Abstract
Background
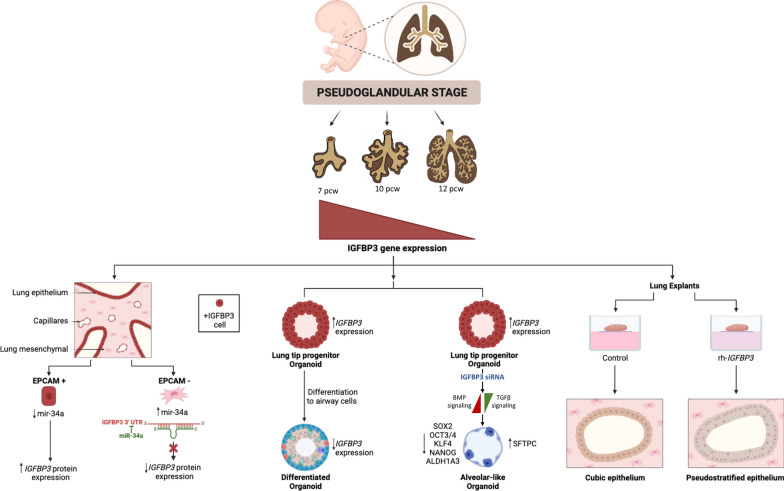
During pseudoglandular stage of the human lung development the primitive bronchial buds are initially conformed by simple tubules lined by endoderm-derived epithelium surrounded by mesenchyme, which will progressively branch into airways and start to form distal epithelial saculles. For first time alveolar type II (AT2) pneumocytes appears. This study aims to characterize the genes and microRNAs involved in this differentiation process and decipher its role in the starting alveolar differentiation.
Methods
Gene and microRNA profiling was performed in human embryonic lungs from 7 to 12 post conception weeks (pcw). Protein expression location of candidate genes were analyzed by immunofluorescense in embryonic lung tissue sections. mRNA/miRNA target pairs were identified using computational approaches and their expression was studied in purified epithelial/mesenchymal cell populations and in isolated tips and stalks from the bronchial tree. Additionally, silencing experiments in human embryonic lung mesenchymal cells and in human embryonic tip-derived lung organoids were performed, as well as organoid differentiation studies. AT2 cell markers were studied by qRT-PCR and by immunofluorescence. The TGFB-β phosphorylated pathways was analyzed with membrane protein arrays. Lung explants were cultured in air/liquid interface with/without peptides.
Results
We identified 88 differentially expressed genes, including IGFBP3. Although IGFBP3 mRNA was detected in both epithelial and mesenchymal populations, the protein was restricted to the epithelium, indicating post-transcriptional regulation preventing IGFBP3 protein expression in the mesenchyme. MicroRNA profiling identified miR-34a as an IGFBP3 regulator. miR-34a was up-regulated in mesenchymal cells, and its silencing in human embryonic lung mesenchymal cells increased IGFBP3 levels. Additionally, IGFBP3 expression showed a marked downregulation from 7 to 12 pcw, suggesting its involvement in the differentiation process. The differentiation of human tip-derived lung embryonic organoids showed a drastic reduction in IGFBP3, supported by the scRNAseq data. IGFBP3 silencing in organoids activated an alveolar-like differentiation process characterized by stem cell markers downregulation and upregulation of AT2 markers. This process was mediated by TGFβ signalling inhibition and BMP pathway activation.
Conclusions
The IGFBP3/miR-34a axis restricts IGFBP3 expression in the embryonic undifferentiated lung epithelium, and the progressive downregulation of IGFBP3 during the pseudoglandular stage is required for alveolar differentiation.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-024-03883-1.
Keywords: Human lung embryogenesis, IGFBP3, miR-34a, Organoids, Mesenchyme, Epithelium, Tips, Stalks, Alveolospheres
Background
Human lung development begins at 3–4 post conception weeks (pcw) and can be divided into five stages according to major histological changes: embryonic (3–4 to 6–8 pcw), pseudoglandular (6–8 to 16 pcw), canalicular (16 to 26–28 pcw), saccular (26–28 to 32–36 pcw) and alveolar (32–36 to pcw to 2–4 years postnatal age) (1–3). During the pseudoglandular stage, a gradual process of bronchial bud branching leads to the formation of successive generations of airways. Histologically, in the early pseudoglandular stage, the primitive bronchial buds are simple tubules lined by a thick layer of endoderm-derived epithelium, surrounded by abundant loose mesenchyme [1, 4]. These bronchial buds undergo progressive branching, resulting in increasing generations of airways initially characterized by a pseudostratified epithelium, which, around 12 pcw (late pseudoglandular stage), becomes lined by a simple columnar respiratory epithelium [4, 5]. Later, the epithelial cells will differentiate into ciliated cells, goblet cells and basal cells, surrounded by mesenchyme with a simple capillary network [6]. This epithelium is notable for prominent subnuclear vacuolization, conferring a “pseudoglandular” appearance to these simple tubules [1].
At molecular level, numerous studies have demonstrated the importance of paracrine signaling coordination between mesenchyme and undifferentiated epithelium for normal cell differentiation and lung morphogenesis [7–10]. Although most studies have been performed in animal models, key differences have been reported recently in human lung development [9, 11] and organoid studies have begun to decipher the role of molecular pathways directly in human samples [12–14]. Single-cell RNAseq has highlighted the existence of a proximal–distal gradient in epithelial differentiation and different niche regions in the mesenchymal compartment with distinct signaling interactions [15–18].
Despite these advancements, the number of studies using human embryonic samples remains limited. Additionally, variability among samples due to differences in gestational age measurement or interindividual differences necessitates further research and validation through functional studies. Moreover, many signaling networks have not been thoroughtly studies, particulary the role of non-coding RNAs, including microRNAs (miRNAs), in regulating human lung development [4, 19, 20]. miRNAs are small non-coding RNA molecules (22–24 nucleotides in length) that negatively regulate the expression of their target genes post-transcriptionally, mainly by blocking the translation of messenger RNA (mRNA) into protein or degrading it [21, 22]. Furthermore, they have been described to play an important role in stem cell regulation [23, 24]. Most miRNA profile studies in lung development have been performed in rats [25, 26] or mouse fetal lungs [27–29], with few in human embryonic samples [4, 27, 30].
For the proper development of the human lung, coordination between epithelial cells and the surrounding mesenchyme is essential. Numerous molecules are involved in cell differentiation and morphological development of the lung, such as SOX2 and SOX9 genes, TGFβ family and the WNT pathway. The insulin-like growth factor binding protein (IGFBP) family comprises six secreted proteins (IGFBP1 to IGFBP6) that can bind and regulate the insulin-like growth factors (IGFs), which regulate cell proliferation, differentiation and survival [31]. IGFBP3 is the most abundant circulating IGFBP and can be found in the uterus, mammary gland, and placenta [32]. The mature IGFBP3 protein plays numerous key roles in the adult human by acting both in an IGF-dependent and -independent manner. It is known to interfere with TGFβ/Smad signaling, inhibiting cell proliferation and inducing apoptosis [33, 34]. IGFBP3 is a soluble protein that can participate in local and distal communication [35], highlighting its interest in development. Although most studies have been focused on the role of the soluble IGFBP3, different functions have been described for cellular IGFBP3, including the control of stemness and differentiation processess [36, 37].
In the present study, we investigated genes and miRNAs involved in the differentiation process during human pseudoglandular stage by studying human embryonic lungs from 8 to 12 pcw of development. During the analysis, we identified the interaction between IGFBP3 and miR-34a and demonstrated that miR-34a mediated regulation restricted the expression of IGFBP3 protein to the epithelium, where it plays a role in the proximal–distal differentiation process. IGFBP3 silencing is essential for allowing alveolar differentiation.
Methods
Human embryonic samples
Twenty-five clinically aborted embryos and fetuses were donated with written informed consent to the Body Donation Service of the Human Anatomy and Embryology Unit of the Faculty of Medicine and Health Sciences of University of Barcelona for morphological and molecular studies. The samples included lungs from the 7th to the 13th pcw of development. Lung samples were obtained under control of an Olympus stereo microscope SZ61. The study was approved by the Bioethics Commission of the University of Barcelona (project approval number IRB00003099).
RNA extraction
Total RNA was extracted using Trizol reagent (Invitrogen) as per the manufacturer’s protocol. The RNA concentration was quantified using a NanoDrop 1000 Spectrophotometer. The quality of total RNAs was estimated by Agilent 2100 Bioanalyzer using the RNA 6000 NANO kit. For the microarray study, only samples with RIN > 8 were included.
mRNA profiling
Global gene expression in 6 whole embryonic lung samples and 3 adult lung normal tissues were analyzed by Affymetrix GeneChip Human Gene 2.1 ST array in the Genomics core facility from IDIBAPS. Raw.cel files were provided which were used for further analysis. Bioinformatic analysis was performed using R version 4.1.1 and Bioconductor packages. Background correction, filtering and normalization were performed with robust multi-array analysis (RMA) method from oligo package (Bioconductor). Differential expression analysis was performed using limma package. Hierarchical cluster analysis of top differentially expressed genes (DEG) were plotted using heatmap.2 from gplots package. Enrichment analysis for Gene Ontology (GO) terms was performed using topGO package from Bioconductor.
miRNA profiling
miRNA expression analysis was performed as previously described using TaqMan® Array Human MicroRNA Set Cards v2.0 (ThermoFisher, Waltham, Massachusetts, USA) [38] that included 758 miRNAs. Real-time quantitative PCR reactions were performed on an ABI 7900 HT Sequence Detection System (ThermoFisher). Relative miRNA expression was calculated using the 2−ΔΔCt method. Normalization was performed with miR-191, based on preliminary analyses comparing the stability of several endogenous controls including RNU6B, RNU24, RNU43, RNU44, RNU48 and MammU6 and potential endogenous miRNAs; miR-191 had the lowest variability. As calibrator sample, we used the mean miRNA value of an adult lung normal tissue. All miRNAs that were expressed in less than 10% of the samples or with an unreliable quantification were excluded from further analysis, leaving a final working set of 257 miRNAs.
miRNA target identification
RmiR package (Bioconductor version 2.6) was used to identify potential miRNA-mRNA pairs as previously performed [38]. Using RmiR we cross-correlate the expression data from the DEG mRNA signature with the whole miRNA expression data in the same embryonic samples (n = 6), based on the predicted targets from TargetScan (http://www.targetscan.org/) database. The identified mRNA/miRNA pairs with a minimum negative correlation of -0.4, were validated in all available embryonic samples (n = 22) using single miRNA and miRNA assays by real time PCR.
