Highlights
-
•
Microstructural properties of white matter tracts are prognostic of slow recovery.
-
•
A prognostic model to predict athletes at risk for slow recovery has been developed.
-
•
The model reaches an AUC of 0.90, a sensitivity of 100%, and a specificity of 79%.
-
•
A fully reproducible pipeline has been used to accelerate scientific discovery.
Keywords: Mild Traumatic Brain Injury (mTBI), Persisting post-concussion symptoms (PPCS), Diffusion tensor analysis, White matter, Collegiate athletes, Prognostic model
Abstract
Background and objectives
After a concussion diagnosis, the most important issue for patients and loved ones is how long it will take them to recover. The main objective of this study is to develop a prognostic model of concussion recovery. This model would benefit many patients worldwide, allowing for early treatment intervention.
Methods
The Concussion Assessment, Research and Education (CARE) consortium study enrolled collegiate athletes from 30 sites (NCAA athletic departments and US Department of Defense service academies), 4 of which participated in the Advanced Research Core, which included diffusion-weighted MRI (dMRI) data collection. We analyzed the dMRI data of 51 injuries of concussed athletes scanned within 48 h of injury. All athletes were cleared to return-to-play by the local medical staff following a standardized, graduated protocol. The primary outcome measure is days to clearance of unrestricted return-to-play. Injuries were divided into early (return-to-play < 28 days) and late (return-to-play >= 28 days) recovery based on the return-to-play clinical records. The late recovery group meets the standard definition of Persisting Post-Concussion Symptoms (PPCS). Data were processed using automated, state-of-the-art, rigorous methods for reproducible data processing using brainlife.io. All processed data derivatives are made available at https://brainlife.io/project/63b2ecb0daffe2c2407ee3c5/dataset. The microstructural properties of 47 major white matter tracts, 5 callosal, 15 subcortical, and 148 cortical structures were mapped. Fractional Anisotropy (FA) and Mean Diffusivity (MD) were estimated for each tract and structure. Correlation analysis and Receiver Operator Characteristic (ROC) analysis were then performed to assess the association between the microstructural properties and return-to-play. Finally, a Logistic Regression binary classifier (LR-BC) was used to classify the injuries between the two recovery groups.
Results
The mean FA across all white matter volume was negatively correlated with return-to-play (r = −0.38, p = 0.00001). No significant association between mean MD and return-to-play was found, neither for FA nor MD for any other structure. The mean FA of 47 white matter tracts was negatively correlated with return-to-play (rμ = −0.27; rσ = 0.08; rmin = −0.1; rmax = −0.43). Across all tracts, a large mean ROC Area Under the Curve (AUCFA) of 0.71 ± 0.09 SD was found. The top classification performance of the LR-BC was AUC = 0.90 obtained using the 16 statistically significant white matter tracts.
Discussion
Utilizing a free, open-source, and automated cloud-based neuroimaging pipeline and app (https://brainlife.io/docs/tutorial/using-clairvoy/), a prognostic model has been developed, which predicts athletes at risk for slow recovery (PPCS) with an AUC=0.90, balanced accuracy = 0.89, sensitivity = 1.0, and specificity = 0.79. The small number of participants in this study (51 injuries) is a significant limitation and supports the need for future large concussion dMRI studies and focused on recovery.
1. Introduction
After a concussion diagnosis, the most important issue for patients and loved ones is how long it will take them to recover. In collegiate and adolescent athletes, the median recovery is 12 and 19.5 days, respectively (Broglio et al., 2022, Purcell et al., 2016), but approximately 15 % and 31 % (respectively) have still not recovered by 1 month (28 days) (Broglio et al., 2022, Zemek et al., 2016). These athletes unfortunately meet the 28-day definition of slow recovery, commonly referred to as either Persistent Post Concussion Symptoms (PPCS) or Post-Concussion Syndrome. In non-sports-medicine environments, the prevalence of PPCS is even higher, with PPCS of 23–59 % among civilians visiting the emergency department (Tator et al., 2016, Wäljas et al., 2015), and a staggering 75 % at 6 months post-injury in active duty warfighters with blast-related mild Traumatic Brain Injury (mTBI) (Walker et al., 2017). Recovering slowly from a concussion is detrimental for all these populations, and a clinical tool that could accurately predict PPCS would be revolutionary in both concussion management and concussion science.
1.1. Related work
Creating a model to predict concussion recovery has been a far-reaching goal within the field of concussion research and medicine for over a decade. The literature can be broken down into two categories: 1) sports-related concussion (SRC) models where the outcome measure is return-to-play clearance, and 2) civilian emergency department (ED) managed mTBI where the outcome measure is symptom resolution (using the Rivermead Post-concussion Questionnaire (RPQ) or the Glasgow Outcome Score Extended (GOSE)) at 6–12 months. The literature can be further subdivided into prognostic models limited to clinical exam variables, or those that use advanced experimental biomarkers, such as neuroimaging.
In the SRC literature, in which patients are generally young (adolescent and young adult) and healthy (few comorbidities), recovery is relatively fast. Broglio and colleagues (Broglio et al., 2022) report a median of 12.8 days, with 85 % of athletes cleared at 28 days. To the best of our knowledge, there are 2 published prognostic models of SRC. Morgan and colleagues (Morgan et al., 2015) studied SRCs in adolescents at a regional concussion clinic. Using clinical variables only, their Logistic Regression (LR) model predicting PPCS had a sensitivity of 55 % and a specificity of 93 %. They did not publish the AUC score. The stronger predictors in the model were history of mood disorder and presence of delayed symptoms. Chu and colleagues (Chu et al., 2022) applied a CatBoost model to over 600 child and adolescent concussed athletes and were able to predict PPCS (defined as 21 days) with an average AUC of 0.81. A companion paper to this manuscript (under review) by Rooks and colleagues utilized the large clinical core of the Concussion Assessment, Research and Education (CARE) dataset. They performed a standard LR model with stepwise variable selection and reported an AUC of 0.72, with a specificity of 11 % and sensitivity of 98 %.
In the more substantially injured civilian ED mTBI literature, where the prevalence of PPSC at 6 months is nearly 50 % (Mikolić et al., 2021), there has been a concerted effort to build and validate prognostic models of recovery (Mikolić et al., 2021, Lingsma et al., 2015, Cnossen et al., 2018). In 2015, Silverberg and colleagues (Silverberg et al., 2015) published a systematic review outlining the lack of multiple clinical variable prognostic models for predicting PPCS. Among the deficiencies was the absence of validation against separate datasets. In response, Mikolic and colleagues (Mikolić et al., 2021) identified 3 prognostic models using 3 different datasets that predict PPCS in mTBI ED patients: Stulemeijer and colleagues (Nijmegen) (Stulemeijer et al., 2008), Cnossen and colleagues (TRACK-TBI Pilot) (Cnossen et al., 2017), and Cnossen & colleagues (UPFRONT) (Cnossen et al., 2018), Mikolic and colleagues (Mikolić et al., 2021) then tested the three models against the CENTER-TBI dataset. They found the best prognostic PPCS models utilized 2-week, post-injury symptom scores and had an AUC=0.75–0.76. Additional machine learning studies of ED mTBI have focused on the GOSE at 1 month, with a prevalence of not recovered (GOSE<8) of ∼ 60 %. Both Falk and colleagues (Falk et al., 2021) and Bittencourt and colleagues (Bittencourt et al., 2021) report AUCs of (0.79–0.80). Babcock and colleagues (Babcock et al., 2013) followed 406 children and adolescents who were evaluated for an mTBI/concussion in the emergency department setting. In a logistic regression model, several clinical variables were associated with PPCS (e.g. headache, age, and admission to the hospital), but the total model had poor predictive ability (AUC=0.66).
In summary, in both the SRC literature and the more severe mTBI ED literature (e.g., LOC∼50 %, positive CT findings ∼ 20 %), clinical variable models have poor prognostic capabilities (AUC=0.66–0.81). Thus, building models using tractography and diffusion MRI metrics appears more promising.
