Abstract
A physical association between the II–III loop of the DHPR (dihydropryidine receptor) and the RyR (ryanodine receptor) is essential for excitation–contraction coupling in skeletal, but not cardiac, muscle. However, peptides corresponding to a part of the II–III loop interact with the cardiac RyR2 suggesting the possibility of a physical coupling between the proteins. Whether the full II–III loop and its functionally important ‘C’ region (cardiac DHPR residues 855–891 or skeletal 724–760) interact with cardiac RyR2 is not known and is examined in the present study. Both the cardiac DHPR II–III loop (CDCL) and cardiac peptide (Cc) activated RyR2 channels at concentrations >10 nM. The skeletal DHPR II–III loop (SDCL) activated channels at ≤100 nM and weakly inhibited at ≥1 μM. In contrast, skeletal peptide (Cs) inhibited channels at all concentrations when added alone, or was ineffective if added in the presence of Cc. Ca2+-induced Ca2+ release from cardiac sarcoplasmic reticulum was enhanced by CDCL, SDCL and the C peptides. The results indicate that the interaction between the II–III loop and RyR2 depends critically on the ‘A’ region (skeletal DHPR residues 671–690 or cardiac 793–812) and also involves the C region. Structure analysis indicated that (i) both Cs and Cc are random coil at room temperature, but, at 5 °C, have partial helical regions in their N-terminal and central parts, and (ii) secondary-structure profiles for CDCL and SDCL are similar. The data provide novel evidence that the DHPR II–III loop and its C region interact with cardiac RyR2, and that the ability to interact is not isoform-specific.
Keywords: cardiac muscle, dihydropyridine receptor, excitation–contraction coupling, ryanodine receptor, protein–protein interaction, skeletal muscle
Abbreviations: Cc, cardiac C region peptide; Cs, skeletal C region peptide; CDCL, cardiac dihydropyridine receptor II–III loop; DHPR, dihydropyridine receptor; EC, excitation–contraction; HSQC, heteronuclear single-quantum coherence; I′c, control mean current; I′t, mean current under test conditions; Po, open probability; Poc, control Po; Pot, Po under test conditions; RyR, ryanodine receptor; SDCL, skeletal DHPR II–III loop; SR, sarcoplasmic reticulum; Tc, mean closed time; To, mean open time
INTRODUCTION
The aim of the present study was to investigate interactions between the native cardiac RyR (ryanodine receptor) (RyR2) Ca2+-release channel and fragments of the DHPR (dihydropyridine receptor) L-type Ca2+ channel. The RyR in the SR (sarcoplasmic reticulum) of cardiac muscle is activated during EC (excitation–contraction) coupling by Ca2+ entering the fibre through L-type Ca2+ channels. In contrast, skeletal muscle RyR (RyR1) activation depends on a protein–protein interaction with DHPR. Although physical coupling between the DHPR and RyR is not required for EC coupling in the heart [1,2], there is evidence that the two proteins, or parts of the proteins, can interact with each other under appropriate conditions. Parts of the DHPR II–III loop bind to RyR2 [3,4]. In intact myocytes (i) a dihydropyridine (BayK) binds to the DHPR and enhances RyR2 activity [5,6], (ii) ryanodine enhances dihydropyridine binding to the cardiac DHPR [7] and (iii) the ten C-terminal residues of the cardiac DHPR ‘A’ sequence (residues 803–812) depress Ca2+ spark frequency [8]. Although most lipid bilayer studies fail to show an interaction between the DHPR II–III loop and RyR2 [9–12], recent reports indicate that A peptides (corresponding to 20 residues in the N-terminus of the II–III loop) can alter RyR2 activity [13,14]. In the light of these studies, we have examined the recombinant cardiac and skeletal DHPR II–III loops (between the second and third transmembrane repeats; CDCL and SDCL respectively) and the cardiac and skeletal C peptides (Cc, cardiac residues 855–891 and Cs, skeletal residues 724–760).
The II–III loop and its ‘C’ region have been studied because they are required for skeletal EC coupling and are thought to contain the parts of the DHPR that physically interact with RyR1 in this process [15–17]. In addition, the corresponding fragments of the DHPR interact with RyR1 in vitro. The recombinant SDCL and the acidic C region [18] are high-affinity activators of RyR1 [12,18–20]. The strongly α-helical basic ‘A’ region peptide [21] also activates RyR1 with high affinity [10–12,20,22]. These functional interactions possibly reflect binding reactions that contribute to the protein–protein interactions in skeletal muscle [18,20]. The fact that the A peptides also interact with RyR2 provides evidence for potential DHPR–RyR interactions in the heart [13]. It is thus important to discover whether the other regions of the DHPR that are functionally active on RyR1s, i.e. the C region of the II–III loop and the recombinant II–III loop, can also modify the activity of RyR2 channels.