Immunofluorescence
Formalin fixed paraffin embedded (FFPE) tissue sections were pre-treatred as follows: 5-μm-thick tissue sections were mounted onto Dako Silanized Slides (Dako, Glostrup, Denmark). For dewaxing and antigen retrieval, the sections were manually immersed in Target Retrieval solution (S1699, Dako) and heated at 95–99 °C for 20 min in Dako PT Link instrument. Then the temperature was allowed to go down until 65 °C and then the slides were washed three times in PBS at room temperature (RT) for 5 min each.
Cultured mesenchymal cells were pre-treated as follows [10]: cells seeded on coverslips were fixed in 4% paraformaldehyde in DPBS at RT followed by permeabilization with 0.5% of Triton X-100 (Santa Cruz Biotechnology, Dallas, TX, USA) in DPBS 1x (PBTx) at RT for 5 min.
Both, FFPE and Fixed cultured cells, were then blocked with 1% Bovine Serum Albumin (BSA) (Sigma-Aldrich) and 5% Normal Donkey Serum (NDS) (Sigma-Aldrich, Sant Luis, Missouri, USA) in PBTx for 60 min at RT. After that they were incubated with primary antibodies (Table S1) diluted in blocking solution overnight at 4 °C in darkness. After primary antibody washing (3 × 5 min PBS), secondary antibodies (Table S1) diluted in 5% NDS in PBTx 0.1% were incubated for 2 h at RT in darkness. Finally, samples were stained and mounted on microscope slides using Fluoroshield with DAPI (Sigma-Aldrich).
Immunofluorescence analysis was performed using Olympus BX51 Fluorescence Microscope (Olympus, Tokyo, Japan) and cell images were acquired by DPController and DPManager software (Olympus).
Isolation of epithelial and mesenchymal enriched populations
Fetal lung samples were mechanically dissociated using a scalpel and enzymatically dissociated using Collagenase/Dispase (Cat N. 10,269,638,001, Roche; Merck Millipore, Burlington, Massachusetts, USA) (45 min at 37 °C) and Trypsin–EDTA (0.25%), phenol red (Cat N. 25,200,056, Life Technologies) (10 min at 37 °C). Then samples were loaded in a 15 mL tube and incubated with DMEM and DNase I (10 mL DMEM + 13.5 μl DNase I) during 10 min in agitation. To remove cell clumps, the samples were filtered using a 100 μm and a 40 μm filter. Filtered samples were centrifuged for 5 min at 550 g at 4 °C and the cell pellet was washed with 1 mL of Buffer (PBS, pH 7.2, 0.5% BSA, 2 mM EDTA) and transferred to a 1.5 mL tube. Then, cells were centrifuged 5 min at 550g at 4 °C and the cell pellet was resuspended in 300 μl of Buffer. After cell counting, 5 × 107 cells were used for further processing.
To perform the magnetic labelling, cells were mixed with 100 μl FcR Blocking Reagent, human (Miltenyi Biotec, Cologne, Germany). Then, 100 μl CD326 (EpCAM) Microbeads (Miltenyi Biotec) were added and incubated during 30 min at 4 °C. After incubation, 1 mL of Buffer was added and samples were centrifuged at 300g for 10 min. The cell pellet was resuspended in 500 μl of buffer.
To proceed with the magnetic separation, the MS columns (Miltenyi Biotec) were placed in the magnetic field and hydrated with 500 μl of Buffer. Cell suspension was applied to the column, and we collected the flow-through containing the EPCAM- cells (mesenchymal enriched population). Then the MS column was washed three times with 500 μl of Buffer.
Finally, we removed the MS column from the magnetic field and placed the column in a 1.5 mL collection tube. Then 500 μl of buffer was added to recover the EPCAM + cells (epithelial enriched population).
mRNA and single miRNA quantification
mRNA and miRNAs were quantified as previously described using TaqMan assays (the arrays ussed are shown Table S1) [39, 40].
Culture and transfection of lung mesenchymal cells
Human embryonic lung mesenchymal cells were obtained from the culture of the mesenchymal enriched population obtained after EPCAM ± microbead purification, and cultured as previously described [10].
One day before transfection, 7 × 104 cells were seeded in 6-well plates. The following day, cells were transfected with 200 nM miR-34a-ASO (5′-ACAACCAGCTAAGACACTGCCA-3′) or ASO control (5′-GATACCAGGGACATACGCTTGATCCTAGC-3′) using Lipofectamine 3000 (Life Technologies). After 24 h incubation, miR-34a levels were quantified to corroborate proper miR-34a inhibition.
Tips and stalks acquirement from human embryonic lungs
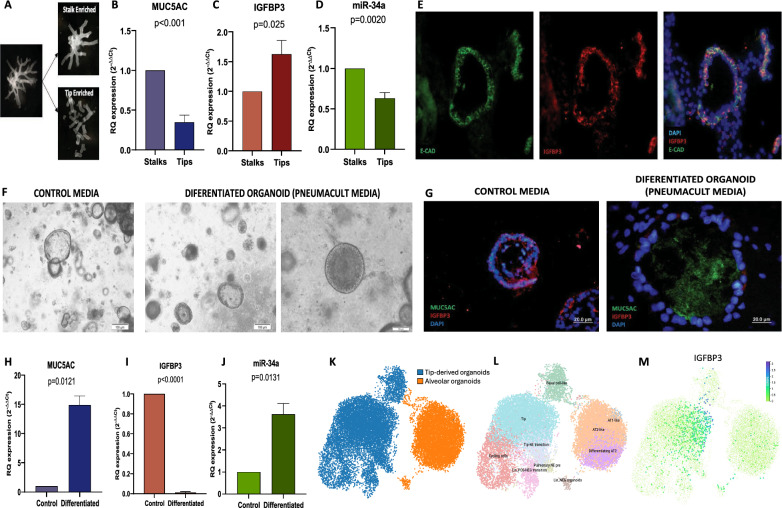
Human embryonic lung lobes were incubated in Dispase II 8U/mL (D4693, Merck Millipore) 30 min in ice followed by 15 min in an orbital mixer at room temperature. Mesenchyme was dissected away under a stereoscopic microscope using sterilized ophthalmic microsurgery instrumental and tungsten needles. Tips and stalks were isolated by cutting the very end of the branching tip and the stalk area more proximally, as it is indicated in Fig. 5a and Video S1. Obtained tips and stalks were processed for organoid culture (tips) and for RNA purification (tips and stalks).
Fig. 5.
IGFBP3 overexpression in tips and in tip-derived organoids becomes downregulated with the differentiation process. a Tip and stalk isolation after treatment of the bronchial tree with dispase and mechanical separation of the mesenchyme from the epithelium under the stereoscopic microscope. b Quantification of MUC5AC, c IGFBP3 and d miR-34a mRNA levels in tip and stalk enriched populations (n = 9 per group). e Immunofluorescence analysis of IGFBP3 and E-CAD in tip-derived organoids. f Microscope images showing gross morphology of tip-derived organoids cultured with control media vs differentiated organoids after 30-day culture with differentiation media (Pneumacult). g Immunofluorescence analysis of IGFBP3 and MUC5Ac in organoids cultured with control media and after 30-day differentiation with Pneumacult media. h Analysis of MUC5AC, i IGFBP3 and j miR-34a mRNA levels in control and differentiated organoids (n = 3 per group). k UMAP visualization of single-cell RNA sequencing data from control and alveolar differentiated tip-derived organoids, with colors representing the cell of origin. l UMAP visualization showing the different cell types identified in the control and alveolar differentiated tip-derived organoids. m UMAP illustrating IGFBP3 transcript expression in the control and alveolar differentiated tip-derived organoids
Organoid culture
Human embryonic lung organoids were obtained from culture of lung tips. Immediately after obtention, tips were transferred into cold liquid Matrigel (Corning Matrigel Growth Factor Reduced Basement Membrane Matrix, Corning, NY, USA), and then 30μL of Matrigel containing the tips were placed in a 12-well plate. The plate was then incubated for 30 min at 37 °C to allow Matrigel polymerize and solidify. Afterwards, each well was filled with 1000μL of organoid medium prepared according to [13]. Organoid medium contained: Advanced DMEM/F12 (ThermoFisher) supplemented with Amphotericin B (2.5ug/ml, ThermoFisher), 1 × Glutamax (ThermoFisher), 1 × N2 supplement (ThermoFisher), 1 × B27 supplement (ThermoFisher), 0.05% BSA (Sigma-Aldrich), 2% PS (ThermoFisher), FGF7 (10 ng/mL, Stemcell Technologies, Grenoble, France), CHIR99021 (3 μM, Stemcell Technologies), Retinoic Acid (50 nM, Stemgent, Cambridge, Massachusetts, USA), 50 μg/mL Ascorbic Acid (Sigma Aldrich) and 0,4 μM 1-Thioglycerol (Sigma Aldrich). The plate was then incubated under standard tissue culture conditions (37 °C, 5% CO2). Tips growth in a patterned structure before first passage and acquired a spheric structure thereafter (Fig. S1). Organoids used in the different experiments were obtained at passage 3–4.
Organoid differentiation
To differentiate human embryonic lung tip-derived organoids to proximal airways, organoid medium was removed from the plate and exchanged by 1000μL of an established human airway differentiating medium, PneumaCult™ (Stem Cell Technologies, Vancouver, BC, Canada). Organoids were grown in this medium for 5 pcw at regular culture conditions (37 °C, 5% CO2).
Organoids recovery and paraffin embedding for immunofluorescence studies
Organoid medium was aspirated from the well and using 1 mL of cold Advanced DMEM/F12 medium we detached the Matrigel drop. Matrigel drop including organoids was transferred into a 15 mL tube and cold consecutive washes (3x) with 10 mL cold DMEM/F12 was required for organoid recovering from Matrigel. Lastly, organoids were further washed with 1 mL Corning Cell Recovery Solution (Corning, NY, USA) and incubated on ice for 30 min, inverting the tube after 15 min. Then, organoids were washed with cold DPBS and centrifuged at 200g for 5 min at 4 °C. Supernatant was discarded and afterwards organoids were fixed in paraformaldehyde 4% for 30 min at RT. Thereafter organoids were washed with DPBS for 3 times and after centrifuging at 200g for 5 min at 4 °C the pellet was included in 100 μl of Histogel (Thermo Fisher Scientific, Waltham, Massachusetts) for 45 min (or until solidification). Once included in Histogel, dehydration procedure in scale-up ethanol concentrations (70%, 80%, 95% and 100%) was performed prior to xylene treatment and paraffin embedding.