1.2. Current study
The National Collegiate Athletic Association and United States Department of Defense Concussion Assessment, Research, and Education (NCAA/DOD CARE) consortium has produced a set of unique neuroimaging and phenotype datasets, publicly available for research purposes. Clinical measures have been collected for thousands of individuals from diverse backgrounds and with a history of concussion, and MRI data has been collected for hundreds. These datasets represent the first opportunity of its kind to investigate critical questions about the pathophysiology and etiology of sports-related concussion. In particular, diffusion tensor analysis has recently started to be used to evaluate microstructural changes after a concussion (Mustafi et al., 2018, Wu et al., 2020, Palacios et al., 2022). This is especially the case as a recent report (Wu et al., 2020) demonstrated an association between diffusion MRI metrics and recovery time (return-to-play).
Using a CARE cohort of 51 injuries diagnosed with sports-related concussion and for whom anatomical (T1w), diffusion-weighted MRI (dMRI) and return to play (RTP) data were collected within 24–48 h post injury, the main aim of this study was to build a prognostic model of concussion recovery by leveraging microstructural properties of the brain. To achieve this, data were processed and analyzed using brainlife.io, the BRAIN Initiative free and secure cloud computing platform for scientific transparency and rigor (Hayashi et al., 2024). Microstructural properties were then extracted from the major brain structures, including subcortical structures, deep white matter tracts, and cortical areas to evaluate whether they were associated with the athletes’ recovery time. Finally, white matter tract features were used to classify individuals at risk for slow recovery (PPCS).
2. Materials and Methods
2.1. Study Participants, data source and data preprocessing
All participants provided informed consent approved by the Medical College of Wisconsin and Human Research Protection Office.
2.1.1. Study participants
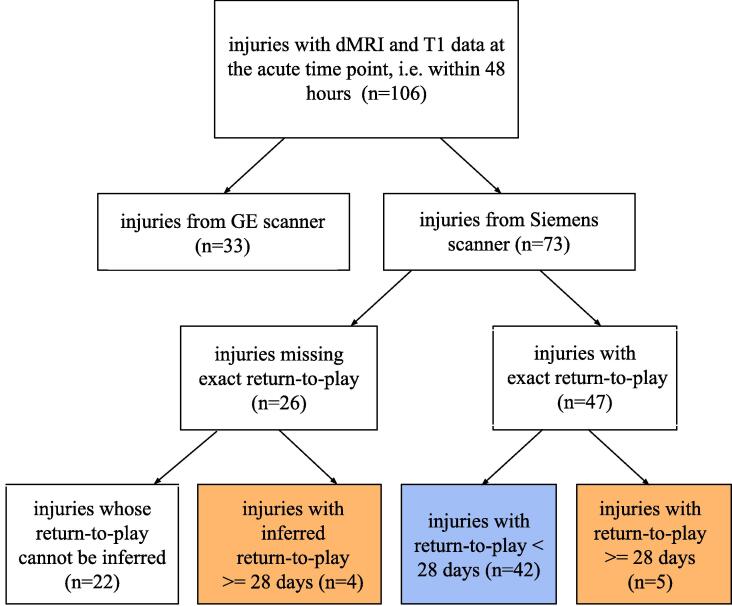
This study began with 106 injuries which were acquired within 48 h and contained both a T1 and dMRI scan (see consort Fig. 1). Thirty three injury scans originated from a GE scanner but were not used in the analyses because the intensity values of derivative data were a factor of 2 smaller than intensity values coming from a Siemens scanner, making them not comparable. Seventy three injury scans originated on Siemens scanners and were used for further analysis. The 73 injuries belonged to 64 unique athletes (7 with 2 injuries and 2 with 3 injuries), of whom 10 were female and the mean age was 18.85 (±0.85 years). Return-to-play, defined as the time interval between the injury and unrestricted return to play, was the primary outcome measure. Twenty-six injuries had missing return-to-play values, hence the correlation analysis was limited to 47 injuries (of whom 8 were female). Missing return to play time points occurs for many reasons, including athletic season ends, athlete transfers, athlete quitting the team, season ending, additional injury (e.g. concussion and torn anterior cruciate ligament), other reasons (e.g. the player is lost to follow-up). Injuries were divided into two groups based on their return-to-play: Early recovery: return-to-play < 28 days, and Late recovery: return-to-play >= 28 days. Given this definition, the number of late recovery injuries was 5. However, since 4 injuries with missing return-to-play had a time to asymptomatic >= 28 days, they were added to the Late recovery group, increasing it to 9. See eTable 1 for the demographics table.
Fig. 1.
Consort diagram of injury selection.
2.1.2. Data source
The CARE dataset, sponsored by the National Collegiate Athletic Association (NCAA) and U.S. Department of Defense (DoD), is the largest concussion database of its kind, comprising data from cadets and NCAA student-athletes. The portion of the CARE dataset used in this study is the first release (CARE 1.0), collected between 2014 and 2018 (Broglio et al., 2017). Participants with diffusion MRI data were recruited from three Advanced Research Core (ARC) sites: Virginia Tech (VT), University of North Carolina (UNC), and University of California Los Angeles (UCLA).
A cutting edge Magnetic Resonance Imaging (MRI) protocol was applied with the goal of identifying advanced biomarkers of concussion. Imaging standardization and quality-control procedures were ensured to enable study and comparison of diverse types of concussion and mTBI (Broglio et al., 2017). T1- weighted images were acquired on a 3 T Siemens using a 3D Magnetization Prepared Rapid Gradient Echo (3D MP-RAGE) acquisition, with isotropic resolution of 1 mm. The diffusion scans were acquired on Siemens MAGNETOM 3 T Prisma (UNC and UCLA) or 3 T Tim Trio (VT, UNC, and UCLA) scanners across the three ARC sites. 30 directions at b-value of 1000 s/mm2 and 8 b0 (b-value = 0 s/mm2) were acquired (one b0 volume with a reversed phase-encoding direction). Other MRI parameters were echo time/repetition time = 98/7900 ms, field of view = 243 mm, matrix size = 90 · 90, whole-brain coverage = 60 slices, slice thickness = 2.7 mm, and isotropic resolution = 2.7 mm. No obvious outliers that require advanced data harmonization approaches were observed (Mustafi et al., 2018). Further details about quality assurance procedures are available in previous works (Broglio et al., 2017, Nencka et al., 2018). The T1 images were rigorously examined for lesions and abnormalities by our group. No abnormalities were found.
2.1.3. Data preprocessing
A fully reproducible neuroimaging sequence of data preprocessing via cloud services (Apps) was implemented on brainlife.io. Table 1. Reports the individual Apps components of the pipeline. Quality check was performed automatically, semi-automatically, or manually after each step.
Table 1.
The brainlife.io processing pipeline used to process the data and extract the features of interest.
| App name | App purpose | App DOI |
|---|---|---|
| QSIPrep – preprocessing workflow | Preprocess dMRI data | brainlife.app.246 (Cieslak et al., 2021) |
| MRTrix3 – WMC Anatomically Constrained Tractography (ACT) | Build an anatomically-constrained, whole brain tractography | brainlife.app.319 (Smith et al., 2012) |
| FreeSurfer 7.1.1 | Create brain parcellations | brainlife.app.462 (Fischl, 2012) |
| Compute summary statistics of diffusion measures from Freesurfer Parcellation (volume) | Compute statistics from diffusion measures inside a parcellation generated by Freesurfer | brainlife.app.554 |
| White Matter Anatomy Segmentation | Segment the tractogram into 61 white matter tracts | brainlife.app.188 (Bullock et al., 2019) |
| Remove Tract Outliers | Remove spurious fibers from the tracts | brainlife.app.195 (Yeatman et al., 2012) |
| Generate figures of white matter tracts overlaid on anatomical image | Check white matter tract quality | brainlife.app.607 |
| Tract Analysis Profiles | Create tract profiles of microstructural white matter properties | brainlife.app.361 (Yeatman et al., 2012) |
2.1.3.1. Neuroimaging data preprocessing
An initial data preprocessing was performed by the ARC neuroimaging team to ensure best practices for quality assurance and image data analysis. For a detailed description of the initial preprocessing procedure, please refer to (Broglio et al., 2017).
