but also in the density and distribution of cone and rod cells10. Thus, the differential rates of generation of rods and cones20 and their programmed cell death in development97,98 may also determine the pattern of each cell type across the retina. In the more local environment, cell–cell signalling may control spatial arrangements between groups of cell types99,100.
Nuclear receptors
Nuclear receptors are ligand-regulated transcription factors. They include TRβ2 and RXRγ, which have known ligands (thyroid hormone and retinoids, respectively), and orphan receptors (such as NR2E3, COUP-TF, RORα and RORβ), which lack known physiological ligands. The potential of these transcription factors to respond to ligands allows an integration of transcriptional control with extrinsic signals that may be crucial for rod versus cone differentiation, cone opsin patterning or other functions during photoreceptor differentiation62–64,71,84,86 (FIGs 3,4). Some orphan receptors, such as RORβ, have a large carboxy-terminal pocket that may bind hydrophobic molecules with lower specificity and affinity than conventional hormonal ligands101. Thus, apart from the endocrine signal of thyroid hormone, other small molecules may provide paracrine or autocrine signals that locally modify nuclear receptor activity during development. Although some nuclear receptors such as nuclear receptor subfamily 2 group E member 1 (NR2E1; also known as TLX) may act early in neurogenesis in retinal progenitor cells102,103, nuclear receptors have a role predominantly at later stages of photoreceptor differentiation. This provides an interesting contrast to the roles of bHLH, homeodomain and other types of transcription factors42 during earlier stages in retinal neurogenesis.
Gene-regulatory networks (GRNs)
We primarily discuss GRNs pertaining to rod differentiation and homeostasis because most studies have focused on the regulation of rod gene expression. Early induction of NRL expression in photoreceptor precursors80 seems to be a decisive step in determining the rod versus cone outcome. We predict that a substantial fraction of photoreceptor precursors express NRL at a given time in development in response to prevailing signals. The long period from final mitosis to the functional maturation of rods (2–4 weeks in mice and 5–10 weeks in humans) can be attributed to step-wise changes in gene expression patterns (influenced by extrinsic factors) that eventually lead to the expression of a specific set of phototransduction genes and a functional rod photoreceptor.
Regulation of NRL expression.
A major regulator of rod development upstream of NRL is RORβ, which is widely expressed in retinal progenitors and in immature photoreceptors. The role of RORβ in driving a precursor into the first phase of the rod state is probably mediated via induction of NRL expression71 in cooperation with other transcription factors. An approximately 3-kb genomic region80 that lies immediately upstream of the transcription start site of the mouse Nrl gene includes many conserved sequence elements with potential binding sites for bHLH proteins, nuclear receptors and other factors, such as myocyte enhancer factor 2C, that are expressed in the developing and mature retina (M.-A. Kautzmann, M. Akimoto and A. Swaroop, unpublished observations). Additional regulatory factors that could promote rod development via the RORβ–NRL pathway include taurine, activin A, sonic hedgehog, fibroblast growth factor and retinoblastoma protein104–110. Retinoic acid increases the number of cells expressing rod marker genes111,112, potentially by elevating Nrl expression113. By contrast, the signal transducer and activation of transcription (STAT) signalling pathway, which is downstream of receptors for ciliary neurotrophic factor (CNTF) and leukaemia inhibitory factor, regulates NRL and CRX expression and therefore rod differentiation114,115. CNTF is a negative regulator of rhodopsin expression, yet it increases photoreceptor cell survival116,117. How the cytokine and janus kinase (JAK)–STAT signalling pathways are integrated in the transcriptional hierarchy of NRL–CRX–NR2E3 requires further investigation.
Three key regulators of rod development — NRL, CRX and NR2E3 — continue to be expressed at high levels in the mature retina, suggesting that they have important roles in rod homeostasis. We hypothesize that these proteins auto-regulate and cross-regulate each other, thereby maintaining their expression levels throughout the life of rods.
GRNs during development and homeostasis.
GRNs118,119 have three key components — transcription factors, cofactors and/or modulators, and transcriptional targets. In such networks, signalling molecules modulate the expression and/or activity of transcription factors that act as key nodes and control multiple cellular targets. One approach for elucidating GRNs is to examine global expression profiles after disrupting key regulatory nodes. Computer-based analysis of cis-acting elements and chromatin immunoprecipitation (ChIP) studies have identified shared as well as specific transcriptional targets of NRL, CRX, TRβ2 and NR2E3 (REFs 60,62,120,121). To elucidate GRNs that underlie photoreceptor development and homeostasis, the approaches must be multi-pronged and include global expression profiling (or RNA sequencing) of purified photoreceptors from different genetic backgrounds (such as Nrl−/−, Crx−/−, Rorb−/− and rd7 mice), global target analyses using sequencing of ChIP DNAs (ChIP-Seq) at different stages of development, in vitro and in vivo strategies for determining direct biological responses (for example, in vivo electroporation122), and network and pathway analyses.
Another important question is, how are NRL, CRX and NR2E3 involved in assembling distinct transcriptional complexes to generate quantitatively distinct and precise gene expression patterns? In addition to unique combinations of cis-acting regulatory sequences, the strength of protein interactions and combinatorial synergy (or antagonism) can produce quantitative differences in the expression of genes that are regulated by a similar set of transcription factors. NRL and CRX can recruit TATA box-binding protein123, CREB-binding protein and STAGA53 to generate transcriptional complexes. The strength and stability of interaction among regulatory factors can be modulated by post-translational modifications that permit the assembly of distinct complexes, which bind to enhancer sequences. For example, the human NRL protein of 237 amino acids contains 18 serine and 9 threonine residues and is highly phosphorylated124. SUMOylation of NRL is also crucial for its function during photoreceptor development125. The modulation of NRL activity by kinases and other signalling molecules might therefore be a key mechanism for controlling the timing and level of expression of rod genes. Notably, disease-causing mutations in NRL affect its phosphorylation state and interaction with CRX and other transcriptional regulators126. Another modulator of photoreceptor development is E3 SUMO-protein ligase PIAS3, which directly interacts with CRX and NR2E3; SUMOylation of NR2E3 by PIAS3 enhances its activity as a repressor of cone genes127.
Although still a work in progress, a simplified GRN for rod differentiation and homeostasis is illustrated in FIG. 5.