Organoid nucleofection
Organoids (cultured according to [14]) were mechanically dissociated using glass Pasteur pipettes. After cell counting, 2 × 105 cells were re-suspended with Lonza P3-Primary cell nucleofection Solution and 1 μl of IGFBP3 DsiRNA (Design id: hs.Ri.IGFBP3.13.1) or Negative Control DsiRNA (IDT, Coralville, Iowa) were added. Then the solution was transferred to 100 μl nucleofection cuvette (V4XP-3012, Lonza, Basel, Switzerland). Nucleofection was performed using Lonza 4D Nucleofector with X unit using the program EA125. After nucleofection, organoid culture medium supplemented with 10 μM Y-27632 (ROCK inhibitor, ROCKi, 72,304, StemCell) was added to the nucleofection cuvette and cell mixture was then recovered and seeded into Matrigel and distributed in 24-well plate wells and cultured with organoid culture medium with 10 μM ROCKi. Sequential photos were obtained using EzScope 101 live cell imaging system (Blue-Ray Biotech, New Taipei City, Taiwan) during 48 h before RNA extraction.
Lung explant culture with IGFBP3
Small pieces of human embryonic lungs were placed on top of a Nucleopore Track-Etch membrane (Whatman, Florham Park, NJ) suspended over culture media (D-MEM/F12 supplemented with Penicillin/Streptomycin) in 12 well plates. 50 nM of Recombinant human IGFBP3 protein (Active) (ab280941, Abcam, Cambridge, UK) or PBS (Control) was added to the medium. The lung explants were cultured in the cell incubator (37 °C, 5% CO2) for 2 days and then the samples were formalin-fixed and paraffin-embedded for histological analysis.
Protein arrays
C-Series Human TGF beta Pathway Phosphorilation Array C1 (cat. No. AAH-TGFB-1, RayBiotech, Georgia, USA) was used to evaluate 8 phosphorilated human proteins from TGFβ/Smad signaling, including ATF2 (P-Thr69/71), c-Fos (P-Thr232), c-Jun (P-Ser73), SMAD1 (P-Ser463/465), SMAD2 (P-Ser245/250/255), SMAD4 (P-Thr277), SMAD5 (P-Ser463/465) and TAK1 (P-Ser412). Organoids were nucleofected with control or IGFBP3 siRNA as previously described and total protein was purified at 24 h. Fifty µg of total protein was used for the analysis. The protein array incubation and protein detection was performed as per the manufacturer’s protocol. Chemiluminescence detection was performed on ChemiDoc (Bio-Rad, California, USA). For protein quantification, intensities were measured using Quantity One software v4.6.3 (Bio-Rad). In total 6 protein Arrays (corresponding to 3 independent replicates) were used to identify differences between control and IGFBP3 siRNA-transfected organoids in three independent replicates.
In silico analysis of single cell RNAseq data on alveolar organoids
Single cell RNAseq data from control and alveolar differentiated organoids were obtained from [12]. The analysis of IGFBP3 expression levels and cell location in the cell populations present in the control and alveolar differentiated organoids were analysed using CELLxGENE portal [41]. Available at https://fetal-lung-organoid.cellgeni.sanger.ac.uk/.
Statistical analysis
T-tests or U-Mann Whitney when appropriate, were used for comparisons between two groups of samples and ANOVA test for comparisons between more than two groups. Pearson correlation was used to correlate mRNA and miRNA expression. For all transfection experiments comparing control vs. condition, one T-test analysis was used. All statistical analyses were performed using GraphPad Prism 9 (La Jolla, CA, USA) or R program version 4.1.1.
Results
Analysis of differential mRNA expression from early to late pseudoglandular stage of human lung development
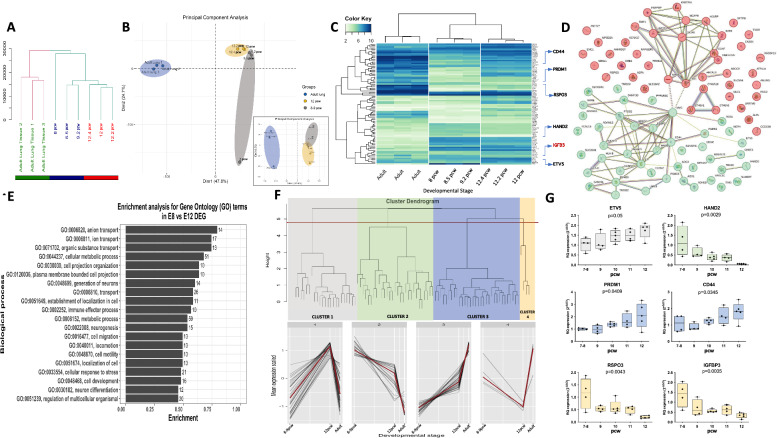
To identify genes involved in the regulation of the differentiation process occurring from 8 to 12 pcw of human embryonic lung development we performed expression arrays in 9 lung samples, including three 8–9 pcw, three 12 pcw embryonic lungs and 3 human adult tissues. The hierarchical cluster based on the whole mRNA expression revealed two well-differentiated groups: adult tissues; and embryonic tissues. Focusing on embryonic tissues we can observe that clearly early pseudoglandular stage samples (8 and 9 pcw samples) grouped together separately from late pseudoglandular stage samples (12 pcw) (Fig. 1a). The principal component analysis reinforced this observation by differentiating 3 basic groups from left to right in the graph (Dimension 1, explaining 47.8% of variance vs. Dimension 2, 24.7% of variance; and Dimension 1 vs. Dimension 3, 6.5% of variance): Adult tissues; 12 pcw; and 8–9 pcw embryonic lungs (Fig. 1b).
Fig. 1.
Gene expression analysis in 8 vs 12 pcw human embryonic lungs and adult lung samples. a Hierarchical cluster analysis showing how the different samples group according to their expression pattern. b Principal component analysis showing that samples group according to their age using Dimension 1 (47.8% of variability) and Dimension 2 (24.7%). In the small box in the right bottom the PCA have been represented using Dimension 1 and Dimension 3 (6.5%) and sows similar aggrupation clusters. c Heat map with the 88 differentially expressed genes (DEG) identified between 8 vs 12 pcw in human embryonic lungs. The hierarchical cluster showed that using this gene signature samples grouped in 3 groups: adult; 8–9 pcw; and 12 pcw. Arrows indicate the location of the genes that we have further analyzed in the following analysis, including IGFBP3 which is highlighted in red. d String analysis (https://string-db.org) showing the relation between the identified genes. Color nodes are based on the results of the k-means clustering analysis (n = 2). e Enrichment analysis for Gene Ontology (GO) terms using the DEG identified. The numbers indicated in the bars indicate the number of genes included in each GO term. f Hierarchical cluster analysis by gene showing 4 gene clusters with different expression trends. The expression trend from 8 pcw to adult of each cluster is shown in the bottom of the graph and the red line is showing the mean expression of all genes included in the cluster. g Expression analysis by real time PCR of selected genes of each cluster in embryonic samples from 7–8 (n = 4), 9 (n = 4), 10 (n = 5), 11 (n = 4) and 12 (n = 5) pcw human lung embryonic tissues
Differential expression analysis showed 88 differentially expressed genes (DEG) (p < 0.001; Table S2) between 8 and 9 pcw samples and 12 pcw samples. Figure 1c shows a heat map of the expression of the DEG in all analyzed samples allowing us to observe the expression trend of the DEG also in the adult tissue.
String analysis was performed to identify the interactome between the 88 DEG identified. Fifty-two genes (59.1%) showed described interactions with other genes from the signature. K-means analysis constated the presence of two main clusters of genes according to their interactions connected by MYC (Fig. 1d).
The enrichment analysis for Gene Ontology (GO) terms (Fig. 1e, and Table S3) showed that the most significantly enriched GO terms were related to “anion”, “ion” and “organic substance” transport (GO:0006820, GO:0006811, GO:0071702). Interestingly, also GO terms related to “generation of neurons”, “neurogenesis” and “neuron differentiation” can be observed (GO:0048699, GO:0022008, GO:0030182) which can be explained by the presence at this time of neural crest populations [16]. Also, more general GO terms related to development such as “cell development” and “regulation of multicellular organismal” can be observed (GO:0048468, GO:0051239).
Next, we investigated the expression trends of the DEG identified. The Hierarchical cluster analysis by genes identified 4 clusters with different expression trends between 8 and 9 pcw to adult. Cluster 1 included genes whose expression increased from 8–9 to 12 pcw, but that decreased in adult lung tissue; Cluster 2 included genes that become downregulated from 8 pcw to adult; Cluster 3 included genes that became upregulated from 8–9 pcw to adult; and finally, Cluster 4 include genes which expression decreases from 8–9 to 12 pcw, but that become upregulated in adult tissue (Fig. 1F). To validate that these trends were continuous during the pseudoglandular stage we added embryonic samples from 10 and eleven pcw to the analysis. We chose several genes of each cluster and analyzed them by real-time PCR in at least 4 samples of each pcw from 7–8 to 12 (4 samples of 7–8 pcw, 4 samples of 9 pcw, 5 samples 10 pcw, 4 samples of 11 pcw and 5 samples of 12 pcw human lung embryonic tissues). For the analysis, we selected genes of interest in each cluster: ETV5 from Cluster 1; HAND2 from Cluster 2; PRDM1 and CD44 from Cluster 3; and RSPO3 and IGFBP3 from Cluster 4. In all genes, we observed a linear trend in the expression level according to the developmental time. The ANOVA analysis showed significant differences between different pcw in all genes (p ≤ 0.05) (Fig. 1g). For further analysis, we decided to focus on the IGFBP3 gene.