From our side, preprocessing was performed using QSIPrep 0.15.3 (Cieslak et al., 2021), which is based on Nipype 1.7.0 (Gorgolewski et al., 2011); (Gorgolewski et al., 2018); RRID:SCR_002502). The brainlife.io App “QSIPrep – preprocessing workflow” was used for this step (brainlife.app.246). A comprehensive description of the anatomical and diffusion data preprocessing, as automatically generated by the QSIPrep software to promote reproducibility, is described in sections Anatomical data preprocessing and Diffusion data preprocessing of the eMethods.
HTML reports as automatically generated by QSIPrep were examined for each participant. No errors were reported. Additionally, after every step of the pipeline, the results were checked manually.
2.1.3.2. Tractography generation
For each subject, probabilistic Anatomically-Constrained Tractography (ACT) (Smith et al., 2012) was performed with MRtrix3 (Tournier et al., 2012) using the brainlife.io App “mrtrix3 – WMC Anatomically Constrained Tractography (ACT)” (brainlife.app.319) (Mcpherson, 2018). First, fiber orientation distributions (FOD) were reconstructed using the Constraint Spherical Deconvolution (CSD) model (Tournier et al., 2007, Jeurissen et al., 2014). Then, a whole brain tractogram was obtained through probabilistic tracking using the second-order Integration over Fiber Orientation Distributions (iFOD2) algorithm (Lmax = 6, maximum curvature = 35 deg, 3 M streamlines, min length = 25 mm, max length = 250 mm) (Tournier and Calamante, 2010).
2.1.3.3. Summary statistics of diffusion measures from freesurfer parcellation
A set of white and gray matter parcels was obtained from the T1w image using the software FreeSurfer (Fischl, 2012), implemented in the brainlife.io App “FreeSurfer 7.1.1” (brainlife.app.462).
Mean FA and MD were computed for each parcel of the Freesurfer parcellation (Destrieux Atlas, aparc.a2009s (Destrieux et al., 2010) using the brainlife.io App “Compute summary statistics of diffusion measures from Freesurfer Parcellation (volume)” (brainlife.app.554).
2.1.3.4. White matter tract segmentation
A set of 61 major white matter tracts were extracted from each whole brain tractogram using the connectivity-based automatic procedure described in (Bullock et al., 2019) and available through the brainlife.io App “White Matter Anatomy Segmentation” (brainlife.app.188). This technique, similar to the White Matter Query Language (WMQL) approach (Wassermann et al., 2013, Wassermann et al., 2016) uses multiple cortical (and sub-cortical) Regions of Interest (ROIs) per tract to derive the segmentation. These ROIs were computed with FreeSurfer (Fischl, 2012).
Spurious streamlines were pruned following the procedure described in (Yeatman et al., 2012), implemented in the brainlife.io App “Remove Tract Outliers” (brainlife.app.195). Specifically, streamlines with a distance of more than 4sd from the tract centroid or with a length of more than 4sd from the streamline mean length were removed.
A final visual quality check was performed tract-by-tract by inspecting the images obtained from the brainlife.io App “Generate figures of white matter tracts overlaid on anatomical image” (brainlife.app.607). Sagittal, coronal, and axial projections of each tract overlaid on its corresponding T1w image were inspected and tract outliers were removed from the consequent analysis. In particular, since in several cases the segmented Cerebellar tracts had a non-plausible anatomical shape, they were removed from the dataset. After this pruning, the number of remaining white matter tracts was 47.
2.1.3.5. Tract profiles analysis
Tract profiles can be described as a summary of certain white matter properties along the length of white matter tracts. Tract profile analysis has been widely used to compare groups or populations (Yeatman et al., 2012, Yeatman et al., 2012, Chandio et al., 2020, Kruper et al., 2021, Vinci-Booher et al., 2022) and it could potentially become a powerful and intuitive diagnostic tool (Yeatman et al., 2012). In this study, we compared FA profiles and MD profiles of the two recovery groups to evaluate whether a change in the microstructural properties of the major human white matter tracts at the time of injury is associated with a late recovery.
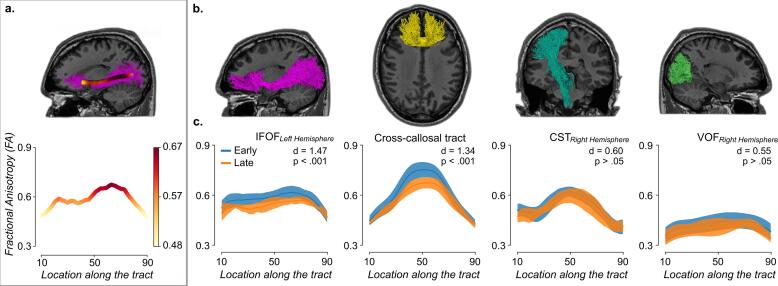
For each tract of interest, which is composed of N fibers, tract profiles are computed as follows: 1) each fiber is resampled into 100 equidistant nodes; 2) the tract’s core is computed by averaging the coordinates (x, y, z) of each fiber at each node (colored tube in Fig. 3a top); 3) microstructural measurements (FA or MD) are calculated for each node of each fiber; 4) the tract profile is finally computed by taking the weighted average of the microstructural measurements of each individual fibers at each node (see Fig. 3a bottom). Fibers are weighted by their distance from the tract’s core: the higher the distance, the lower the probability that the fiber belongs to the tract, and thus the lower its weight. Tract profiles were computed with the brainlife.io App “Tract Analysis Profiles” (brainlife.app.361), which implements the method in (Yeatman et al., 2012).
Fig. 3.
White matter tract profilometry. a. The core tract is computed for each tract (inner tube). An example is provided for the left IFOF in the top panel. The corresponding Fractional Anisotropy (FA) profile is extracted by taking a weighted average of the FA measurements of each individual fiber (see bottom panel). b. Visualization of the anatomy of some of the white matter tracts of interest. From left to right: the left inferior fronto-occipital fasciculus (left IFOF), the Cross-callosal tract (anterior Frontal CC), the right corticospinal tract (right CST), and the right vertical occipital fasciculus (right VOF). c. FA profile analysis. FA profile mean ± sd are plotted for each tract and for the early vs. late recovery groups (blue and orange, respectively). To minimize partial volume effects when computing the mean by averaging the FA measurements along the tract, only nodes from 10 to 90 were taken into account, disregarding the extremities of the tracts. The two tracts on the left have a high effect size (d > 1), while the two tracts on the right have a poor effect size (d < 1). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Individual mean. For each tract profile, we also computed its mean by averaging the microstructural measurements along the tract. In this case, to minimize partial volume effects, only nodes from 10 to 90 are taken into account, disregarding the extremities of the tracts, which have a higher probability of containing different populations of neurons.
Group mean. To compare the two recovery groups, the mean and standard deviation (sd) within each group and each tract were computed (see Fig. 3b).
2.2. Statistical analyses
Correlation analysis, effect sizes analysis, and ROC AUC analysis were performed to assess whether differences in the microstructural properties were associated with return-to-play. The Pearson correlation coefficient (r) was computed between microstructural measurements (mean fractional anisotropy [FA] and mean diffusivity [MD]) and the continuous variable return-to-play in multiple brain regions. For each test, a p-value was estimated using Wald Test with a null-hypothesis r = 0 and an alternative hypothesis of r < 0 when comparing the FA values. An alternative hypothesis of r > 0 was used when comparing MD values. p-values were corrected for multiple comparisons using False Discovery Rate (FDR; (Benjamini and Hochberg, 1995, Genovese et al., 2002). To ensure that a correlation was still present even without outliers, the analysis was repeated after having discarded participants with a return-to-play outside the range mean ± 3sd. All analyses were completed in a Jupyter Notebook served by brainlife.io, Python 3.8 and scipy 1.7.1 (Virtanen et al., 2020), and are available at: https://brainlife.io/project/63b2ecb0daffe2c2407ee3c5/groupanalysis and (https://github.com/Nicholas-Port/Predicting-Persistent-Post-Concussion-Symptoms).