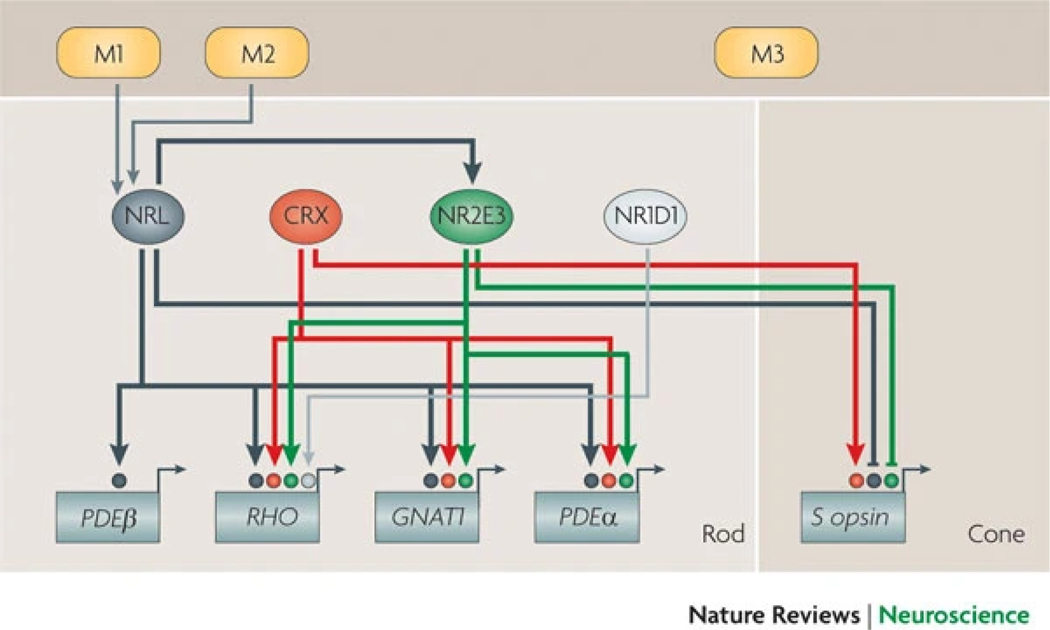
Figure 5: A simplified representation of rod gene-regulatory networks.
Neural retina leucine zipper protein (NRL) acts synergistically with cone–rod homeobox protein (CRX) and/or photoreceptor-specific nuclear receptor (NR2E3) in many distinct complexes that bind to enhancer sequences and dictate the expression of various rod genes. The rhodopsin (RHO) promoter is likely to be regulated by additional factors, such as nuclear receptor subfamily 1 group D member 1 (NR1D1)68, to maintain high but precisely controlled levels in rods. Other promoters (such as that for rod cGMP-specific 3′,5′-cyclic phosphodiesterase subunit-β (PDEβ)) may bind the widely expressed transcription factors SP1 and SP4 (Ref. 153), together with NRL. NRL and NR2E3 repress the activity of cone genes (such as S opsin), whereas CRX activates both rod and cone genes. M1, M2 and M3 indicate modulators (such as fibroblast growth factor 2, retinoic acid and post-translational modifiers) that control or fine-tune the activity of key transcription factors. Thin lines denote experiment-based suggested links, whereas thick lines denote confirmed regulatory connections. GNAT1, guanine nucleotide-binding protein G(t) subunit α1.
Retinal and macular diseases
Photoreceptor function and homeostasis are stringently controlled at many levels — regulation of gene expression, transport of selected proteins to the base of cilia, outer segment biogenesis and signal transmission (FIG. 1b,c). Mutations in over 100 genes that encode proteins with different roles in photoreceptor function have been associated with retinal degeneration128 (see the RetNet website listed in Further information). Interestingly, distinct mutations can lead to a similar clinical phenotype, or disease caused by similar mutations may have variable severity. Unlike the retina of many fish and amphibians129,130, the adult mammalian retina cannot generate new photoreceptors; hence, dysfunction or death of photoreceptors results in the loss of vision.
Mutations in NRL, NR2E3 and CRX cause varying retinal disease phenotypes (TABLE 1). All NRL mutations reported so far are missense or frameshift, and most seem to have a dominant impact on transcription-regulating activity126,131. NR2E3 mutations, by contrast, result in phenotypes such as enhanced s cone syndrome (ESCS)132–135, which can be explained by the primary role of NR2E3 as a repressor of cone genes in photoreceptor precursors63,136. CRX mutations generate an array of clinical phenotypes, from more severe congenital blindness in LCA to late-onset retinitis pigmentosa50,137. The retinal disease genes associated with photoreceptor dysfunction or death (based on the RetNet database; see Further information) are listed in Supplementary information S1 (table). A substantial fraction of these disease genes seem to be direct or indirect targets of NRL, CRX or NR2E3, as revealed by gene expression profiles of knockout mice and/or ChIP-Seq analysis60,63,64,138 (H. Hao, D. Kim and A. Swaroop, unpublished observations).
Table 1 |.
Clinical phenotypes caused by mutations in key transcriptional regulators of photoreceptor development
| Gene | Transcription factor type | Human diseases | Mouse mutant phenotypes |
|---|---|---|---|
| CRX | Paired-type homeodomain | Autosomal dominant cone–rod dystrophy; autosomal dominant and recessive Leber’s congenital amaurosis; autosomal dominant retinitis pigmentosa | Lack of outer segments in rods and cones; circadian entrainment defect; impaired bipolar cell differentiation and maintenance |
| NRL | Maf family basic leucine zipper | Autosomal dominant and recessive retinitis pigmentosa | Complete loss of rods; transformation of rods into S cones, with shorter outer segments than normal; enhanced S cone function; photoreceptor layer rosettes |
| NR2E3 | Orphan nuclear receptor | Enhanced S cone syndrome; autosomal dominant retinitis pigmentosa | Appearance of rod photoreceptors expressing cone genes; photoreceptor layer rosettes; retinal degeneration |
| OTX2 | Paired-type homeodomain | Microphthalmia (syndromic) | Loss of rods and cones and increase in amacrine-like neurons; loss of pinealocytes; impaired bipolar cell differentiation and maintenance; patterning abnormalities in the midbrain and hindbrain |
| RORB | Orphan nuclear receptor | None discovered | Loss of rods and over-production of S cones; lack of all outer segments; retinal disorganization; retarded induction of S opsin expression in cones; circadian behaviour abnormalities; abnormal gait |
| THRB | Thyroid hormone-activated nuclear receptor | Resistance to thyroid hormone, typically autosomal dominant; endocrine and neurological dysfunction; rare colour vision abnormalities | Loss of M opsin and extended distribution of S opsin in cones across the retina instead of normal bias in the ventral region; endocrine dysfunction; deafness |
CRX, cone–rod homeobox protein; NR2E3, photoreceptor-specific nuclear receptor; NRL, neural retina leucine zipper protein; THRB, thyroid hormone receptor β. Data are from the RetNet database (http://www.sph.uth.tmc.edu/retnet/), the OMIM database (http://www.ncbi.nlm.nih.gov/omim)) and from references in the main text.