IGFBP3 protein expression was restricted to the epithelium
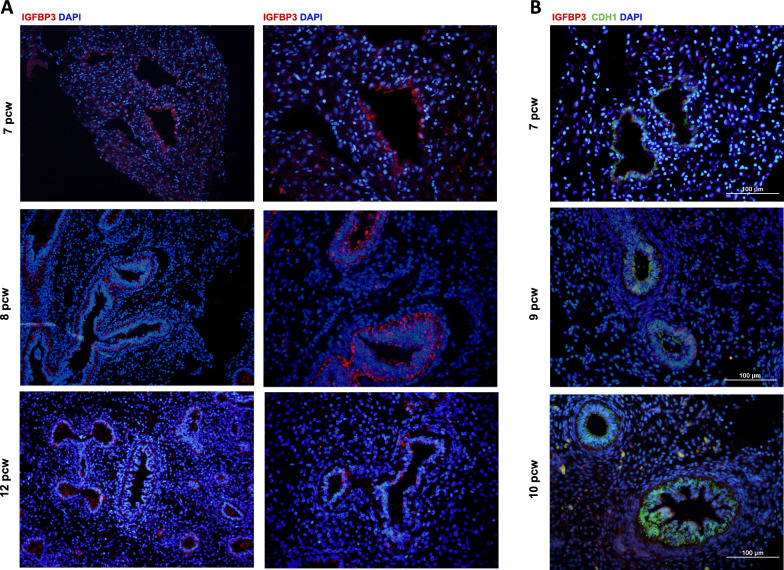
The analysis by immunofluorescence of IGFBP3 protein expression in lung sections from 7 to 12 pcw of development showed that the protein expression was enriched in the epithelium and highly reduced in the surrounding mesenchyme (Fig. 2a). Especially, in 8- and 9-pcw samples the IGFBP3 expression was restricted in the epithelium and not detected in the surrounding mesenchyme. Colocalization of IGFBP3 and the epithelial marker E-CAD corroborated the epithelial expression (Fig. 2b).
Fig. 2.
Immunofluorescence detection of IGFBP3 in human lung embryonic and fetal tissues. a IGFBP3 protein expression in human embryonic lungs from 7 to 12 pcw showing preferential expression in epithelial cells. b IGFBP3 and E-CAD protein expression in 7–10 pcw lung embryonic tissues showing co-expression of both markers in epithelial cells
IGFBP3 mRNA expression in epithelial vs mesenchymal enriched populations
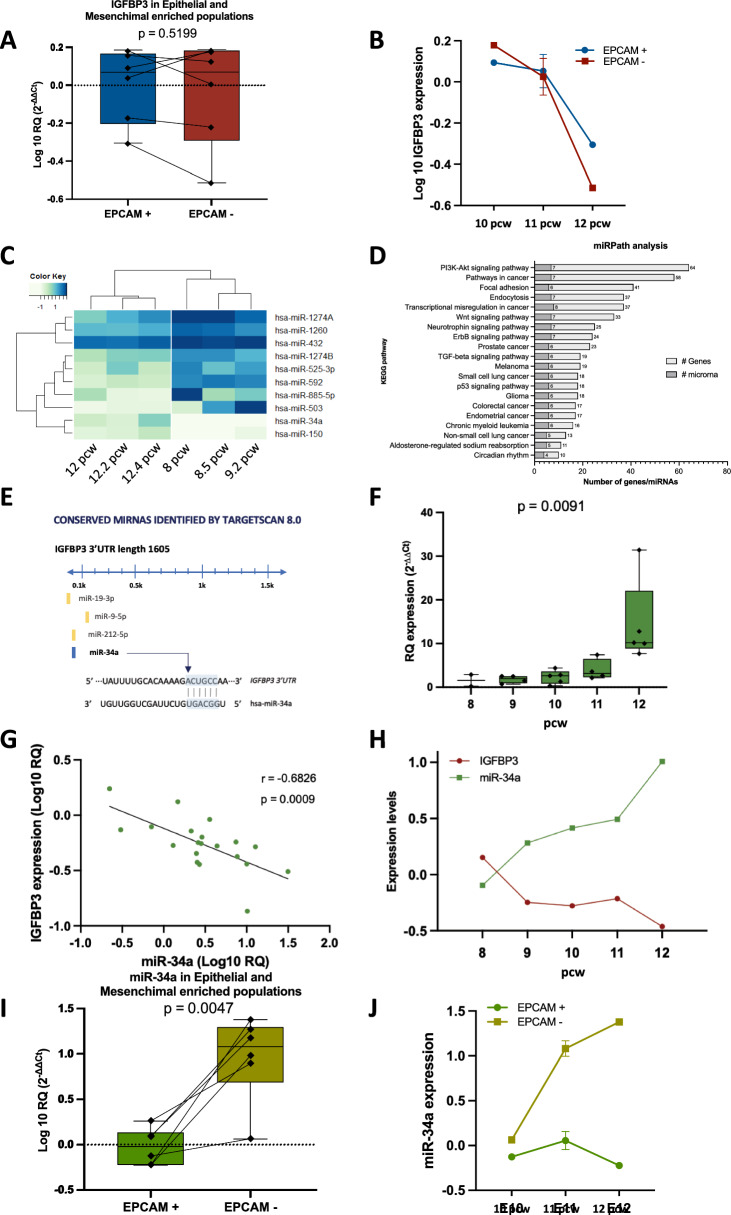
To determine whether the observed differences at the protein level between epithelium and mesenchyme were related to differences in mRNA expression between the different compartments, we decided to dissociate the tissue and isolate using an EPCAM antibody epithelial enriched populations (EPCAM + population), and mesenchyme enriched population (EPCAM- population) and analyzed IGFBP3 expression separately. We could not observe significant differences in IGFBP3 mRNA levels between both populations that could explain the differences observed at the protein level (Fig. 3a). In fact, the expression in both populations decreased from 10 to 12 pcw (Fig. 3b) as previously observed when analyzing the whole lung (Fig. 1g). This result suggested the involvement of a potential post-transcriptional regulation, such as miRNAs, to explain the differences in IGFBP3 protein expression between epithelium and mesenchyme.
Fig. 3.
IGFBP3 is negatively regulated by miR-34a in the mesenchymal compartment. a IGFBP3 mRNA expression analysis in EPCAM + (epithelial enriched) vs EPCAM- (mesenchymal enriched) purified cells not showing significant differences between both populations. b IGFBP3 mRNA expression by pcw in EPCAM + vs EPCAM- cell populations showing that in both groups of cells IGFBP3 mRNA expression decreases over time. c Heatmap of the 10 differentially expressed miRNAs identified between 8 and 12 pcw. d DIANA-miRPath v2.0 analysis (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index) using the 10 differentially expressed miRNAs identified. The KEGG signaling pathways identified are ordered by number of genes targeted by the selected miRNAs. The x-axis indicates the number of genes and miRNAs identified in the miRNA/target interaction analysis which are included in each KEGG pathway identified showed in the y-axis. e Schematic representation of the 3’UTR region of IGFBP3 and the predicted conserved miRNAs targeting it, including miR-34a which base to base pairing is represented in the bottom of the figure. f Analysis by real time PCR of miR-34a expression in embryonic samples from 8 (n = 2), 9 (n = 4), 10 (n = 5), 11 (n = 4) and 12 (n = 5) pcw human lung embryonic tissues showing that miR-34a expression increases over time. g Correlation analysis in paired samples of miR-34a and IGFBP3 expression showing a significant negative correlation between both genes. h Quantification of IGFBP3 and miR-34a shows an opposing pattern of expression. IGFBP3 becomes downregulated over time while miR-34a becomes upregulated. i miR-34a expression analysis in EPCAM + vs EPCAM- purified cells showing significant differences between both populations. miR-34a is upregulated in the mesenchymal enriched population. j miR-34a expression by pcw in EPCAM + vs EPCAM- cell populations showing that while in the EPCAM- group of cells miR-34a expression increases over time, in the EPCAM + group the miR-34a expression remains stable at low levels
Analysis of differential miRNA expression from early to late pseudoglandular stage of human lung development
To identify miRNAs involved in the pseudoglandular stage differentiation process we analyzed the expression of 758 miRNAs in the same samples previously used in the mRNA profile (n = 6, Fig. 1). Differential expression analysis identified 10 miRNAs differentially expressed between 8 and 9 pcw samples and 12 pcw lung samples (Fig. 3c and Table S4).
We performed an exploratory analysis using miRPath v2.0 to identify potential pathways regulated by the differentially expressed miRNAs. Twenty-one KEGG pathways were identified with a p ≤ 0.001, with several genes being potential targets of the identified miRNAs (Fig. 3d and Table S3). The “ErbB signaling pathway” was the most significant pathway identified, including 24 genes targeted by 7 of the identified miRNAs. Other key lung pathways identified in the analysis included the “PI3K-Akt signaling pathway”, the “Wnt signaling pathway”, the “TGF-beta signaling pathway” and the “p53 signaling pathway”.
Identification of miRNAs regulating IGFBP3
To identify potential miRNAs targeting IGFBP3 in the miRNA signature, we cross-correlated the miRNA expression with DEG identified in the same embryonic sample using RmiR program, which uses predicted target information from TargetScan (http://www.targetscan.org/). RmiR identified several gene-miRNAs pairs that showed an inverse expression correlation (minimum considered − 0.4) and that TargetScan identified as potential candidates: TNXB and miR-148a (r = − 0.4154632); RSPO3 and miR-16 (r = − 0.4288525); PRDM1 and miR-186 (r = − 0.6858153); RSPO3 and miR-195 (r = − 0.6850341); HAND2 and miR-25 (r = − 0.4435238); and the highest inverse correlation was identified between IGFBP3 and miR-34a (r = − 0.7947488).