2.2.1. Correlation analysis: Deep white matter, subcortical nuclei, and cortex
In particular, the goal was to understand whether a similar effect was detectable in both white matter and cortical regions. The brain regions considered as a whole were: (i) deep white matter (all white matter voxels), (ii) subcortical nuclei, i.e. Cerebellum-Cortex, Thalamus-Proper, Caudate, Putamen, Pallidum, Hippocampus, and Amygdala, and (iii) 148 cortical regions (all from a single FreeSurfer parcellation tool (Fischl, 2012), Destrieux Atlas (Destrieux et al., 2010). In all three regions, mean FA and MD were computed for the left and right hemispheres separately.
2.2.2. Correlation analysis: Corpus callosum and white matter tracts
A second step was to focus only on FA and individual white matter regions, and specifically on the corpus callosum and white matter tracts. The correlation coefficient was computed between mean FA and return-to-play for each of the 5 regions of the corpus callosum (the anterior and posterior horn, and two mid-anterior, mid-posterior regions, and the central callosal regions) and for each of the 47 white matter tracts. For each white matter tract, mean FA (FAtract) was estimated through profilometry (Yeatman et al., 2012).
2.2.3. Effect size analysis
A common measure used in clinical studies to discriminate between two subject groups is Cohen’s d coefficient. For each white matter tract, the mean () and the standard deviation () of the FA tract profiles were computed for the early and late recovery groups. Cohen’s d was computed for each of the 47 tracts individually:
A d < 0.40 is considered poor, 0.40 < d < 0.80 moderate, and d > 0.80 large (Bakker et al., 2019, Ferris et al., 2022). Mean d coefficients were estimated via bootstrap (resampling with replacement, 10,000 iterations) (Pestilli et al., 2011)
2.2.4. Receiver Operating Characteristic (ROC) curve analysis
An ROC curve analysis was performed to determine the predictive utility of the group FA means for each of the individual 47 white matter tracts (Southall et al., 2000). ROC Area Under the Curve (AUCFA) scores were estimated using the scikit-learn package version 1.0 (Pedregosa et al., 2012). An AUCFA score > 0.80 indicates that, given two samples belonging to two different classes, there is an 80 % chance that the model can distinguish them correctly, and thus is usually considered clinically useful (Ferris et al., 2022, Mandrekar, 2010). Mean AUCFA were estimated via bootstrap (resampling with replacement, 10,000 iterations) (Pestilli et al., 2011). The statistical significance of the AUCFA of each one of the 47 tracts was tested using bootstrapping and FDR-corrected.
2.3. Logistic Regression classification
A Logistic Regression Binary Classifier (LR-BC) was used over individual mean FA tract profiles to discriminate between the two recovery classes. The dataset was split into a training set (⅔ of the samples) and a test set (⅓ of the samples) using stratified random splits to preserve the proportion between the two classes. First, the LR-BC was trained using the mean FA of all tracts as features. Hyperparameter tuning and a 5-fold Stratified Cross Validation (CV) strategy were implemented in scikit-learn 1.0 (Pedregosa et al., 2012) to select the best model. Then, using the same procedure, the LR-BC was trained also using the mean FA of only the statistically significant tracts identified by the Receiver Operating Characteristic (ROC) curve analysis (see paragraph 2.2.4). Finally, the performances of the two classifiers were evaluated on the held-out test set using the recommended metrics when dealing with unbalanced datasets, namely Area Under the Curve (AUC), balanced accuracy, sensitivity, and specificity. In addition, for visualization purposes, the model was trained also using only the top two statistically-significant white matter tracts identified by the ROC curve analysis. A standard linear regression predicting return-to-play in days was also performed.
2.4. Data Sharing, scientific transparency, and rigor
The original dataset can be accessed at https://fitbir.nih.gov/. The ezBIDS tool (Levitas et al., 2024) was used to convert DICOM data to NIFTI and to upload data to brainlife.io using the Brain Imaging Data Structure (BIDS) standard. The set of features extracted from the original dataset and used for this study, as well as the data processing pipeline used to extract the features and the Jupyter Notebooks used for the statistical analysis, are accessible on brainlife.io at https://brainlife.io/project/63b2ecb0daffe2c2407ee3c5/pipeline.
3. Results
3.1. Correlation analysis
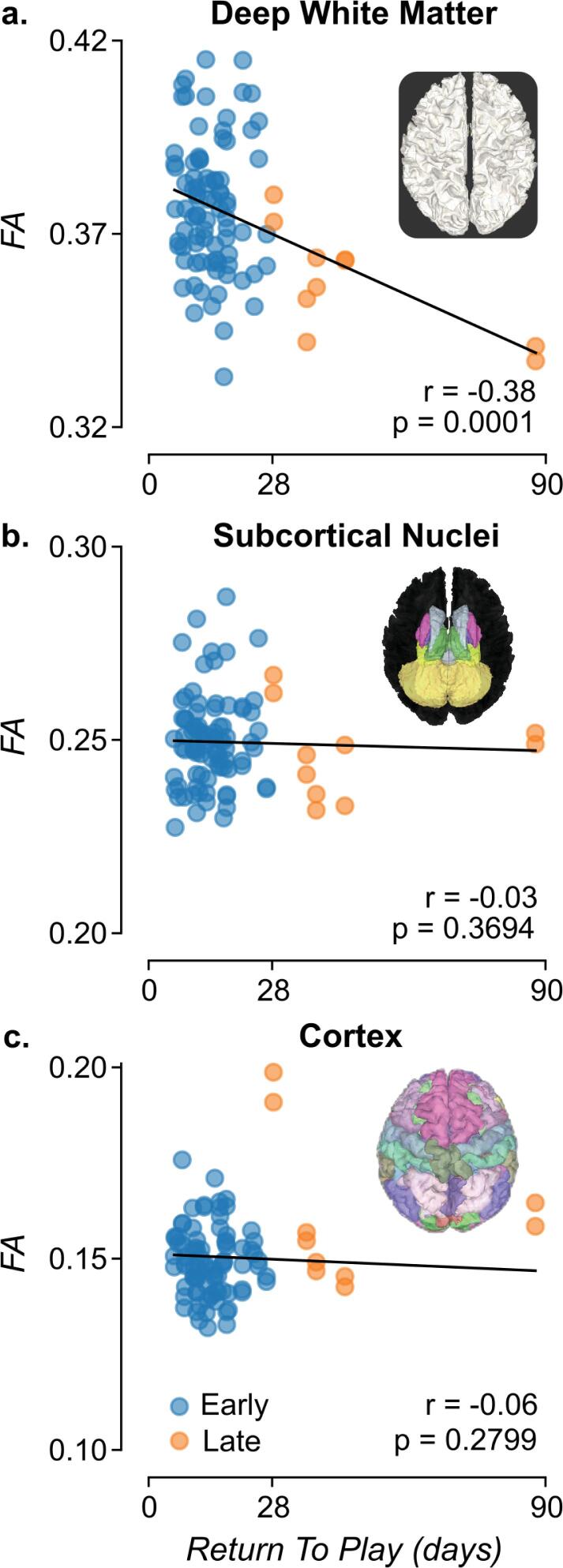
For the correlation analysis, the dataset consisted of 47 injuries of 42 student athletes (8 female), with a mean age of 18.77 (SD=0.84). The association between the microstructural properties of the whole white matter within the entire brain volume of individual study participants was investigated. FA was estimated using the Diffusion Tensor Model (Basser et al., 1994, Pierpaoli et al., 1996) in each white matter voxel, identified using a multi-tissue segmentation (Fischl, 2012). On average, 486,613 (±48,790 sd) white matter voxels were measured for each participant. Mean FA was estimated across all voxels in the brain and correlated with each individual’s return-to-play. A moderate but statistically significant correlation was found (r = −0.38, p = 0.0001; one-sided Wald test; Fig. 2a), with a stronger correlation in the left (r = −0.42, p = 0.0013) than in the right (r = −0.34, p = 0.0092) hemispheres. To ensure that a correlation was still present even without outliers, the analysis was repeated without them (only one participant did not meet the criterion, i.e. only the two data points for which return-to-play = 87.7 days were excluded). A smaller but still statistically significant correlation was found (r = −0.25, p = 0.0094). Finally, the analysis was repeated using MD, but no significant correlation was found for return-to-play with (r = 0.02, p = 0.4352) or without (r = −0.07, p = 0.7315) the outlier. A qualitative inspection of possible confounding variables of site, sex, number of injuries, and sport type are plotted in eFigure 1.