THRB mutations are associated with the syndrome of resistance to thyroid hormone (see the OMIM entry for THRB), but photoreceptor dysfunction has been unexplored in this disease. Further studies of this syndrome and other congenital thyroid disorders are warranted to ascertain the requirement of human photoreceptors for thyroid hormone.
Designing knowledge-based treatments
Molecular details of transcription-regulatory networks underlying photoreceptor development, combined with high throughput genomic and genetic technologies, have facilitated the design of potential new treatment strategies for retinal degenerative diseases.
Gene discovery.
Disease-causing mutations have so far been identified in only approximately 50% of patients with inherited retinal degenerative diseases, owing to their extensive clinical and genetic heterogeneity. Comprehensive transcriptome analyses of photoreceptors and the retina have led to the identification of genetic defects in retinal diseases, particularly for cases in which the chromosomal region of a disease locus has been determined. Positional cloning strategies are inadequate in families with a low number of members affected by retinal degenerative disease. The knowledge of expression during photoreceptor development is also valuable in interpreting data from exome (capturing and sequencing all exons) and whole-genome sequencing methods, which are now being used for discovering novel disease mutations139,140.
Determining gene-regulatory networks.
Temporal ChIP-Seq analyses may allow identification of direct targets (the ‘targetome’) of the six key transcription factors and help establish regulatory networks that dictate photoreceptor morphology and function. Elucidation of such gene-regulatory networks should have a major impact on delineating the pathways of photoreceptor degeneration (BOX 1). Proteomics analysis of the developing retina141 and of purified photoreceptors may also yield novel insights into regulatory pathways and provide disease-specific biomarkers.
Box 1 |. Pathways of photoreceptor degeneration and targets for drug discovery.
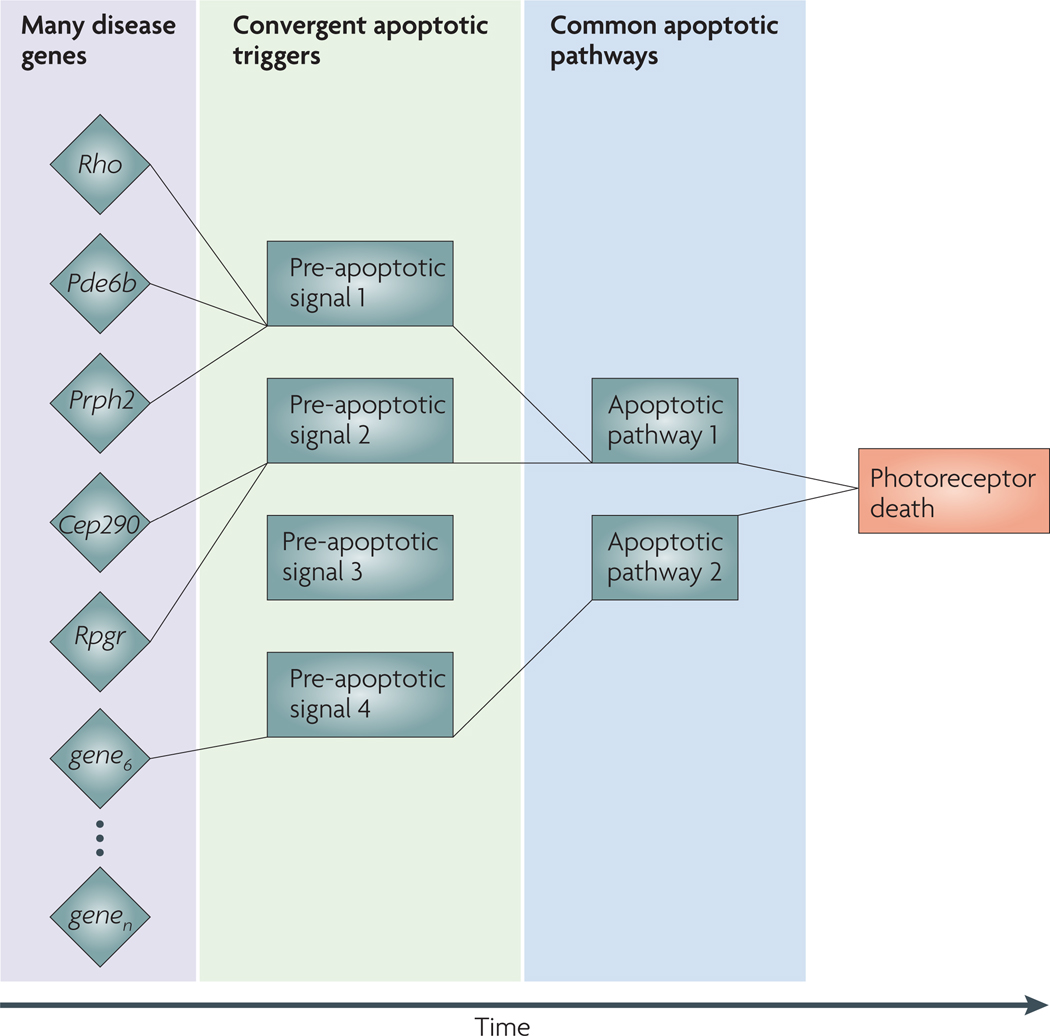
Leber’s congenital amaurosis, retinitis pigmentosa and other Mendelian retinal diseases constitute a genetically heterogeneous spectrum of clinical phenotypes. Given that mutations in over 100 genes can lead to photoreceptor cell death and most are limited to few patients and/or families, interventions that target these genes and mutations are less likely to be developed than those that target more common diseases because of cost and efficiency considerations. Alternative approaches are therefore needed to develop therapies for orphan diseases. We postulate a convergence of the impact of mutations on a select group of ‘pre-apoptotic signals’ that induce apoptotic pathways (see the figure). Mouse mutants with retinal degeneration are excellent models of human retinal degenerative diseases including those caused by mutations in rhodopsin (Rho), rod cGMP-specific 3′,5′-cyclic phosphodiesterase subunit-β (Pde6b), peripherin 2 (Prph2; also known as Rds), centrosomal protein of 290 kDa (Cep290) and X-linked retinitis pigmentosa GTPase regulator (Rpgr) (left column). We suggest that temporal profiling of changes in gene expression in mutant photoreceptors will allow the identification of a limited number of pre-apoptotic signalling molecules (middle column). Targeting terminal effectors of apoptosis (right column) for therapy may be ineffective and lead to undesirable side effects. As the photoreceptor degeneration process is associated with the remodelling of the inner retina, global gene profiling of purified rods and cones80 should reduce the background noise149. Gene profiling or RNA sequencing, followed by network or pathway analysis, should lead to identification of common pre-apoptotic signals. These can then be targeted to screen small molecules to intervene in retinal degenerative diseases.