In Fig. 3e are represented the 3’UTR region of IGFBP3 and the binding sites of the conserved miRNAs predicted by TargetScan 8.0, which includes miR-34a, which was also one of the miRNAs identified differentially expressed between 8–9 and 12 pcw of lung development. To validate the identified correlation between miR-34a and IGFBP3 during the pseudoglandular stage of lung development. We analyzed the expression of miR-34a using single TaqMan assays in all the embryonic samples available (n = 20, Fig. 3f). miR-34a gradually increases its expression from 8 to 12 pcw of lung development (ANOVA p = 0.0091). Moreover, the correlation analysis in all samples validated the negative correlation identified between IGFBP3 and miR-34a (r = − 0.6826; p = 0.0009, Fig. 3g). In Fig. 3h, we can observe how IGFBP3 shows an opposite pattern of expression with miR-34a, with the highest IGFBP3 and the lowest miR-34a levels in the early pseudoglandular stage (8 pcw) and with the lowest IGFBP3 and highest miR-34a levels in late pseudoglandular stage (12 pcw). During 9, 10 and 11 pcw we can observe a gradual downregulation and upregulation of IGFBP3 and miR-34a, respectively.
miR-34a is upregulated in the mesenchyme
When we analyzed the expression of miR-34a in epithelial enriched populations (EPCAM +) and mesenchyme enriched population (EPCAM-) we observed that it was significantly upregulated in EPCAM- cells (p = 0.0047, Fig. 3i). Of note, the miR-34a expression in EPCAM- populations increased from 10 to 12 pcw (Fig. 3j) as previously observed when analyzing the whole lung (Fig. 3f), while in the EPCAM + population, the miR-34a expression was maintained at constant low levels during these pcw.
miR-34a is inhibiting IGFBP3 translation in lung embryonic mesenchymal cells
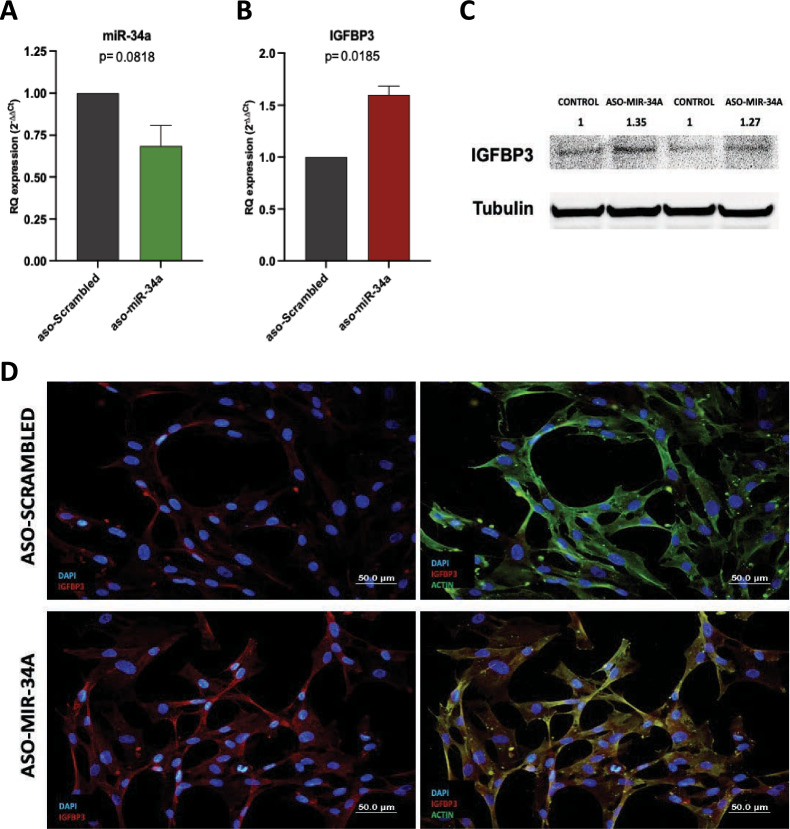
To confirm that miR-34a is regulating IGFBP3 protein levels in the embryonic lung mesenchyme, and therefore is responsible of the lack of IGFBP3 protein expression in the mesenchymal compartment during pseudoglandular stage, we decided to culture primary lung embryonic mesenchymal cells where to inhibit miR-34a levels and study IGFBP3 expression. We inhibited miR-34a using an antisense oligonucleotide (ASO) that reduced miR-34a expression a mean of 31% (p = 0.0818, Fig. 4a) and this was associated with a significant increment of IGFBP3 mRNA levels in the mesenchymal cells transfected with the miR-34a ASO (p = 0.0185, Fig. 4b) in comparison to control cells transfected with a scrambled oligo. Interestingly, this increase in the mRNA levels was also observed at the protein level where we observed a 30% increase in the mesenchymal cells where we silenced miR-34a (Fig. 4c). To further validate this observation, we performed immunofluorescence analysis of IGFBP3 in the cultured embryonic lung mesenchymal cells after silencing miR-34a, and as shown in Fig. 4d, IGFBP3 protein levels increased in the cytoplasm of silenced cells and colocalized with Actin expression in these cells.
Fig. 4.
miR-34a inhibition increases IGFBP3 levels in human lung embryonic mesenchymal cells. Transfection of human lung embryonic mesenchymal cells with an antisense oligonucleotide (ASO) against miR-34a or with a scrambled ASO and analysis of a miR-34a and b IGFBP3 mRNA levels (n = 4 per group). c Analysis of IGFBP3 protein levels by western blot in human lung embryonic mesenchymal cells transfected with ASO-miR-34a or ASO-scrambled. The image showed is a cropped gel focused in the proteins of interest. Full-length blots are presented in Additional file 10. d Immunofluorescence analysis of IGFBP3 and Actin in human lung embryonic mesenchymal cells transfected with ASO-scrambled or with ASO-miR-34a
Differential expression of IGFBP3 in the proximal-distal airway and overexpression in tip-derived organoids
To study whether IGFBP3 expression in epithelial cells could be involved in proximal-distant patterning, we decided to analyze its expression in different locations of the bronchial tree. For this purpose, we isolated the bronchial tree by separating enzymatically first and then manually the mesenchyme, and while working under the stereoscopic microscope, we isolated tips and stalks (Fig. 5a, Video S1). After RNA purification from both isolated tissues, the analysis of MUC5AC mRNA levels, a gene overexpressed in the proximal embryonic airway (stalks) [14], confirmed that we correctly purified enriched populations since MUC5AC were significantly overexpressed in stalk in comparison to the tip enriched population (p < 0.001, Fig. 5b). The analysis of IGFBP3 mRNA levels in both populations showed an upregulation in the distal epithelial population, in the tips (p = 0.025, Fig. 5c). Moreover, miR-34a showed an inverse expression, with downregulation in the tip population (p = 0.002, Fig. 5d). The protein analysis of IGFBP3 in tip-derived organoids confirmed at the protein level the upregulation of IGFBP3 in the tip population since IGFBP3 protein was highly expressed in the epithelial cells of tip-derived organoids (Fig. 5e). E-cadherin was used as an epithelial marker to confirm that IGFBP3 expression was in epithelial cells from the tip-derived organoids.
Organoid differentiation drastically reduces IGFBP3 expression
To study if IGFBP3 expression is lost due to the differentiation process occurring during the branching morphogenesis occurring during pseudoglandular stage we decided to differentiate tip-derived organoids to the proximal airway by culturing them in differentiating medium. Differentiated organoids acquire morphological changes (Fig. 5f) derived of the appearance of differentiated cells such as ciliated and goblet cells. The presence of goblet cells produces an accumulation of mucus in the lumen of the organoid, positive for MUC5AC, which was used as a control for the organoid differentiation process in this article. Thus, it was observed that both MUC5AC protein expression (Fig. 5g) and mRNA expression (Fig. 5h) levels were elevated in differentiated organoids compared to controls. Analysis of IGFBP3 in control and differentiated organoids showed an obvious reduction of IGFBP3 levels in the epithelium of differentiated organoids (Fig. 5g, i), while miR-34a exhibited increased mRNA expression in the differentiated organoids compared to undifferentiated controls (Fig. 5j).
Single-cell RNAseq analysis of alveolar-differentiated organoids showed a drastic reduction of IGFBP3 levels
Using scRNA data from [12], we analyzed the expression of IGFBP3 in tip-derived organoids and alveolar-differentiated organoids. Confirming our previous results, we observed that IGFBP3 expression was drastically reduced after organoid differentiation (Fig. 5k–m). In control tip- derived organoids, IGFBP3 expression was mostly located in tip cells. Additionally, some high IGFBP3 expression was observed in basal cell-like cells. After differentiation, a few dispersed cells of the alveolar-differentiated organoid expressed IGFBP3 at low levels, with a signal concentrated in the lineage negative (Lin_NEG) organoid cell population (Fig. 5l, m).
IGFBP3 silencing induces alveolar differentiation in organoids
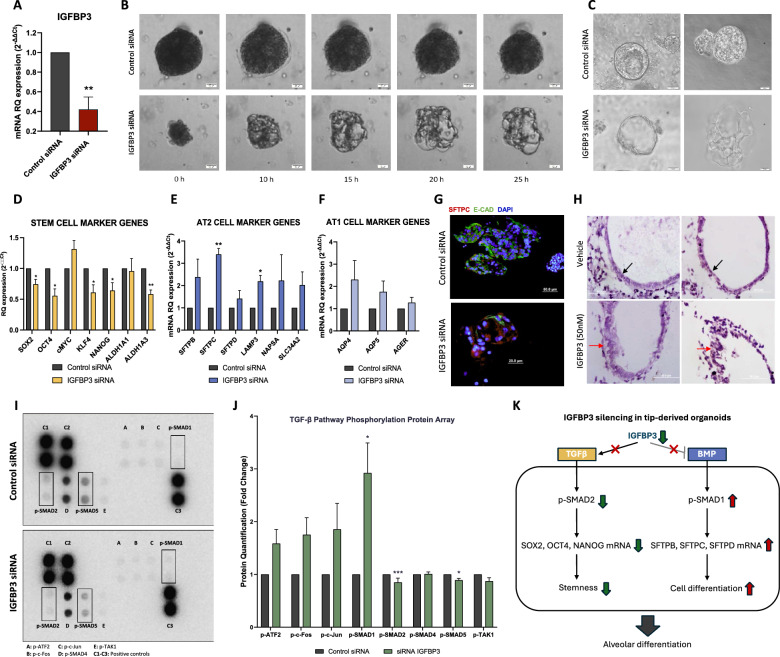
Based on the previous observation that showed the loss of IGFBP3 after differentiation, we aimed to assess its role in the differentiation process. Using nucleofection technology and DsiRNAs we inhibited the expression of IGFBP3 in 12 pcw tip-derived organoids (Fig. 6a). Time-lapse study after transfection capturing one frame every 5 h for 48 h showed that IGFBP3 silenced organoids become differentiated in alveolar-like structures (Fig. 6b, Video S2 and S3). In Fig. 6c can be observed the alveolar-like structures obtained after 48 h of IGFBP3 silencing in tip-derived organoids which resemble alveolospheres.
Fig. 6.