Fig. 2.
Correlation between brain microstructure and time to return-to-play (return-to-play). a. Deep white matter. Distribution of mean Fractional Anisotropy (FA) across the deep white matter voxels versus return-to-play. b. Subcortical nuclei. Distribution of mean FA across 14 subcortical nuclei versus return-to-play. c. Cortex. Distribution of mean FA across 148 cortical parcels versus return-to-play. Left and right hemisphere structures are plotted separately, resulting in two data points per injury. Blue is early recovery and orange is late recovery. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To understand whether the association reported in the previous section is significant only for the deep white matter, the correlation between FA (and MD) and return-to-play either within the volume of seven non-white matter subcortical nuclei (e.g., Thalamus, Hippocampus, Amygdala, etc) or across the cortical volume was investigated. No correlation was found between subcortical FA and return-to-play (r = −0.03, p = 0.3694; one-sided Wald test; Fig. 2b) or between subcortical MD and return-to-play (r = −0.12, p = 0.8652).
No correlation was found between cortical FA and return-to-play (r = −0.06, p = 0.2799; one-sided Wald test; Fig. 2c) or between MD and return-to-play (r = −0.17, p = 0.9480). As these results suggested that deep white matter FA is uniquely associated with return-to-play, all subsequent analyses focussed on white matter and FA.
Previous reports indicate that white matter tissue within the corpus callosum is different between concussed and control individuals (Wu et al., 2020). The association between FA and return-to-play was estimated by subdividing the callosum into 5 regions: the anterior and posterior horn, and the mid-anterior, mid-posterior, and central callosal regions. The strongest correlation was estimated for the mid-posterior corpus callosum (r = −0.37, p = 0.00108, corrected for false discovery rate, FDR), followed by the anterior horn (r = −0.29, p = 0.00983, FDR), the mid-anterior segment (r = −0.28, p = 0.01573, FDR), and the central segment (r = −0.20, p = 0.06732, FDR). The weakest correlation was estimated in the posterior horn (r = −0.11, p = 0.22370).
3.2. Tract profiles: Correlation
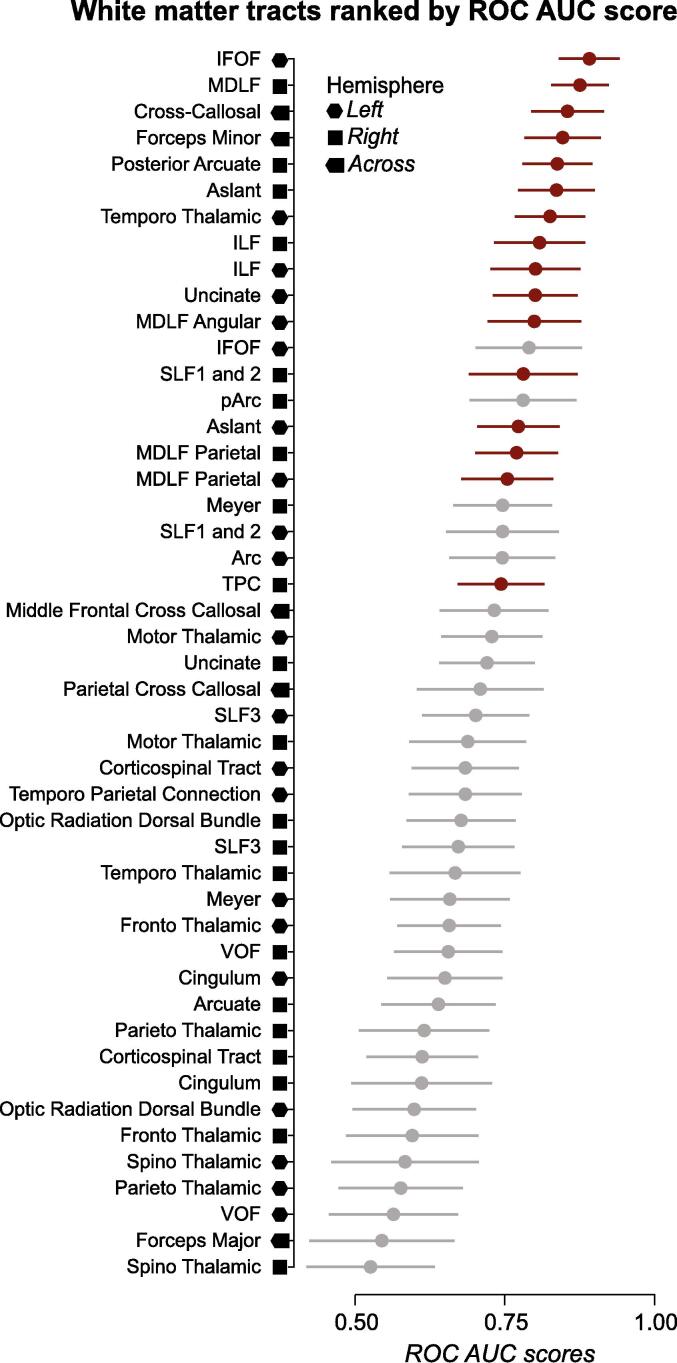
To further understand the components of the whole white matter associated with return-to-play, 47 white matter tracts were segmented using an anatomically-informed method (Bullock et al., 2019) (see Fig. 4 for a comprehensive list of tract names). FA was estimated using profilometry (Yeatman et al., 2012) (see Fig. 3) for each white matter tract individually. The mean FA for each tract (FAtract) was used to estimate 47 correlation values with return-to-play. FA was negatively correlated with return-to-play (rμ = −0.27; rσ = 0.08; rmin = −0.1; rmax = −0.43). The strongest correlation was estimated for the cross-callosum tract connecting the left and right parietal lobes (r = −0.43, p = 0.00003, FDR), and the weakest was for the right-hemisphere Temporal Thalamic Connection (r = −0.1, p = 0.25232, FDR). See eTable 2 for all correlation values. The correlations between cortical areas and return-to-play is listed in eTable 4.
Fig. 4.
Receiver-Operating Characteristics (ROC) Area Under the Curve (AUC) for each of the 47 white matter tracts. Mean ± SD ROC AUC scores estimated to discriminate between the early- and late-recovery groups. AUC scores for each of the 47 tracts were ranked and plotted in decreasing order. Red: Statistically significant tracts at p < 0.001 (FDR). Gray: non significant tracts. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Tract profiles: Effect sizes
Injuries were subdivided into the Early (return-to-play < 28 days) and Late (return-to-play >= 28 days) recovery groups.
In the following sections, the dataset consisted of 51 injuries of 45 student athletes (8 female), with a mean age of 18.75 (SD=0.82) and a mean days to scan from injury of 2.07 days (SD=1.6).
Tract profiles: Cohen’s d. For each tract, the mean FA profile was computed for each subject in the early and late groups. Cohen’s d was computed using the FA values for each of the 47 tracts:
Across all the tracts, the mean dFA was 0.80 ± 0.35 SD. The left Inferior Fronto-Occipital Fasciculus had a very high dFA (IFOF; dFA=1.52 ± 0.31, bootstrap with replacement). See eTable 3 for all Cohen’s d values.
3.4. Tract profiles: ROC analysis
The most common measure used to evaluate a predictive model is the Area Under a Receiver Operator Characteristic Curve (AUC (Southall et al., 2000); see Methods). The AUC was computed to measure how well the FA of each tract predicted athletes in the early vs. late recovery group. Across all tracts, a large mean AUCFA of 0.71 ± 0.09 SD was found. For the left IFOF, a very large mean AUCFA of 0.89 ± 0.05 SD was found. The statistical significance of the AUCFA of each one of the 47 tracts was tested using bootstrapping and a False Discovery Rate (FDR). Results show that the AUCFA of 16 tracts was statistically different from 0.5 at p < 0.001 after FDR correction (see Fig. 4). See eTable 3 for all ROC AUC values.