Gene- and small-molecule-based interventions.
Modulation of transcription factor activity can be used to control the expression of downstream targets and consequently cellular phenotypes. For example, a short 37-amino-acid residue region of NRL that is sufficient for transactivation123 can be used in combination with a specific DNA-binding domain to modulate the expression of downstream genes. Agonists of NR2E3 may be useful in retinal diseases including an enhanced S cone phenotype142. Additionally, global patterns of gene expression in photoreceptors at distinct stages of differentiation and ageing can be used as a reference point to test the effects of candidate compounds for the treatment of specific degenerative diseases.
Photoreceptor precursors and cell-based therapies.
Cell-based strategies to replace defective or dying cells are among the promising approaches for the treatment of diverse retinal degenerative diseases. Numerous studies have attempted to design stem cell-based therapies for producing new photoreceptors. The feasibility of one such approach was demonstrated by transplantation of NRL-expressing committed rod precursors tagged with green flourescent protein into wild-type and degenerating mouse retina143. The photoreceptor precursors differentiated into mature rods and formed synaptic connections with interneurons, resulting in light responsiveness. The age of the donor cells was important, as only the postmitotic committed precursors — and not the dividing cells or the mature photoreceptors — survived and were integrated effectively. Many groups have now demonstrated successful differentiation of photoreceptors from human embryonic stem cells or from induced pluripotent stem cells and their transplantation into the mouse retina144–146. However, appropriate cell surface markers will be needed for sorting and purification of photoreceptor precursors that can be used for transplantation research in humans. CD73, a recently identified marker of photoreceptor precursors147 and other cell surface markers may assist in making the cell-based therapy feasible for retinal neurodegeneration.
Conclusions
We have summarized the current status of research on the genesis of photoreceptors, with a focus on transcription-regulatory factors that control the differentiation of rods and cones. We note that additional mechanisms (such as cellular interactions, microenvironment and epigenetic modifications) also influence the final pattern of the photoreceptor mosaic27,99,100, although we are only beginning to understand how such mechanisms guide the development of the mammalian retina. Despite considerable advances during the past decade, several important questions remain. What controls the timing, number and ratios of different photoreceptor cell types? What determines the expression of only one opsin in a human cone photoreceptor? What sets the morphological and functional polarity of a photoreceptor? How is the spatial and functional relationship of rods and cones with the RPE and interneurons organized in different regions of the retina? How does a cone or a rod photoreceptor form synaptic connections with its cognate bipolar or horizontal cell? How is the photoreceptor mosaic arranged? How is cone survival linked to rod death? The answers to at least some of these questions may lie in delineating GRNs that determine the temporal sequence of events of photoreceptor development and homeostasis. Such insights will also help in designing treatment strategies for retinal degenerative diseases, including the development of new drugs and photoreceptor replacement therapy.
Studies of RORβ, NRL and NR2E3 in mammals have provided particularly strong support for the common photoreceptor precursor hypothesis that is based on an S cone default state. However, it is worth mentioning that a mutation of the zebrafish Tbx2b transcription factor gene has been associated with excess rods and a loss of cones that are sensitive to ultraviolet light148. Although fish have a more complex set of cones than mammals, it is possible that additional developmental plasticity from rods to cones may be independent of the cone to rod plasticity directed by RORβ, NRL and NR2E3 discussed in this article.
Numerous challenges exist, yet we are confident that technological advances in genetics and high-throughput genomics will further our understanding of neurodevelopment and neurodegeneration of retinal cells and intensify the development of novel treatment paradigms.
Supplementary Material
Acknowledegments
This Review is dedicated to the memory of R. Adler, an outstanding scientist and a generous mentor and colleague. We are grateful to P. Raymond for constructive suggestions, T. Cogliati for productive discussions, and L. Ng, D. Sharlin, Alok Swaroop, S. Veleri and L. Kibiuk for help with the figures. We apologize to colleagues whose papers have not been cited because of page limitations. Our research is supported by intramural programmes of the National Eye Institute and National Institute of Diabetes, Digestive and Kidney Diseases.
Glossary
- Leber’s congenital amaurosis
A congenital form of early-onset blindness caused by mutations at many genetic loci
- Retinitis pigmentosa
An inherited progressive degeneration of photoreceptors, generally beginning in the peripheral retina with rod cell dysfunction
- Macular degeneration
A progressive dystrophy initially affecting photoreceptors in the 5–6 mm area around the fovea (the macula), which contains a higher ratio of cones to rods than the peripheral retina. Juvenile forms exhibit a Mendelian inheritance pattern, whereas age-related macular degeneration is a complex multifactorial disease
- Syndromic
Related to a pathology or disease involving multiple organs
- Retinal pigment epithelium
A polarized sheet of epithelial cells between the choroidal capillaries and the photoreceptor cells
- Retinal progenitor cell
A proliferating cell that can give rise to mature retinal cells
- Lineage tracing
An experimental method to identify the origin (progenitor) of a differentiated cell
- Competence
The ability of a retinal progenitor or precursor cell to produce specific cell types
- bHLH transcription factors
A family of transcription factors that contain a characteristic basic region and a helix–loop–helix domain
- Homeodomain transcription factors
A family of transcription factors that contain a characteristic DNA recognition domain, called the homeodomain. They are often involved in patterning spatial domains of developing tissues
- Specification
The developmental process that biases an immature cell to adopt a particular fate; the specified cell is not yet committed to the fate and retains developmental plasticity
- Final mitosis
The last mitotic division of a cell
- Photoreceptor precursor
A post-mitotic cell that is not yet differentiated and does not have a mature functional phenotype of a rod or a cone
- Paired-type homeodomain transcription factor