Silencing of IGFBP3 in tip-derived organoids induces alveolar-like differentiation. a IGFBP3 mRNA levels in tip-derived organoids after siRNA silencing (n = 6 per group). b Time-lapse images of control and IGFBP3 silenced tip-derived organoids during 25 h after transfection. c Representative images of Control and IGFBP3 silenced tip-derived organoids at 48 h post-transfection showed differentiation in alveolar-like structures in the IGFBP3 silenced group. d Expression analysis of stem cell markers in control and IGFBP3 siRNA-transfected organoids (n = 4 per group). e Expression analysis of AT2 cell markers in control and IGFBP3 siRNA-transfected organoids (n = 4 per group). f Expression analysis of AT1 cell markers in control and IGFBP3 siRNA-transfected organoids (n = 4 per group). g Immunofluorescence analysis of SFTPC and E-CAD in control and IGFBP3 silenced organoids. h Hematoxylin staining of tissue sections of lung explants from 10 pcw embryos cultured for 48 h with or without IGFBP3 peptide. A distinctive epithelial morphology can be observed between both groups: the control group showed a cubic epithelium (black arrows) while the IGFBP3 group remained in a more undifferentiated stage (red arrows). i Representative images of one of the 3 independent replicates of TGFβ protein arrays showing differences in p-SMAD1, p-SMAD2 and p-SMAD5 between control and IGFBP3 siRNA-transfected organoids. j Protein quantification in 3 independent replicates of TGFβ related phosphorylated proteins using the protein Arrays in control and IGFBP3 siRNA-transfected organoids (n = 3 per group). k Schematic representation of the effect of IGFBP3 silencing in tip-derived organoid on TGFβ and BMP signaling. *p < 0.05; **p < 0.01; ***p < 0.001
To explore the molecular changes occurring after IGFBP3 silencing we analyzed the expression of six stem cell markers and nine alveolar markers. Compared to control organoids, the IGFBP3 silenced organoids displayed a significant downregulation of stem cell markers including OCT4, KLF4, NANOG and ALDH1A3 (Fig. 6d), which denotes differentiation and loss of stemness potential in line with the observed morphological changes. Interestingly, a significant upregulation of the alveolar type 2 (AT2) cell marker SFTPC and LAMP3 (Fig. 6e) was observed denoting alveolar differentiation. Although not significant, other AT2 (Fig. 6e) and alveolar type 1 (AT1) (Fig. 6f) cell markers also showed upregulation 48 h after IGFBP3 silencing. Immunofluorescence analysis of SFTPC in control and silenced organoids confirmed the overexpression of the AT2 marker SFTPC in the IGFBP3 silenced organoids (Fig. 6g).
High IGFBP3 levels decelerate the differentiation process in lung explants
Lung explants from 10 pcw embryos cultured for 48 h with IGFBP3 peptide had a distinctive epithelial morphology compared to control explants (Fig. 6h). In the histological analysis (hematoxylin staining), while a more cuboidal epithelium was observed in control explants after 48 h of culture, the IGFBP3 explants remained in a previous step showing a more pseudostratified epithelium that had not completed the differentiation to a cubic endodermic epithelium yet.
IGFBP3 silencing induces TGFβ inhibition and BMP activation
To understand the signaling events that control IGFBP3-mediated alveolar differentiation, we decided to profile the levels of 8 phosphorylated proteins related to TGFβ signaling pathway, a key pathway involved in several differentiation processes that have been described to interact with IGFBP3 [33, 34]. Twenty-four hours after silencing IGFBP3 in the tip-derived organoids, we observed a significant inhibition of TGFβ signaling, indicated by the downregulation of p-SMAD2 (p = 0.0001), and an increase in BMP signaling, demonstrated by the significant upregulation of p-SMAD1 (p = 0.0197). Although not significant, there was also an upregulation of p-ATF2 (p = 0.0793), p-c-Fos (p = 0.0692) and p-c-Jun (p = 0.1441). Additionally, a discrete downregulation of p-SMAD5 was noted (p = 0.0203) (Fig. 6i, j). Interestingly, the downregulation of p-SMAD2 correlates with the downregulation of previously described transcriptionally activated target genes, including SOX2, OCT4 and NANOG (Fig. 6d, k). Also, the upregulation of p-SMAD correlates with the upregulation of activated target genes, including SFTPB, SFTPC and SFTPD (Fig. 6e, k), confirming the functional effect of IGFBP3 silencing on the downregulation and upregulation of the TGFβ and BMP signaling, respectively (Fig. 6k).
Discussion
In this study, we conducted an in-depth analysis of the genes and miRNAs involved in the differentiation process from the early to late pseudoglandular stages of human lung development. Before branching initiation, the endodermal bud, surrounded by mesenchymal cells, begins to transform in a pseudostratified epithelium around 10 pcw, and by 12 pcw, a cuboidal epithelium can be observed. These epithelial modifications are closely related to changes in the mesenchymal compartment, which, through its signals, it is the main promoter of the epithelial transformations. We identified 88 DEG that, according to its interactome, could be grouped mainly into two distinct clusters connected by the proto-oncogene MYC, whose expression slightly decreases during the analyzed period. The MYC expression has been shown to be more generalized across different embryonic organs during mouse development compared to other family members [42]. Despite transcriptomic heterogeneity between different studies due to technical, analytical or sample differences [43], we identified genes in our signatures previously described such as CD44, which expression increases and has been shown to be expressed in the epithelial cells of the tips [12], and ETV5, a transcription factor associated with the promotion of epithelial cell differentiation and inhibition of proliferation [44], which becomes upregulated from 8 to 12 pcw in our study. GO analysis allowed us to explore the pathways regulated by the identified genes in more detail. The DEGs could be grouped in several functional categories (Table S3), including “cell development”, which is inherent to the process studied as it describes the progress of a specific cell from an immature to a mature state. This category contains 16 genes, including ETV5 and HAND2. Additionally, other important functional groups identified were related to transport of anions and ions, which include 14 and 17 genes, respectively, such as ATP11A or SLC10A2. The importance of ion and anion transport during lung development has been studied and associated with the lungs as secretory organs during this period. Before birth, the placenta is in charge of the respiratory gas exchange, and the lungs are filled with liquid [45]. To produce the liquid that moves from the basal to the apical part of the epithelium and is eventually released into the lumen, robust control of the anions and ions such as Na+, K+, or Cl− is necessary. Most of these studies have been performed in animal models, but alteration in these molecules have been associated with respiratory diseases in the newborns [46] and even with lung malformations [45]. With the emergence of organoids models we can envision that the results obtained in animal models can be translated to human embryology. Using human embryonic organoids we could study most of these processes directly in human embryonic samples.
Finally, it is noteworthy that we identified 15 genes grouped in GO categories associated with neuron generation and differentiation. This can be explained by the presence at this time of neural crest populations identified in recent single cell RNA studies [16].
To further analyze the gene signature, we grouped genes by expression trends accross three time points: 8–9 pcw, 12 pcw, and adult. Four clusters were identified. A selected group of genes, including the epithelial transcription factor ETV5 (cluster 1), the mesodermal transcription factors HAND2 (cluster 2) and PRDM1 (cluster 3), the cell surface receptor CD44 (cluster 3), the soluble protein pro-angiogenic factor RSPO3 (cluster 4) and the soluble protein IGFBP3 (cluster 4). We validated that their expression trends were gradual by studying them in all available embryonic samples, which allowed us to include additional time points (8, 9, 10, 11 and 12 pcw). Most of these genes were chosen because they showed interactions with the miRNA signature: RSPO3-miR-16; HAND2-miR-25; PRDM1-miR-186 and IGFBP3-miR-34a. Further studies on these gene-miRNA pairs are warranted; however, in the present paper, we have only focused on one of these interactions. During the pseudoglandular stage, the lung is filled with liquid, and several molecules cross the epithelium to be secreted into the lumen. Subsequently, they circulate from the distal to the proximal respiratory airway, eventually passing through the larynx to be expelled out by the mouth and mixed with the amniotic liquid. Therefore, we decided to focus on the study of IGFBP3, a soluble protein previously detected in amniotic liquid [47], as we hypothesized its involvement in distal–proximal communication through the lumen. We observed that the IGFBP3 protein was restricted to epithelium, as miR-34a targeted it in the mesenchyme. These results underscore the necessity for current single-cell RNA studies to validate the results by analyzing the protein levels. As we observed, despite similar mRNA levels between different cell populations such as stromal and epithelial cells, the action of a miRNA can restrict the final protein to one compartment or the other, leading to important functional consequences.
Moreover, considering the KEGG pathways identified enriched in our miRNA signature, the miR-34a/IGFBP3 axis is directly related with “p53 signaling pathway”. miR-34a have been described previously as a transcriptional target of p53 that collaborates in the induction of p53-mediated apoptosis [48]. In this line, IGFBP3 regulates the scape from apoptosis, which is a characteristic of undifferentiated stem cells. Therefore, the downregulation of miR-34a and upregulation of IGFP3 in the epithelium seem to allow the scape from growth inhibition, senescence and apoptosis required for the maintenance of an undifferentiated stage during the first stages of lung developoment. Although acting independently they regulates cell death, it has also been described that working together regulates autophagy in a patient asthma model [49].
Finally, since distal–proximal differentiation begins during the pseudoglandular stage, we decided to evaluate the role of IGFBP3 in this process. IGFBP3 was upregulated in the tips, where a lung stem cell population resides. This observation was validated using human tip-derived organoids, where high IGFBP3 levels were observed. Interestingly, IGFBP3 expression became drastically reduced when we differentiate the tip-derived organoids. IGFBP3 was mostly expressed in a stem cell population, which comprises most of the cells in the epithelium of the tip-derived organoid. The differentiation process drastically reduced their expression restricting it to only a few cells retaining the stem cell phenotype. Using single-cell RNAseq data from [12] we confirmed that in alveolar-differentiated tip-derived organoids, the expression of IGFBP3 became drastically reduced. Initially, IGFBP3 expression was mainly located in tip and basal-like cell populations of the undifferentiated organoid, but after differentiation, only a few cells retained IGFBP3 expression. These cells were mainly located in linage-negative cell populations in the differentiated organoid. This lineage-negative cell population were characterized by the expression of tip progenitors but not AT2 markers, confirming that IGFBP3 after differentiation remains in cell populations not completely differentiated.