3.5. Tract profiles: Logistic regression
Finally, the ability to use white matter tract mean FA to predict recovery groups was evaluated using an LR-BC with a 5-fold Stratified CV strategy. The performance of the LR-BC was evaluated using ROC AUC scores.
First, we trained the model using all 47 white matter tracts. This experiment returned an AUC score of 0.86 on the held-out test set, with balanced accuracy = 0.89, sensitivity = 1.0, and specificity = 0.79. Second, the model was trained using only the 16 statistically-significant white matter tracts identified using the Receiver Operating Characteristic (ROC) curve analysis (i.e. the red-colored tracts in Section 3.4 and Fig. 4). With this model, a higher AUC score was obtained on the held-out test set, specifically an AUC score of 0.90, with balanced accuracy = 0.89, sensitivity = 1.0, and specificity = 0.79.
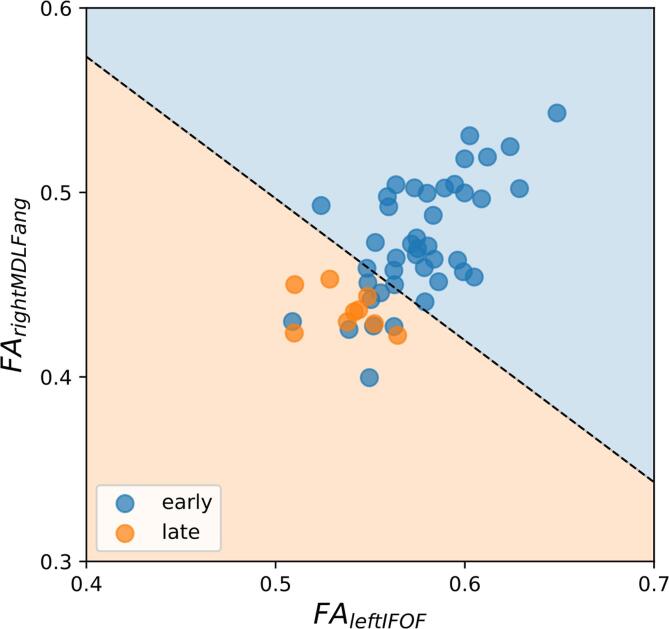
For visualization purposes, the model was trained also using only the top two statistically-significant white matter tracts identified from Fig. 4, i.e. the left IFOF and the right Angular MDLF. The 2D classification plot is shown in Fig. 5. All three classifiers (trained on 47, 16, and 2 tracts), well classify the positive (late recovery) class, with no false negatives (see Fig. 5 and sensitivity = 1.0). On the other hand, a few false positives are present, i.e. a few negative samples (early recovery) were misclassified as positive (late recovery).
Fig. 5.
Classification of late and early recovery using a Logistic Regression classifier. A representative classification plot reporting the Fractional Anisotropy (FA) of the two top-performing white matter tracts (from Fig. 4). Each symbol indicates a study participant. Individual symbols are color-coded by indicating whether a participant was part of the late- or early-recovery group. The decision boundary (black dotted line) divides the plane into the two classes. There are no false negatives, at the expenses of a few false positives.
A standard linear regression predicting return-to-play in days can be found in eTable 5.
4. Discussion
A data analysis approach was developed using established pipelines and diffusion tensor parameters of the human white matter tracts to predict early versus late (PPCS) return-to-play of young adult athletes from the CARE dataset. Best practices in both white matter tractography and machine learning resulted in a prognostic model of PPCS that, utilizing just the mean FA of 16 white matter tracts, demonstrates an AUC of 0.90 (balanced accuracy = 0.89, sensitivity = 1.0, specificity = 0.79). The LR model is capturing the underlying relationship between return-to-play and the white matter tracts’ metric FA (r = −0.38, p = 0.0001).
A non-significant correlation was found instead between MD and return-to-play in the white matter. This is in contrast with two previous studies that examined the association between microstructural metrics with return-to-play (Wu et al., 2020) and with the GOSE outcome measure (Palacios et al., 2022). Wu and colleagues (Wu et al., 2020) reported that higher MD values were associated with a longer time to return-to-play. Palacios and colleagues (Palacios et al., 2022) showed that MD in specific white matter tracts was associated with six-month incomplete recovery (GOSE<8). However, the discrepancies in the results between the different studies could be due to different sample sizes, patient populations, acquisition protocols, and/or data preprocessing pipelines (Wu et al., 2020). Specifically, Wu and colleagues (Wu et al., 2020) did not have in their sample any participants with time to return-to-play > 28 days. Palacios and colleagues (Palacios et al., 2022) analyzed a civilian ED mTBI dataset (TRACK-TBI). Moreover, both studies used the tract-based spatial statistical (TBSS) approach to test for group differences, whereas in this study a more personalized tract profile analysis was adopted.
Compared to previous prognostic models developed in the SRC literature using clinical data, our microstructural-based model overcomes their prognostic capabilities (AUC<0.81 vs AUC>0.90). To the best of our knowledge, only a recently published machine learning model obtained predictive performance above 0.80 (Chu and colleagues (Chu et al., 2022), average AUC=0.81). However, their dataset was composed only of children and adolescents, and their cut-off point for PPCS was 21 days instead of 28 days. Despite differences in the datasets, this is an indication that the use of state-of-the-art machine learning techniques combined with diffusion tensor analysis can boost the performance of a prognostic model of PPCS. Our results, with an AUC of 0.90, indicate white matter integrity may be a clinically useful tool. The choice of the 16 tracts used in the prognostic model are data driven, future research with much larger data will hopefully yield explanations to why these tracts are prognostic.
A key aspect of this study is the use of a free, open science platform for data analysis, namely brainlife.io (Avesani et al., 2019, Hayashi et al., 2024). Open science analysis platforms were created to allow for well-governed, compliant management of extremely large datasets with 100 % transparency and complete reproducibility in the computationally intensive field of neuroimaging. The use of brainlife.io has the additional future advantage of being able to host and offer a free brainlife.io portal that clinicians could use as an experimental tool for predicting – and advising patients regarding – their likely recovery times.
4.1. Limitations
Although our model makes a relatively strong prognostic prediction, there are some limitations. The significant limitation of this study is its sample size. The number of injuries from the Siemens scanner was 73, unfortunately 26 of them were missing the return-to-play outcome measure, which led to a reduced sample suitable for our analysis. PPCS generally occurs in 15–20 % of athletes (Broglio et al., 2022), thus our PPCS sample size (n = 9) was expected. Because the useable dataset of 51 subjects is small, our results must be interpreted carefully and be considered preliminary. A larger open dataset is needed to validate these results and make these results clinically useful. A second limitation is that our neuroimaging pipeline described in Table 1 was validated only with scans acquired from a Siemens scanner. For this reason, the portion of data acquired from a GE scanner of the CARE dataset was not included in the analysis. Tailored scanner harmonization techniques should be applied to address this issue.
5. Conclusions
Overall, this study demonstrates the possibility of using the microstructural properties of white matter tracts to develop a prognostic model for PPCS, which outperforms current predictive models based on clinical data. This result highlights the importance of neuroimaging analysis for concussion research and serves as a first step toward identifying a PPCS neuroimaging biomarker. A clinical tool that could accurately predict PPCS would be revolutionary in both concussion management and concussion science. Moreover, sharing processing pipelines and data derivatives would accelerate scientific discovery in this field.