A DNA-binding transcription-regulating protein that contains a homeodomain with the characteristic amino acid residues of the homeodomain of the Drosophila melanogaster Paired transcription factor
- Basic motif–leucine zipper transcription factor
A transcription factor that contains a characteristic basic motif for DNA binding and a leucine zipper domain for dimerization
- Nuclear receptor
A ligand-regulated transcription factor that includes members with known ligands such as thyroid hormone receptor and retinoid X receptor, and those lacking a known physiological ligand such as retinoid-related orphan receptor
- Superior retina
The dorsal region of the light-sensing tissue at the back of the eye
- Cis-acting elements
DNA sequences that affect the transcription of a gene and are present nearby, on the same chromosome
- Chromatin immunoprecipitation
Often abbreviated as ChIP, this is an experimental technique used to identify DNA sequences that bind to a specific DNA-binding protein in vivo
- Enhanced S cone syndrome
An inherited autosomal recessive retinal disease associated with greater sensitivity to blue light, night blindness and eventual photoreceptor degeneration
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/gene
Opn1sw | Opn1mw
OMIM: http://www.ncbi.nlm.nih.gov/omim
THRB
UniProtKB: http://www.uniprot.org
CRX | L opsin | M opsin | Notch | NR2E3 | NRL | OTX2 | RORβ | S opsin | TRβ2
FURTHER INFORMATION
Anand Swaroop’s homepage: http://www.nei.nih.gov/intramural/nnrl.asp
Douglas Forrest’s homepage: http://www2.niddk.nih.gov/NIDDKLabs/IntramuralFaculty/ForrestDouglas.htm
The Webvision website: http://webvision.med.utah.edu/
References
- 1.Rodieck RW The First Steps in Seeing (Sinauer Associates Publishers, Sunderland, Massachusetts,1998). [Google Scholar]
- 2.Dowling JE The Retina: An Approachable Part of the Brain. (Belknap Press, Harvard Univ. Press, 1987). [Google Scholar]
- 3.Masland RH The fundamental plan of the retina. Nature Neurosci. 4, 877–886 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Wassle H. Parallel processing in the mammalian retina. Nature Rev. Neurosci. 5, 747–757 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Luo DG, Xue T. & Yau KW How vision begins: an odyssey. Proc. Natl Acad. Sci. USA 105, 9855–9862 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter-Dawson LD & LaVail MM Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J. Comp. Neurol. 188, 245–262 (1979). [DOI] [PubMed] [Google Scholar]
- 7.Roorda A. & Williams DR The arrangement of the three cone classes in the living human eye. Nature 397, 520–522 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Xiao M. & Hendrickson A. Spatial and temporal expression of short, long/medium, or both opsins in human fetal cones. J. Comp. Neurol. 425, 545–559 (2000). [PubMed] [Google Scholar]
- 9.Galli-Resta L. Putting neurons in the right places: local interactions in the genesis of retinal architecture. Trends Neurosci. 25, 638–643 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Curcio CA, Sloan KR, Kalina RE & Hendrickson AE Human photoreceptor topography. J. Comp. Neurol. 292, 497–523 (1990). [DOI] [PubMed] [Google Scholar]
- 11. Nathans J, Thomas D. & Hogness DS Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science 232, 193–202 (1986). Together with another paper in the same issue of Science, Nathans and colleagues establish the fundamental basis for colour vision and associated inherited variations in humans.
- 12.Deeb SS Genetics of variation in human color vision and the retinal cone mosaic. Curr. Opin. Genet. Dev. 16, 301–307 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Szel A, Rohlich P, Mieziewska K, Aguirre G. & van Veen T. Spatial and temporal differences between the expression of short- and middle-wave sensitive cone pigments in the mouse retina: a developmental study. J. Comp. Neurol. 331, 564–577 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Szel A, Lukats A, Fekete T, Szepessy Z. & Rohlich P. Photoreceptor distribution in the retinas of subprimate mammals. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 17, 568–579 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Applebury ML et al. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron 27, 513–523 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Nikonov SS, Kholodenko R, Lem J. & Pugh EN Jr. Physiological features of the S- and M-cone photoreceptors of wild-type mice from single-cell recordings. J. Gen. Physiol. 127, 359–374 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adler R. & Raymond PA Have we achieved a unified model of photoreceptor cell fate specification in vertebrates? Brain Res. 1192, 134–150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livesey FJ & Cepko CL Vertebrate neural cell-fate determination: lessons from the retina. Nature Rev. Neurosci. 2, 109–118 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Marquardt T. & Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 25, 32–38 (2002). [DOI] [PubMed] [Google Scholar]
- 20. Carter-Dawson LD & LaVail MM Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J. Comp. Neurol. 188, 263–272 (1979). An elegant delineation of the period of rod and cone photoreceptor genesis in the mouse retina, validated over 25 years later by studies on Nrl and Thrb as specific genetic markers of newly generated rods and cones, respectively.
- 21.Young RW Cell differentiation in the retina of the mouse. Anat. Rec. 212, 199–205 (1985). [DOI] [PubMed] [Google Scholar]
- 22.Holt CE, Bertsch TW, Ellis HM & Harris WA Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron 1, 15–26 (1988). [DOI] [PubMed] [Google Scholar]
- 23.Turner DL, Snyder EY & Cepko CL Lineage-independent determination of cell type in the embryonic mouse retina. Neuron 4, 833–845 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Rapaport DH, Rakic P. & LaVail MM Spatiotemporal gradients of cell genesis in the primate retina. Perspect. Dev. Neurobiol. 3, 147–159 (1996). [PubMed] [Google Scholar]
- 25. Adler R. & Hatlee M. Plasticity and differentiation of embryonic retinal cells after terminal mitosis. Science 243, 391–393 (1989). An early demonstration of developmental plasticity in post-mitotic cells in the chick embryo retina.
- 26.Cayouette M, Poggi L. & Harris WA Lineage in the vertebrate retina. Trends Neurosci. 29, 563–570 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Agathocleous M. & Harris WA From progenitors to differentiated cells in the vertebrate retina. Annu. Rev. Cell Dev. Biol. 25, 45–69 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Reh TA & Cagan RL Intrinsic and extrinsic signals in the developing vertebrate and fly eyes: viewing vertebrate and invertebrate eyes in the same light. Perspect. Dev. Neurobiol. 2, 183–190 (1994). [PubMed] [Google Scholar]
- 29. Cepko CL, Austin CP, Yang X, Alexiades M. & Ezzeddine D. Cell fate determination in the vertebrate retina. Proc. Natl Acad. Sci. USA 93, 589–595 (1996). A comprehensive model that helped to integrate the diverse experimental observations on lineage and cell fate determination in the developing retina.