In this line, when we inhibited IGFBP3 expression using siRNAs in tip-derived organoids, an alveolar-like differentiation was triggered with a significant reduction of stemness. The silencing of IGFBP3 downregulated the expression of pluripotent stem cell markers OCT4, KLF4, Nanog and ALDH1A3. The most established role of IGFBP3 in stemness suggests that soluble IGFBP3, either directly through binding its receptor TMEM219 or indirectly by sequestering IGF-1, can inhibit stem cell proliferation and induce differentiation, as seen in colonic stem cells [50]. In contrast, in lung cancer stem cells, it has been reported that the neutralization of IGFBP3 reduces stemness potential, evidenced by the downregulation of ALDH1A1 and ALDH1A3. BIRC5 and IGFBP3 have an autocrine regulatory relationship, activating signaling mechanisms that regulate cancer stem cells and epithelial-mesenchymal transition [36]. BIRC5 regulates the secretion of IGFBP3, and the loos of IGFBP3 causes a downregulation of BIRC5 levels. Moreover, some authors have described that cellular IGFBP3 can be associated with a more stem-like state, with its function directly related to its cellular location. For instance, in osteosarcoma, it has been reported that nuclear localization of IGFBP3 is associated with a highly differentiated phenotype, while high cytoplasmic IGFBP3 levels are consistent with dedifferentiation [37]. Consistent with this, our immunofluorescence analysis showed high IGFBP3 levels in the cytoplasm of lung epithelial cells from both embryos and tip-derived organoids.
Upon observing morphological changes indicative of alveolar differentiation, we decided to study AT2 and AT1 cell markers whose upregulation supported the observed alveolar differentiation. Notably, the most remarkable upregulation was observed in the AT2 gene SFTPC, at both mRNA and protein levels. SFTPC, described to be exclusively expressed in the lung, serves as a highly specific marker for identifying AT2 cells [51, 52], thus validating the alveolar-like differentiation observed in our morphological analysis. Subsequently, employing an embryonic lung explant model cultured with IGFBP3 peptide, we corroborated the role of this protein in maintaining a more undifferentiated phenotype. Control lung explants exhibited a more differentiated cubic epithelium compared to explants cultured with high doses of IGFBP3. These findings collectively support the role of IGFBP3 as a stem cell gene whose loos is necessary for completing the differentiation process during lung pseudoglandular stage epithelium development.
Finally, to interrogate the mechanisms involved in IGFBP3 mediated alveolar differentiation, we analyzed the Transforming Growth Factor (TGF) family-related proteins after silencing IGFBP3 in tip-derived organoids. The TGF superfamily consists of several ligands, including TGFβ signaling proteins (including SMAD2) and BMP signaling proteins (including SMAD1). TGFβ and BMP pathways have been described to regulate stemness and differentiation through opposing and cooperative activities [53]. Interestingly, after silencing IGFBP3 we observed a downregulation of the TGFβ signaling mediated by a decrease in p-SMAD2, which leads to a reduction of SOX2, OCT4 and NANOG mRNA levels, resulting in diminished stem cell self-renewal. SOX2, OCT4 and NANOG has been previously demonstrated to be transcriptional targets of SMAD2 [54, 55]. At the same time, we noted an upregulation of BMP signaling, indicated by an increased levels of p-SMAD1, p-c-Jun and p-c-Fos. Notably, in mice, it has been demonstrated that the activation of BMP by p-SMAD1, helps to maintain the AT2 identity and allows these cells to differentiate into either AT2 or AT1 cells [56]. It is described that activation of BMP-SMAD1 promotes the expression of Nfatc3, an important transcription activator for surfactant proteins [57], which correlates with our results where we observed an upregulation of SFTPB, SFTPC and SFTPC. Moreover, Frum et al. have recently demonstrated that during alveolar differentiation, inhibithion of TGFβ and simultaneous activation of BMP are required during the differentiation process in humans [58]. Our results show that IGFBP3 activates the necessary crosstalk between TGFβ and BMP signaling that ends in alveolar differentiation. TGFβ signaling, responsible for mantaininig a stem cell phenotype (undifferentiated state), needs to be inhibithed, and at the same time BMP signaling needs to be upregulated to induce/mantein the specific linage differentiation, first to AT2 and subsequently to AT1.In this line, supporting our results (Fig. 6k) it has been reported that in human embryonic stem cells SMAD2 through its transcriptional target Nanog can also repress BMP signaling to maintain the undifferentiated state [59].
Conclusions
In conclusion, our investigation has described a distinctive mRNA and miRNA signature that delineates the critical shift from the early to late pseudoglandular stage in human lung development highlighting the interplay between IGFBP3 and miR-34a which is critical for alveolar differentiation.
Supplementary Information
Additional file 1. Table S1. Primary and secondary antibodies and qPCR probes used in the study.
Additional file 2. Video S1. Isolation of lung tips under the stereoscopic miscroscope using tungsten needles.
Additional file 3. Fig. S1. Follow-up of tip-derived organoid growth before and after the first passage. A Tip-derived organoid growth from day 0 (tip inclusion into matrigel) to 14 showing patterned structure. B Patterned structure of tip-derived organoids resembling a disorganized bronchial tree. C and D Organoids generate spheric structures after the first passage losing the patterned structure.
Additional file 4. Table S2. List of differential expressed genes between 8- and 12-pcw embryonic lungs.
Additional file 5. Table S3. Gene ontology terms identified in the analysis of the differentially expressed genes between 8 and 12 pcw embryonic lungs and the list of genes included in each identified gene ontology term.
Additional file 6. Table S4. List of differential expressed miRNAs between 8 and 12 pcw embryonic lungs.
Additional file 7. Video S2. Time-lapse video where can be observed the morphological changes of organoids transfected with control siRNA during the first 48h after nucleofection.
Additional file 8. Video S3. Time-lapse video where can be observed the morphological changes of organoids transfected with IGFBP3 siRNA during the first 48h after nucleofection.
Acknowledgements
The authors thank: Dr. S. Barambio for comments on embryology. D. Fuster for technical help. The Body Donation Service of the Human Anatomy and Embryology unit of the Faculty of Medicine and Health Sciences of the University of Barcelona for providing funds for the purchase of laboratory materials. The authors declare that they have not used Artificial Intelligence in this study.
Abbreviations
- AT1
Alveolar type 1
- AT2
Alveolar type 2
- ASO
Antisense oligonucleotide
- BSA
Bovine serum albumin
- DEG
Differentially expressed genes
- FFPE
Formalin fixed paraffin embedded
- GO
Gene ontology
- microRNAs
MicroRNAs
- NDS
Normal donkey serum
- pcw
Post conception weeks
- RMA
Multi-array analysis
- RT
Room temperature
Author contributions
M.A-P. and J.J.C. performed the research, the molecular analysis, and the statistical analysis, and wrote the manuscript. T.D., Y.H. R.M.M. collaborated on the research. A.N. designed the study, wrote the manuscript, performed the bioinformatic analysis and took responsibility for the integrity of the data and the accuracy of the molecular and data analysis. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the Ministry of Economy and Competition (MINECO), co-financed with the European Union FEDER funds (SAF2017-88606-P, 2017); SEPAR-AstraZeneca Ayudas Investigación PII Oncología 2021; Becas SEPAR 2022 (n° de Proyecto 1326). Melissa Acosta-Plasencia is a FI-SDUR fellow from AGAUR.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and supplementary materials.
Declarations
Ethics approval and consent to participate
Written informed consent for the use of the clinically aborted embryos and fetuses was obtained from the legally authorized representatives. The representatives were thoroughly informed about the study and were assured of the confidentiality of their personal data. The study was approved by the Bioethics Commission of the University of Barcelona (Approval number: IRB00003099). Title of the approved project: “Characterization of piwiRNAs in Embryonic Lung Stem Cells and Their Importance in Non-Small Cell Lung Cancer” (Approval date: March 20, 2018).
Consent for publication
All authors read and approved the final manuscript for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Melissa Acosta-Plasencia and Joan Josep Castellano have contributed equally to this work.