CRediT authorship contribution statement
Giulia Bertò: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Lauren T. Rooks: Writing – review & editing, Visualization, Methodology, Formal analysis, Data curation, Conceptualization. Steven P. Broglio: Project administration, Funding acquisition. Thomas A. McAllister: Project administration, Funding acquisition. Michael A. McCrea: Project administration, Funding acquisition, Conceptualization. Paul F. Pasquina: Project administration, Funding acquisition. Christopher Giza: Data curation. Alison Brooks: Data curation. Jason Mihalik: Data curation. Kevin Guskiewicz: Data curation. Josh Goldman: Data curation. Stefan Duma: Data curation. Steven Rowson: Data curation. Nicholas L. Port: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Franco Pestilli: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Dr Broglio has current or past research funding from the National Institutes of Health; Centers for Disease Control and Prevention; Department of Defense – USA Medical Research Acquisition Activity, National Collegiate Athletic Association; National Athletic Trainers’ Association Foundation; National Football League/Under Armour/GE; Simbex; and ElmindA. He has consulted for US Soccer (paid), US Cycling (unpaid), University of Calgary SHRed Concussions external advisory board (unpaid), medico-legal litigation, and received speaker honorarium and travel reimbursements for talks given. He is co-author of “Biomechanics of Injury (3rd edition)” and has a patent on “Brain Metabolism Monitoring Through CCO Measurements Using All-Fiber-Integrated Super-Continuum Source” (U.S. 11,529,091 B2). He is on the and is/was on the editorial boards (all unpaid) for Journal of Athletic Training (2015 to present), Concussion (2014 to present), Athletic Training & Sports Health Care (2008 to present), British Journal of Sports Medicine (2008 to 2019). Thomas McAllister has Concussion Research Grants from NIH; US Dept. of Defense; US Dept. of Energy; and the NCAA. He also has Royalties from American Psychiatric Assoc. Publishing for Textbook of TBI. He is part of the Concussion Scientific Advisory Committee; Australian Football League – uncompensated. Michael McCrea has Research funding to Medical College of Wisconsin from US Dept. of Defense (DoD) and NCAA; Research funding to Medical College of Wisconsin from NIH, VA, DoD, CDC, NFL, NCAA, Abbott Laboratories; Book royalties from Oxford University Press. He is Consultant, Neurotrauma Sciences, Inc.; Consultant, Green Bay Packers; Medical legal consulting. He receives Honoraria and travel support for professional speaking engagements and for professional meetings.
Acknowledgements
This research was supported with a grant by the Department of Defense to Nicholas Port and Franco Pestilli (W81XWH-20-1-0717). The brainlife.io project was was funded by grants from the National Institutes of Health (NIH, NIMH R01MH126699, NIBIB R01EB030896, NIBIB R01EB029272), the National Science Foundation (OAC-1916518, IIS-1912270, IIS-1636893, BCS-1734853), and the Wellcome Trust (226486/Z/22/Z), and by a Microsoft Investigator Fellowship and a gift from the Kavli Foundation to Franco Pestilli. Additional support to brainlife.io was provided by BRAIN CONNECTS: Center for Mesoscale Connectivity, grant NIH NINDS UM1NS132207 to Kamil Ugurbil and grant NIH NIMH R01MH133701 to Christopher Rorden. Nicholas Port was also funded by the Indiana Spinal Cord and Brain Injury Research Fund and unrestricted funds from StateSpace Inc. This research was also made possible, in part, with support from the Grand Alliance Concussion Assessment, Research, and Education (CARE) Consortium, funded in part by the National Collegiate Athletic Association (NCAA) and the DoD. The US Army Medical Research Acquisition Activity is the awarding and administering acquisition office, and this work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Psychological Health and Traumatic Brain Injury Program under Award No. W81XWH-14-2-0151. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2024.103646.
Contributor Information
Nicholas L. Port, Email: nport@iu.edu.
Franco Pestilli, Email: pestilli@utexas.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The authors do not have permission to share data.
References
- Avesani P., McPherson B., Hayashi S., et al. The open diffusion data derivatives, brain data upcycling via integrated publishing of derivatives and reproducible open cloud services. Sci. Data. 2019;6(1):69. doi: 10.1038/s41597-019-0073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock L., Byczkowski T., Wade S.L., Ho M., Mookerjee S., Bazarian J.J. Predicting Postconcussion Syndrome After Mild Traumatic Brain Injury in Children and Adolescents Who Present to the Emergency Department. JAMA Pediatr. 2013;167(2):156–161. doi: 10.1001/jamapediatrics.2013.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A., Cai J., English L., Kaiser G., Mesa V., Van Dooren W. Beyond small, medium, or large: points of consideration when interpreting effect sizes. Educ. Stud. Math. 2019;102(1):1–8. [Google Scholar]
- Basser P.J., Mattiello J., LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. B. 1994;103(3):247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57(1):289–300. [Google Scholar]
- Bittencourt M., Balart-Sánchez S.A., Maurits N.M., van der Naalt J. Self-Reported Complaints as Prognostic Markers for Outcome After Mild Traumatic Brain Injury in Elderly: A Machine Learning Approach. Front. Neurol. 2021;12 doi: 10.3389/fneur.2021.751539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broglio S.P., McCrea M., McAllister T., et al. A National Study on the Effects of Concussion in Collegiate Athletes and US Military Service Academy Members: The NCAA–DoD Concussion Assessment, Research and Education (CARE) Consortium Structure and Methods. Sports Med. 2017;47(7):1437–1451. doi: 10.1007/s40279-017-0707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broglio S.P., McAllister T., Katz B.P., et al. The Natural History of Sport-Related Concussion in Collegiate Athletes: Findings from the NCAA-DoD CARE Consortium. Sports Med. 2022;52(2):403–415. doi: 10.1007/s40279-021-01541-7. [DOI] [PubMed] [Google Scholar]
- Bullock D., Takemura H., Caiafa C.F., et al. Associative white matter connecting the dorsal and ventral posterior human cortex. Brain Struct. Funct. 2019;224(8):2631–2660. doi: 10.1007/s00429-019-01907-8. [DOI] [PubMed] [Google Scholar]
- Chandio B.Q., Risacher S.L., Pestilli F., et al. Bundle analytics, a computational framework for investigating the shapes and profiles of brain pathways across populations. Sci. Rep. 2020;10(1):17149. doi: 10.1038/s41598-020-74054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y., Knell G., Brayton R.P., Burkhart S.O., Jiang X., Shams S. Machine learning to predict sports-related concussion recovery using clinical data. Ann. Phys. Rehabil. Med. 2022;65(4) doi: 10.1016/j.rehab.2021.101626. [DOI] [PubMed] [Google Scholar]
- Cieslak M., Cook P.A., He X., et al. QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nat. Methods. 2021;18(7):775–778. doi: 10.1038/s41592-021-01185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnossen M.C., Winkler E.A., Yue J.K., et al. Development of a prediction model for post-concussive symptoms following mild Traumatic Brain Injury: A TRACK-TBI Pilot study. J. Neurotrauma. 2017;34(16):2396–2409. doi: 10.1089/neu.2016.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnossen M.C., van der Naalt J., Spikman J.M., et al. Prediction of Persistent Post-Concussion Symptoms after Mild Traumatic Brain Injury. J. Neurotrauma. 2018;35(22):2691–2698. doi: 10.1089/neu.2017.5486. [DOI] [PubMed] [Google Scholar]
- Destrieux C., Fischl B., Dale A., Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53(1):1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk H., Bechtold K.T., Peters M.E., et al. A prognostic model for predicting one-month outcomes among emergency department patients with mild traumatic brain injury and a presenting Glasgow Coma Scale of fifteen. J. Neurotrauma. 2021;38(19):2714–2722. doi: 10.1089/neu.2021.0137. [DOI] [PubMed] [Google Scholar]
- Ferris L.M., Kontos A.P., Eagle S.R., et al. Utility of VOMS, SCAT3, and ImPACT baseline evaluations for acute concussion identification in collegiate athletes: Findings from the NCAA-DoD concussion Assessment, research and education (CARE) consortium. Am. J. Sports Med. 2022;50(4):1106–1119. doi: 10.1177/03635465211072261. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese C.R., Lazar N.A., Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15(4):870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Gorgolewski K., Burns C.D., Madison C., et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front. Neuroinf. 2011;5:13. doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski, Krzysztof J., Oscar Esteban, Christopher J. Markiewicz, Erik Ziegler, David Gage Ellis, Michael Philipp Notter, Dorota Jarecka, et al. 2018. “Nipype.” Software. Zenodo. 10.5281/zenodo.596855. [DOI]
- Hayashi S., Caron B.A., Heinsfeld A.S., et al. brainlife.io: a decentralized and open-source cloud platform to support neuroscience research. Nat. Methods. 2024;21:809–813. doi: 10.1038/s41592-024-02237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeurissen B., Tournier J.D., Dhollander T., Connelly A., Sijbers J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage. 2014;103:411–426. doi: 10.1016/j.neuroimage.2014.07.061. [DOI] [PubMed] [Google Scholar]
- Kruper J., Yeatman J.D., Richie-Halford A., et al. Evaluating the Reliability of Human Brain White Matter Tractometry. Apert Neuro. 2021;1(1) doi: 10.52294/e6198273-b8e3-4b63-babb-6e6b0da10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitas D., Hayashi S., Vinci-Booher S., et al. ezBIDS: Guided standardization of neuroimaging data interoperable with major data archives and platforms. Sci. Data. 2024;11(1):179. doi: 10.1038/s41597-024-02959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingsma H.F., Yue J.K., Maas A.I.R., Steyerberg E.W., Manley G.T. TRACK-TBI Investigators. Outcome prediction after mild and complicated mild traumatic brain injury: external validation of existing models and identification of new predictors using the TRACK-TBI pilot study. J. Neurotrauma. 2015;32(2):83–94. doi: 10.1089/neu.2014.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010;5(9):1315–1316. doi: 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- Mcpherson B. MRtrix3 ACT.; 2018. https://doi.org/10.25663/BL.APP.101.