- 30.Oliver G. et al. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development 121, 4045–4055 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Mathers PH, Grinberg A, Mahon KA & Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature 387, 603–607 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Zuber ME, Gestri G, Viczian AS, Barsacchi G. & Harris WA Specification of the vertebrate eye by a network of eye field transcription factors. Development 130, 5155–5167 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Tetreault N, Champagne MP & Bernier G. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev. Biol. 327, 541–550 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Livne-Bar I. et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc. Natl Acad. Sci. USA 103, 4988–4993 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapaport DH, Wong LL, Wood ED, Yasumura D. & LaVail MM Timing and topography of cell genesis in the rat retina. J. Comp. Neurol. 474, 304–324 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Morrow EM, Belliveau MJ & Cepko CL Two phases of rod photoreceptor differentiation during rat retinal development. J. Neurosci. 18, 3738–3748 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornish EE, Hendrickson AE & Provis JM Distribution of short-wavelength-sensitive cones in human fetal and postnatal retina: early development of spatial order and density profiles. Vision Res. 44, 2019–2026 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Hendrickson A. et al. Rod photoreceptor differentiation in fetal and infant human retina. Exp. Eye Res. 87, 415–426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yaron O, Farhy C, Marquardt T, Applebury M. & Ashery-Padan R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development 133, 1367–1378 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Jadhav AP, Mason HA & Cepko CL Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133, 913–923 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Wall DS et al. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell Biol. 184, 101–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatakeyama J. & Kageyama R. Retinal cell fate determination and bHLH factors. Semin. Cell Dev. Biol. 15, 83–89 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Le TT, Wroblewski E, Patel S, Riesenberg AN & Brown NL Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev. Biol. 295, 764–778 (2006). [DOI] [PubMed] [Google Scholar]
- 44. Nishida A. et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nature Neurosci. 6, 1255–1263 (2003). This paper puts OTX2 upstream of NRL and CRX in the transcriptional hierarchy controlling photoreceptor differentiation.
- 45.Koike C. et al. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol. Cell. Biol. 27, 8318–8329 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen S. et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19, 1017–1030 (1997). Refs 46–48 independently identified the homeodomain transcription factor CRX, which plays a key part in photoreceptor development.
- 47.Furukawa T, Morrow EM & Cepko CL Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91, 531–541 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Freund CL et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91, 543–553 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Swaroop A. et al. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum. Mol. Genet. 8, 299–305 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Sohocki MM et al. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am. J. Hum. Genet. 63, 1307–1315 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furukawa T, Morrow EM, Li T, Davis FC & Cepko CL Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nature Genet. 23, 466–470 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Mitton KP et al. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J. Biol. Chem. 275, 29794–29799 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Hennig AK, Peng GH & Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 1192, 114–133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swaroop A. et al. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc. Natl Acad. Sci. USA 89, 266–270 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pittler SJ et al. Functional analysis of the rod photoreceptor cGMP phosphodiesterase α-subunit gene promoter: Nrl and Crx are required for full transcriptional activity. J. Biol. Chem. 279, 19800–19807 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Yoshida S. et al. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum. Mol. Genet. 13, 1487–1503 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Mears AJ et al. Nrl is required for rod photoreceptor development. Nature Genet. 29, 447–452 (2001). [DOI] [PubMed] [Google Scholar]
- 58. Oh EC et al. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc. Natl Acad. Sci. USA 104, 1679–1684 (2007). Refs 57 and 58 are key papers that demonstrated and established the essential and instructive role of NRL in photoreceptor differentiation.
- 59.Kobayashi M. et al. Identification of a photoreceptor cell-specific nuclear receptor. Proc. Natl Acad. Sci. USA 96, 4814–4819 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oh EC et al. Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res. 1236, 16–29 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bumsted O’Brien KM et al. Expression of photoreceptor-specific nuclear receptor NR2E3 in rod photoreceptors of fetal human retina. Invest. Ophthalmol. Vis. Sci. 45, 2807–2812 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Peng GH, Ahmad O, Ahmad F, Liu J. & Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 14, 747–764 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Cheng H. et al. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum. Mol. Genet. 15, 2588–2602 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Rattner A. & Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J. Neurosci. 25, 118–129 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J, Rattner A. & Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum. Mol. Genet. 15, 2146–2156 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Akhmedov NB et al. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc. Natl Acad. Sci. USA 97, 5551–5556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corbo JC & Cepko CL A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 1, e11 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng H. et al. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet. 13, 1563–1575 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Andre E. et al. Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 17, 3867–3877 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chow L, Levine EM & Reh TA The nuclear receptor transcription factor, retinoid-related orphan receptor β, regulates retinal progenitor proliferation. Mech. Dev. 77, 149–164 (1998). [DOI] [PubMed] [Google Scholar]
- 71. Jia L. et al. Retinoid-related orphan nuclear receptor RORβ is an early-acting factor in rod photoreceptor development. Proc. Natl Acad. Sci. USA 106, 17534–17539 (2009). This report refines the transcriptional regulatory networks and places RORβ upstream of NRL in rod differentiation.
- 72.Srinivas M, Ng L, Liu H, Jia L. & Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor β. Mol. Endocrinol. 20, 1728–1741 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Sjoberg M, Vennstrom B. & Forrest D. Thyroid hormone receptors in chick retinal development: differential expression of mRNAs for α and N-terminal variant β receptors. Development 114, 39–47 ( 1992). [DOI] [PubMed] [Google Scholar]
- 74. Ng L. et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nature Genet. 27, 94–98 (2001). A demonstration that a thyroid hormone receptor, TRβ2, is a key factor in directing differential patterning of M and S opsins in cones in the mammalian retina.
- 75.Ng L, Ma M, Curran T. & Forrest D. Developmental expression of thyroid hormone receptor β2 protein in cone photoreceptors in the mouse. Neuroreport 20, 627–631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu A. et al. Retarded developmental expression and patterning of retinal cone opsins in hypothyroid mice. Endocrinology 150, 1536–1544 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brzezinski J, Lamba DA & Reh TA Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137, 619–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katoh K. et al. Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J. Neurosci. 30, 6515–6526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szel A, van Veen T. & Rohlich P. Retinal cone differentiation. Nature 370, 336 (1994). [DOI] [PubMed] [Google Scholar]
- 80.Akimoto M. et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl Acad. Sci. USA 103, 3890–3895 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bowmaker JK Evolution of vertebrate visual pigments. Vision Res. 48, 2022–2041 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Nathans J. The evolution and physiology of human color vision: insights from molecular genetic studies of visual pigments. Neuron 24, 299–312 (1999). [DOI] [PubMed] [Google Scholar]
- 83.Mollon JD & Bowmaker JK The spatial arrangement of cones in the primate fovea. Nature 360, 677–679 (1992). [DOI] [PubMed] [Google Scholar]
- 84.Roberts MR, Hendrickson A, McGuire CR & Reh TA Retinoid X receptor γ is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest. Ophthalmol. Vis. Sci. 46, 2897–2904 (2005). [DOI] [PubMed] [Google Scholar]
- 85.Satoh S. et al. The spatial patterning of mouse cone opsin expression is regulated by bone morphogenetic protein signaling through downstream effector COUP-TF nuclear receptors. J. Neurosci. 29, 12401–12411 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fujieda H, Bremner R, Mears AJ & Sasaki H. Retinoic acid receptor-related orphan receptor α regulates a subset of cone genes during mouse retinal development. J. Neurochem. 108, 91–101 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Jones I, Ng L, Liu H. & Forrest D. An intron control region differentially regulates expression of thyroid hormone receptor β2 in the cochlea, pituitary, and cone photoreceptors. Mol. Endocrinol. 21, 1108–1119 (2007). [DOI] [PubMed] [Google Scholar]
- 88.Liu H. et al. NeuroD1 regulates expression of thyroid hormone receptor 2 and cone opsins in the developing mouse retina. J. Neurosci. 28, 749–756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G. & Reh TA Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc. Natl Acad. Sci. USA 103, 6218–6223 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pessoa CN et al. Thyroid hormone action is required for normal cone opsin expression during mouse retinal development. Invest. Ophthalmol. Vis. Sci. 49, 2039–2045 (2008). [DOI] [PubMed] [Google Scholar]
- 91. Wang Y. et al. A locus control region adjacent to the human red and green visual pigment genes. Neuron 9, 429–440 (1992). The first identification of a locus control region that helps determine the selective expression of red-sensitive and green-sensitive photopigment in a specific cone photoreceptor.