References
- 1.Schittny JC. Development of the lung. Cell Tissue Res. 2017;367:427–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rackley CR, Stripp BR. Building and maintaining the epithelium of the lung. J Clin Investig. 2012;122(8):2724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ernst LM, Ruchelli ED, Carreon CK, Huff DS. Color atlas of human fetal and neonatal histology. Berlin: Springer; 2019. [Google Scholar]
- 4.Navarro A, Marrades RM, Viñolas N, Quera A, Agustí C, Huerta A, et al. MicroRNAs expressed during lung cancer development are expressed in human pseudoglandular lung embryogenesis. Oncology. 2009;76(3):162–9. [DOI] [PubMed] [Google Scholar]
- 5.Joshi S, Kotecha S. Lung growth and development. Early Human Dev. 2007;83(12):789–94. [DOI] [PubMed] [Google Scholar]
- 6.Mullassery D, Smith NP, editors. Lung development. Seminars in pediatric surgery. Amsterdam: Elsevier; 2015. [DOI] [PubMed] [Google Scholar]
- 7.Mendelson CR. Role of transcription factors in fetal lung development and surfactant protein gene expression. Annu Rev Physiol. 2000;62(1):875–915. [DOI] [PubMed] [Google Scholar]
- 8.Shannon JM, Hyatt BA. Epithelial-mesenchymal interactions in the developing lung. Annu Rev Physiol. 2004;66:625–45. [DOI] [PubMed] [Google Scholar]
- 9.Nikolić MZ, Sun D, Rawlins EL. Human lung development: recent progress and new challenges. Development. 2018;145(16):dev163485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canals J, Navarro A, Vila C, Canals JM, Díaz T, Acosta-Plasencia M, et al. Human embryonic mesenchymal lung-conditioned medium promotes differentiation to myofibroblast and loss of stemness phenotype in lung adenocarcinoma cell lines. J Exp Clin Cancer Res. 2022;41(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danopoulos S, Alonso I, Thornton ME, Grubbs BH, Bellusci S, Warburton D, et al. Human lung branching morphogenesis is orchestrated by the spatiotemporal distribution of ACTA2, SOX2, and SOX9. Am J Physiol-Lung Cell Mol Physiol. 2018;314(1):L144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim K, Donovan AP, Tang W, Sun D, He P, Pett JP, et al. Organoid modeling of human fetal lung alveolar development reveals mechanisms of cell fate patterning and neonatal respiratory disease. Cell Stem Cell. 2023;30(1):20–37. [DOI] [PubMed] [Google Scholar]
- 13.Miller AJ, Hill DR, Nagy MS, Aoki Y, Dye BR, Chin AM, et al. In vitro induction and in vivo engraftment of lung bud tip progenitor cells derived from human pluripotent stem cells. Stem cell Rep. 2018;10(1):101–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolić MZ, Caritg O, Jeng Q, Johnson J-A, Sun D, Howell KJ, et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. Elife. 2017;6:e26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He P, Lim K, Sun D, Pett JP, Jeng Q, Polanski K, et al. A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates. Cell. 2022;185(25):4841–60. [DOI] [PubMed] [Google Scholar]
- 16.Cao S, Feng H, Yi H, Pan M, Lin L, Zhang YS, et al. Single-cell RNA sequencing reveals the developmental program underlying proximal–distal patterning of the human lung at the embryonic stage. Cell Res. 2023;33:421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sountoulidis A, Marco Salas S, Braun E, Avenel C, Bergenstråhle J, Theelke J, et al. A topographic atlas defines developmental origins of cell heterogeneity in the human embryonic lung. Nat Cell Biol. 2023;25(2):351–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller AJ, Yu Q, Czerwinski M, Tsai Y-H, Conway RF, Wu A, et al. In vitro and in vivo development of the human airway at single-cell resolution. Dev Cell. 2020;53(1):117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoshgoo N, Kholdebarin R, Iwasiow BM, Keijzer R. MicroRNAs and lung development. Pediatr Pulmonol. 2013;48(4):317–23. [DOI] [PubMed] [Google Scholar]
- 20.Gross N, Kropp J, Khatib H. MicroRNA signaling in embryo development. Biology. 2017;6(3):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallory AC, Vaucheret H. MicroRNAs: something important between the genes. Curr Opin Plant Biol. 2004;7(2):120–5. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32(4):189–97. [DOI] [PubMed] [Google Scholar]
- 23.Navarro A, Monzó M. MicroRNAs in human embryonic and cancer stem cells. Yonsei Med J. 2010;51(5):622–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng Z-L, Lin X-l, Tan L-L, Liu Y-M, Qu K, Wang Z. MicroRNAs: important regulators of induced pluripotent stem cell generation and differentiation. Stem Cell Rev Rep. 2018;14:71–81. [DOI] [PubMed] [Google Scholar]
- 25.Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, et al. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37(3):268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Kai G, Pu X-D, Qing K, Guo X-R, Zhou X-Y. Expression profile of microRNAs in fetal lung development of Sprague-Dawley rats. Int J Mol Med. 2012;29(3):393–402. [DOI] [PubMed] [Google Scholar]
- 27.Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn Off Publ Am Assoc Anat. 2007;236(2):572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouhaddioui W, Provost PR, Tremblay Y. Expression profile of androgen-modulated microRNAs in the fetal murine lung. Biol Sex Differ. 2016;7(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mujahid S, Logvinenko T, Volpe MV, Nielsen HC. miRNA regulated pathways in late stage murine lung development. BMC Dev Biol. 2013;13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin NW, Liu C, Yang IV, Maier LA, DeMeo DL, Wood C, et al. Sex-specific differences in microRNA expression during human fetal lung development. Front Genet. 2022;13:762834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daza DO, Sundström G, Bergqvist CA, Duan C, Larhammar D. Evolution of the insulin-like growth factor binding protein (IGFBP) family. Endocrinology. 2011;152(6):2278–89. [DOI] [PubMed] [Google Scholar]
- 32.Ranke MB. Insulin-like growth factor binding-protein-3 (IGFBP–3). Best Pract Res Clin Endocrinol Metab. 2015;29(5):701–11. [DOI] [PubMed] [Google Scholar]
- 33.Baxter RC. Insulin-like growth factor binding protein-3 (IGFBP-3): novel ligands mediate unexpected functions. J Cell Commun Signal. 2013;7:179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varma Shrivastav S, Bhardwaj A, Pathak KA, Shrivastav A. Insulin-like growth factor binding protein-3 (IGFBP-3): unraveling the role in mediating IGF-independent effects within the cell. Front Cell Dev Biol. 2020;8:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada PM, Lee K-W. Perspectives in mammalian IGFBP-3 biology: local vs. systemic action. Am J Physiol-Cell Physiol. 2009;296(5):C954–76. [DOI] [PubMed] [Google Scholar]
- 36.Kahm Y-J, Jung U, Kim R-K. BIRC5 regulates IGFBP-3-induced autocrine loop in cancer stem cells. bioRxiv. 2023:2023.03.09.531969.
- 37.Ressler S, Radhi J, Aigner T, Loo C, Zwerschke W, Sergi C. Insulin-like growth factor-binding protein-3 in osteosarcomas and normal bone tissues. Anticancer Res. 2009;29(7):2579–87. [PubMed] [Google Scholar]
- 38.Diaz-Beya M, Navarro A, Ferrer G, Diaz T, Gel B, Camos M, et al. Acute myeloid leukemia with translocation (8; 16)(p11; p13) and MYST3-CREBBP rearrangement harbors a distinctive microRNA signature targeting RET proto-oncogene. Leukemia. 2013;27(3):595–603. [DOI] [PubMed] [Google Scholar]
- 39.Moisés J, Navarro A, Santasusagna S, Viñolas N, Molins L, Ramirez J, et al. NKX2–1 expression as a prognostic marker in early-stage non-small-cell lung cancer. BMC Pulm Med. 2017;17(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallardo E, Navarro A, Vinolas N, Marrades RM, Diaz T, Gel B, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30(11):1903–9. [DOI] [PubMed] [Google Scholar]
- 41.Megill C, Martin B, Weaver C, Bell S, Prins L, Badajoz S, et al. Cellxgene: a performant, scalable exploration platform for high dimensional sparse matrices. bioRxiv. 2021:2021.04. 05.438318.
- 42.Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, et al. Differential expression of myc family genes during murine development. Nature. 1986;319(6056):780–3. [DOI] [PubMed] [Google Scholar]
- 43.Mooney M, McWeeney S. Data integration and reproducibility for high-throughput transcriptomics. Int Rev Neurobiol. 2014;116:55–71. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Hogan BL. Differential gene expression in the distal tip endoderm of the embryonic mouse lung. Gene Expr Patterns. 2002;2(3–4):229–33. [DOI] [PubMed] [Google Scholar]
- 45.Hooper S, Harding R. Fetal lung liquid: a major determinant of the growth and functional development of the fetal lung. Clin Exp Pharmacol Physiol. 1995;22(4):235–41. [DOI] [PubMed] [Google Scholar]
- 46.Pitkänen OM, O’Brodovich HM. Significance of ion transport during lung development and in respiratory disease of the newborn. Ann Med. 1998;30(2):134–42. [DOI] [PubMed] [Google Scholar]
- 47.Murisier-Petetin G, Gremlich S, Damnon F, Reymondin D, Hohlfeld P, Gerber S. Amniotic fluid insulin-like growth factor binding protein 3 concentration as early indicator of fetal growth restriction. Eur J Obstet Gynecol Reprod Biol. 2009;144(1):15–20. [DOI] [PubMed] [Google Scholar]
- 48.He L, He X, Lim LP, De Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin H, Zhang S, Sun Y, Li S, Ning Y, Dong Y, et al. MicroRNA-34/449 targets IGFBP-3 and attenuates airway remodeling by suppressing Nur77-mediated autophagy. Cell Death Disease. 2017;8(8):e2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D’Addio F, La Rosa S, Maestroni A, Jung P, Orsenigo E, Nasr MB, et al. Circulating IGF-I and IGFBP3 levels control human colonic stem cell function and are disrupted in diabetic enteropathy. Cell Stem Cell. 2015;17(4):486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalina M, Mason RJ, Shannon JM. Surfactant protein C is expressed in alveolar type II cells but not in Clara cells of rat lung. Am J Respir Cell Mol Biol. 1992;6(6):594–600. [DOI] [PubMed] [Google Scholar]
- 52.Mulugeta S, Beers MF. Surfactant protein C: its unique properties and emerging immunomodulatory role in the lung. Microbes Infect. 2006;8(8):2317–23. [DOI] [PubMed] [Google Scholar]
- 53.Sakaki-Yumoto M, Katsuno Y, Derynck R. TGF-β family signaling in stem cells. Biochim et Biophys Acta (BBA)-Gen Subj. 2013;1830(2):2280–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyazono K, Ehata S, Koinuma D. Tumor-promoting functions of transforming growth factor-β in progression of cancer. Upsala J Med Sci. 2012;117(2):143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu R-H, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, et al. NANOG is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3(2):196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung M-I, Bujnis M, Barkauskas CE, Kobayashi Y, Hogan BL. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145(9):dev163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo Y, Chen H, Ren S, Li N, Mishina Y, Shi W. BMP signaling is essential in neonatal surfactant production during respiratory adaptation. Am J Physiol-Lung Cell Mol Physiol. 2016;311(1):L29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frum T, Hsu PP, Hein RF, Conchola AS, Zhang CJ, Utter OR, et al. Opposing roles for TGFβ-and BMP-signaling during nascent alveolar differentiation in the developing human lung. NPJ Regen Med. 2023;8(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakaki-Yumoto M, Liu J, Ramalho-Santos M, Yoshida N, Derynck R. Smad2 is essential for maintenance of the human and mouse primed pluripotent stem cell state. J Biol Chem. 2013;288(25):18546–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Primary and secondary antibodies and qPCR probes used in the study.
Additional file 2. Video S1. Isolation of lung tips under the stereoscopic miscroscope using tungsten needles.
Additional file 3. Fig. S1. Follow-up of tip-derived organoid growth before and after the first passage. A Tip-derived organoid growth from day 0 (tip inclusion into matrigel) to 14 showing patterned structure. B Patterned structure of tip-derived organoids resembling a disorganized bronchial tree. C and D Organoids generate spheric structures after the first passage losing the patterned structure.
Additional file 4. Table S2. List of differential expressed genes between 8- and 12-pcw embryonic lungs.
Additional file 5. Table S3. Gene ontology terms identified in the analysis of the differentially expressed genes between 8 and 12 pcw embryonic lungs and the list of genes included in each identified gene ontology term.
Additional file 6. Table S4. List of differential expressed miRNAs between 8 and 12 pcw embryonic lungs.
Additional file 7. Video S2. Time-lapse video where can be observed the morphological changes of organoids transfected with control siRNA during the first 48h after nucleofection.
Additional file 8. Video S3. Time-lapse video where can be observed the morphological changes of organoids transfected with IGFBP3 siRNA during the first 48h after nucleofection.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and supplementary materials.