- Mikolić A., Polinder S., Steyerberg E.W., et al. Prediction of global functional outcome and post-concussive symptoms after mild Traumatic Brain Injury: External validation of prognostic models in the Collaborative European NeuroTrauma Effectiveness Research in Traumatic Brain Injury (CENTER-TBI) study. J. Neurotrauma. 2021;38(2):196–209. doi: 10.1089/neu.2020.7074. [DOI] [PubMed] [Google Scholar]
- Morgan C.D., Zuckerman S.L., Lee Y.M., et al. Predictors of postconcussion syndrome after sports-related concussion in young athletes: a matched case-control study. J. Neurosurg. Pediatr. 2015;15(6):589–598. doi: 10.3171/2014.10.PEDS14356. [DOI] [PubMed] [Google Scholar]
- Mustafi S.M., Harezlak J., Koch K.M., et al. Acute White-Matter Abnormalities in Sports-Related Concussion: A Diffusion Tensor Imaging Study from the NCAA-DoD CARE Consortium. J. Neurotrauma. 2018;35(22):2653–2664. doi: 10.1089/neu.2017.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nencka A.S., Meier T.B., Wang Y., et al. Stability of MRI metrics in the advanced research core of the NCAA-DoD concussion assessment, research and education (CARE) consortium. Brain Imaging Behav. 2018;12(4):1121–1140. doi: 10.1007/s11682-017-9775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios E.M., Yuh E.L., Mac Donald C.L., et al. Diffusion tensor imaging reveals elevated diffusivity of white matter microstructure that is independently associated with long-term outcome after mild Traumatic Brain Injury: A TRACK-TBI study. J. Neurotrauma. 2022;39(19–20):1318–1328. doi: 10.1089/neu.2021.0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. arXiv [csLG]. Published online January 2, 2012. Accessed November 5, 2022. https://www.jmlr.org/papers/volume12/pedregosa11a/pedregosa11a.pdf?ref=https://githubhelp.com.
- Pestilli F., Carrasco M., Heeger D.J., Gardner J.L. Attentional enhancement via selection and pooling of early sensory responses in human visual cortex. Neuron. 2011;72(5):832–846. doi: 10.1016/j.neuron.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli C., Jezzard P., Basser P.J., Barnett A., Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201(3):637–648. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- Purcell L., Harvey J., Seabrook J.A. Patterns of Recovery Following Sport-Related Concussion in Children and Adolescents. Clin. Pediatr. 2016;55(5):452–458. doi: 10.1177/0009922815589915. [DOI] [PubMed] [Google Scholar]
- Silverberg N.D., Gardner A.J., Brubacher J.R., Panenka W.J., Li J.J., Iverson G.L. Systematic review of multivariable prognostic models for mild traumatic brain injury. J. Neurotrauma. 2015;32(8):517–526. doi: 10.1089/neu.2014.3600. [DOI] [PubMed] [Google Scholar]
- Smith R.E., Tournier J.D., Calamante F., Connelly A. Anatomically-constrained tractography: improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage. 2012;62(3):1924–1938. doi: 10.1016/j.neuroimage.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Southall B., Buxton B.F., Marchant J.A., Hague T. Computer Vision — ECCV 2000. Springer; Berlin Heidelberg: 2000. On the Performance Characterisation of Image Segmentation Algorithms: A Case Study; pp. 351–365. [Google Scholar]
- Stulemeijer M., van der Werf S., Borm G.F., Vos P.E. Early prediction of favourable recovery 6 months after mild traumatic brain injury. J. Neurol. Neurosurg. Psychiatry. 2008;79(8):936–942. doi: 10.1136/jnnp.2007.131250. [DOI] [PubMed] [Google Scholar]
- Tator C.H., Davis H.S., Dufort P.A., et al. Postconcussion syndrome: demographics and predictors in 221 patients. J. Neurosurg. 2016;125(5):1206–1216. doi: 10.3171/2015.6.JNS15664. [DOI] [PubMed] [Google Scholar]
- Tournier J.D., Calamante F. Proceedings of the International Society for Magnetic Resonance in Medicine. Vol. 1670. John Wiley & Sons, Inc; New Jersey, USA: 2010. Connelly A, Others. Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions. [Google Scholar]
- Tournier J.D., Calamante F., Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: Non-negativity constrained super-resolved spherical deconvolution. Neuroimage. 2007;35(4):1459–1472. doi: 10.1016/j.neuroimage.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Tournier J.D., Calamante F., Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol. 2012;22(1):53–66. doi: 10.1002/ima.22005. [DOI] [Google Scholar]
- Vinci-Booher S., Caron B., Bullock D., James K., Pestilli F. Development of white matter tracts between and within the dorsal and ventral streams. Brain Struct. Funct. 2022;227(4):1457–1477. doi: 10.1007/s00429-021-02414-5. [DOI] [PubMed] [Google Scholar]
- Virtanen P., Gommers R., Oliphant T.E., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17(3):261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäljas M., Iverson G.L., Lange R.T., et al. A prospective biopsychosocial study of the persistent post-concussion symptoms following mild traumatic brain injury. J. Neurotrauma. 2015;32(8):534–547. doi: 10.1089/neu.2014.3339. [DOI] [PubMed] [Google Scholar]
- Walker W.C., Franke L.M., Sima A.P., Cifu D.X. Symptom Trajectories After Military Blast Exposure and the Influence of Mild Traumatic Brain Injury. J. Head Trauma Rehabil. 2017;32(3):E16–E26. doi: 10.1097/HTR.0000000000000251. [DOI] [PubMed] [Google Scholar]
- Wassermann D., Makris N., Rathi Y., et al. On describing human white matter anatomy: the white matter query language. Med Image Comput Comput Assist Interv. 2013;16(Pt 1):647–654. doi: 10.1007/978-3-642-40811-3_81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann D., Makris N., Rathi Y., et al. The white matter query language: a novel approach for describing human white matter anatomy. Brain Struct. Funct. 2016;221(9):4705–4721. doi: 10.1007/s00429-015-1179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurology. 2020;95(7):e781-e792. [DOI] [PMC free article] [PubMed]
- Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, Feldman HM. Tract profiles of white matter properties: automating fiber-tract quantification. . 2012;7(11):e49790. [DOI] [PMC free article] [PubMed]
- Proc. Natl. Acad. Sci. USA 2012;109(44):E3045-E3053. [DOI] [PMC free article] [PubMed]
- Zemek R., Barrowman N., Freedman S.B., et al. Clinical Risk Score for Persistent Postconcussion Symptoms Among Children With Acute Concussion in the ED. J. Am. Med. Assoc. 2016;315(10):1014–1025. doi: 10.1001/jama.2016.1203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors do not have permission to share data.