- 92.Smallwood PM, Wang Y. & Nathans J. Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc. Natl Acad. Sci. USA 99, 1008–1011 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y. et al. Mutually exclusive expression of human red and green visual pigment-reporter transgenes occurs at high frequency in murine cone photoreceptors. Proc. Natl Acad. Sci. USA 96, 5251–5256 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Deeb SS, Liu Y. & Hayashi T. Mutually exclusive expression of the L and M pigment genes in the human retinoblastoma cell line WERI: resetting by cell division. Vis. Neurosci. 23, 371–378 (2006). [DOI] [PubMed] [Google Scholar]
- 95.Tsujimura T, Chinen A. & Kawamura S. Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc. Natl Acad. Sci. USA 104, 12813–12818 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu XL et al. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell 137, 1018–1031 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Young RW Cell death during differentiation of the retina in the mouse. J. Comp. Neurol. 229, 362–373 (1984). [DOI] [PubMed] [Google Scholar]
- 98.Voyvodic JT, Burne JF & Raff MC Quantification of normal cell death in the rat retina: implications for clone composition in cell lineage analysis. Eur. J. Neurosci. 7, 2469–2478 (1995). [DOI] [PubMed] [Google Scholar]
- 99.Malicki J. Cell fate decisions and patterning in the vertebrate retina: the importance of timing, asymmetry, polarity and waves. Curr. Opin. Neurobiol. 14, 15–21 (2004). [DOI] [PubMed] [Google Scholar]
- 100.Hayashi T. & Carthew RW Surface mechanics mediate pattern formation in the developing retina. Nature 431, 647–652 (2004). [DOI] [PubMed] [Google Scholar]
- 101.Stehlin C. et al. X-ray structure of the orphan nuclear receptor RORβ ligand-binding domain in the active conformation. EMBO J. 20, 5822–5831 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu RT et al. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc. Natl Acad. Sci. USA 97, 2621–2625 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang CL, Zou Y, Yu RT, Gage FH & Evans RM Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 20, 1308–1320 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Young TL & Cepko CL A role for ligand-gated ion channels in rod photoreceptor development. Neuron 41, 867–879 (2004). [DOI] [PubMed] [Google Scholar]
- 105.Davis AA, Matzuk MM & Reh TA Activin A promotes progenitor differentiation into photoreceptors in rodent retina. Mol. Cell. Neurosci. 15, 11–21 (2000). [DOI] [PubMed] [Google Scholar]
- 106.Levine EM, Roelink H, Turner J. & Reh TA Sonic hedgehog promotes rod photoreceptor differentiation in mammalian retinal cells in vitro. J. Neurosci. 17, 6277–6288 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McFarlane S, Zuber ME & Holt CE A role for the fibroblast growth factor receptor in cell fate decisions in the developing vertebrate retina. Development 125, 3967–3975 (1998). [DOI] [PubMed] [Google Scholar]
- 108.Zhang J. et al. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nature Genet. 36, 351–360 (2004). [DOI] [PubMed] [Google Scholar]
- 109.Chen D. et al. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5, 539–551 (2004). [DOI] [PubMed] [Google Scholar]
- 110.Siffroi-Fernandez S, Felder-Schmittbuhl MP, Khanna H, Swaroop A. & Hicks D. FGF19 exhibits neuroprotective effects on adult mammalian photoreceptors in vitro. Invest. Ophthalmol. Vis. Sci. 49, 1696–1704 (2008). [DOI] [PubMed] [Google Scholar]
- 111.Hyatt GA, Schmitt EA, Fadool JM & Dowling JE Retinoic acid alters photoreceptor development in vivo. Proc. Natl Acad. Sci. USA 93, 13298–13303 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kelley MW, Williams RC, Turner JK, Creech-Kraft JM & Reh TA Retinoic acid promotes rod photoreceptor differentiation in rat retina in vivo. Neuroreport 10, 2389–2394 (1999). [DOI] [PubMed] [Google Scholar]
- 113.Khanna H. et al. Retinoic acid regulates the expression of photoreceptor transcription factor NRL. J. Biol. Chem. 281, 27327–27334 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rhee KD, Goureau O, Chen S. & Yang XJ Cytokine-induced activation of signal transducer and activator of transcription in photoreceptor precursors regulates rod differentiation in the developing mouse retina. J. Neurosci. 24, 9779–9788 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Graham DR, Overbeek PA & Ash JD Leukemia inhibitory factor blocks expression of Crx and Nrl transcription factors to inhibit photoreceptor differentiation. Invest. Ophthalmol. Vis. Sci. 46, 2601–2610 (2005). [DOI] [PubMed] [Google Scholar]
- 116.Ezzeddine ZD, Yang X, DeChiara T, Yancopoulos G. & Cepko CL Postmitotic cells fated to become rod photoreceptors can be respecified by CNTF treatment of the retina. Development 124, 1055–1067 (1997). [DOI] [PubMed] [Google Scholar]
- 117.LaVail MM et al. Protection of mouse photoreceptors by survival factors in retinal degenerations. Invest. Ophthalmol. Vis. Sci. 39, 592–602 (1998). [PubMed] [Google Scholar]
- 118.Davidson EH & Levine MS Properties of developmental gene regulatory networks. Proc. Natl Acad. Sci. USA 105, 20063–20066 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Levine M. & Davidson EH Gene regulatory networks for development. Proc. Natl Acad. Sci. USA 102, 4936–4942 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hsiau TH et al. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE 2, e643 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Qian J. et al. Identification of regulatory targets of tissue-specific transcription factors: application to retina-specific gene regulation. Nucleic Acids Res. 33, 3479–3491 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Matsuda T. & Cepko CL Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl Acad. Sci. USA 104, 1027–1032 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Friedman JS et al. The minimal transactivation domain of the basic motif-leucine zipper transcription factor NRL interacts with TATA-binding protein. J. Biol. Chem. 279, 47233–47241 (2004). [DOI] [PubMed] [Google Scholar]
- 124.Swain PK et al. Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J. Biol. Chem. 276, 36824–36830 (2001). [DOI] [PubMed] [Google Scholar]
- 125.Roger JE, Nellissery J, Kim DS & Swaroop A. Sumoylation of bZIP transcription factor NRL modulates target gene expression during photoreceptor differentiation. J. Biol. Chem. 15 Jun 2010. (doi: 10.1074/jbc.M110.142810). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kanda A, Friedman JS, Nishiguchi KM & Swaroop A. Retinopathy mutations in the bZIP protein NRL alter phosphorylation and transcriptional activity. Hum. Mutat. 28, 589–598 (2007). [DOI] [PubMed] [Google Scholar]
- 127.Onishi A. et al. Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron 61, 234–246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wright AF, Chakarova CF, Abd El-Aziz MM & Bhattacharya SS Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nature Rev. Genet. 11, 273–284 (2010). [DOI] [PubMed] [Google Scholar]
- 129.Hitchcock PF & Raymond PA The teleost retina as a model for developmental and regeneration biology. Zebrafish 1, 257–271 (2004). [DOI] [PubMed] [Google Scholar]
- 130.Vergara MN & Del Rio-Tsonis K. Retinal regeneration in the Xenopus laevis tadpole: a new model system. Mol. Vis. 15, 1000–1013 (2009). [PMC free article] [PubMed] [Google Scholar]
- 131.Nishiguchi KM et al. Recessive NRL mutations in patients with clumped pigmentary retinal degeneration and relative preservation of blue cone function. Proc. Natl Acad. Sci. USA 101, 17819–17824 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Haider NB et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nature Genet. 24, 127–131 (2000). An important paper that identified NR2E3 as the gene responsible for enhanced S cone phenotype in humans.
- 133.Wright AF et al. Mutation analysis of NR2E3 and NRL genes in enhanced S cone syndrome. Hum. Mutat. 24, 439 (2004). [DOI] [PubMed] [Google Scholar]
- 134.Jacobson SG et al. Nuclear receptor NR2E3 gene mutations distort human retinal laminar architecture and cause an unusual degeneration. Hum. Mol. Genet. 13, 1893–1902 (2004). [DOI] [PubMed] [Google Scholar]
- 135.Sharon D, Sandberg MA, Caruso RC, Berson EL & Dryja TP Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann-Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch. Ophthalmol. 121, 1316–1323 (2003). [DOI] [PubMed] [Google Scholar]
- 136.Kanda A. & Swaroop A. A comprehensive analysis of sequence variants and putative disease-causing mutations in photoreceptor-specific nuclear receptor NR2E3. Mol. Vis. 15, 2174–2184 (2009). [PMC free article] [PubMed] [Google Scholar]
- 137.Jacobson SG et al. Retinal degenerations with truncation mutations in the cone-rod homeobox (CRX) gene. Invest. Ophthalmol. Vis. Sci. 39, 2417–2426 (1998). [PubMed] [Google Scholar]
- 138.Corbo JC, Myers CA, Lawrence KA, Jadhav AP & Cepko CL A typology of photoreceptor gene expression patterns in the mouse. Proc. Natl Acad. Sci. USA 104, 12069–12074 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tucker T, Marra M. & Friedman JM Massively parallel sequencing: the next big thing in genetic medicine. Am. J. Hum. Genet. 85, 142–154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ng SB et al. Exome sequencing identifies the cause of a mendelian disorder. Nature Genet. 42, 30–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Barnhill AE et al. Characterization of the retinal proteome during rod photoreceptor genesis. BMC Res. Notes 3, 25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wolkenberg SE et al. Identification of potent agonists of photoreceptor-specific nuclear receptor (NR2E3) and preparation of a radioligand. Bioorg. Med. Chem. Lett. 16, 5001–5004 (2006). [DOI] [PubMed] [Google Scholar]
- 143. MacLaren RE et al. Retinal repair by transplantation of photoreceptor precursors. Nature 444, 203–207 (2006). This report used NRL-positive rod photoreceptor precursors to show the feasibility and challenges of cell-based replacement therapies for retinal degenerative diseases.
- 144.Meyer JS et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc. Natl Acad. Sci. USA 106, 16698–16703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lamba DA, Gust J. & Reh TA Transplantation of human embryonic stem cell-derived photoreceptors restores some visual function in Crx-deficient mice. Cell Stem Cell 4, 73–79 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Osakada F. et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nature Biotech. 26, 215–224 (2008). [DOI] [PubMed] [Google Scholar]
- 147.Koso H. et al. CD73, a novel cell surface antigen that characterizes retinal photoreceptor precursor cells. Invest. Ophthalmol. Vis. Sci. 50, 5411–5418 (2009). [DOI] [PubMed] [Google Scholar]
- 148.Alvarez-Delfin K. et al. Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc. Natl Acad. Sci. USA 106, 2023–2028 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Parapuram S. & Swaroop A. Eye, Retina, and Visual System of the Mouse (eds Chalupa LM & Williams RW) 675–683 (MIT Press, Cambridge, Massachusetts; London, UK: 2008). [Google Scholar]
- 150.Yu J. et al. From disease genes to cellular pathways: a progress report. Novartis Found. Symp. 255, 147–160; discussion 160–144, 177–148 (2004). [DOI] [PubMed] [Google Scholar]
- 151.Hansen RM & Fulton AB The course of maturation of rod-mediated visual thresholds in infants. Invest. Ophthalmol. Vis. Sci. 40, 1883–1886 (1999). [PubMed] [Google Scholar]
- 152.Hansen RM & Fulton AB Development of the cone ERG in infants. Invest. Ophthalmol. Vis. Sci. 46, 3458–3462 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lerner LE, Gribanova YE, Ji M, Knox BE & Farber DB Nrl and Sp nuclear proteins mediate transcription of rod-specific cGMP-phosphodiesterase β-subunit gene: involvement of multiple response elements. J. Biol. Chem. 276, 34999–35007 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.