The results show that the full II–III loop and the C region peptides do alter RyR2 activity in an isoform-independent interaction. We suggest that the DHPR and RyR do not interact significantly in the heart in vivo because the DHPR is (i) expressed in relatively small amounts [23] and (ii) not targeted to positions opposite RyRs [24]. However, the proteins have the potential to interact and to transmit signals between the external environment and the SR.
EXPERIMENTAL
Materials
The following peptides were synthesized and purified (by the John Curtin School of Medical Research Biomolecular Resource Facility) as described previously [21,22,25]. Peptide Cs, E724FESNVNEVKDPYPSADFPGDDEEDEPEIPVSPRPRP760 and peptide Cc, D855LQPNESEDKSPYPNPETTGEEDEEEPEMPVGPRPRP891.
Expression of the DHPR II–III loop
The 391 or 405 bp cDNA fragments encoding SDCL and CDCL respectively were amplified by PCR and cloned in-frame downstream of a poly-histidine-tagged ubiquitin sequence in the plasmid pHUE [26]. The construct was checked by sequencing to exclude amplification errors. The plasmid was transferred into Escherichia coli BL21, and expression of fusion protein was induced by addition of 0.1 mM IPTG (isopropyl β-D-thiogalactoside) to the culture medium. The His-tagged protein was purified by chromatography on Ni–agarose [26,27]. Ubiquitin was removed from the N-terminal end of the II–III loop by digestion with a His-tagged ubiquitin-dependent protease [26]. The ubiquitin protease and cleaved ubiquitin were removed by re-chromatography on Ni–agarose. The recombinant II–III loop, without additional residues, was purified further by preparative electrophoresis under native conditions using a Bio-Rad model 491 prep cell. The sample was eluted in 25 mM Tris/HCl and 192 mM glycine, pH 8.3. Uniformly 15N-labelled protein was produced by growing the expression strain in M9 minimal medium with 15NH4Cl as the sole nitrogen source.
Single channel measurements [21,22,25]
Recording solutions were (cis): 230 mM caesium methanesulphonate, 20 mM CsCl and 10 mM Tes (pH 7.4 with CsOH) and (trans): 230 mM caesium methanesulphonate, 20 mM CsCl, 1 mM CaCl2 and 10 mM Tes (pH 7.4). The cis Ca2+ concentration was normally either 10−4 or 10−7 M, buffered using 1 mM BAPTA [bis-(o-aminophenoxy)ethane-N, N, N′, N′-tetra-acetic acid]. The cis chamber was held at ground and the voltage of the trans chamber controlled. Bilayer potential, expressed as Vcis−Vtrans, was changed between +40 and −40 mV every 30 s. We recorded 2 min of activity under control conditions and after each addition of peptide or II–III loop.
Analysis of channel activity
Currents were analysed over one to two 30 s periods of continuous activity at +40 and −40 mV. Slow fluctuations in the baseline were corrected using an in-house baseline-correction program (written by Dr D. R. Laver, University of Newcastle, Newcastle, Australia). Channel activity was measured either as ‘mean current’ (average of all data points in a record) or as open probability (Po), using a threshold analysis with the program Channel 2, (developed by P. W. Gage and M. Smith, John Curtin School of Medical Research). Measurements of mean current, performed on records from experiments containing one to four channels, included all channel activity from the smallest subconductance level to maximum openings. Po, mean open time (To) and mean closed time (Tc) measurements were restricted to records in which the opening of a single channel only was detected. This analysis does not detect openings that fall within the baseline noise. Threshold levels for channel opening and closing were set to exclude baseline noise, at approx. 20% of the maximum single channel conductance. Channel activity is expressed as relative Po to include data in which activity varied from ∼0.01 to ∼0.6 and data from bilayers containing more than one channel. Relative Po was calculated either (i) from I′t/I′c, where I′t is the mean current under test conditions and I′c is the control mean current or (ii) from Pot/Poc, where Pot is the open probability under test conditions and Poc is the control open probability. Since mean current divided by the maximum current approximates open probability, I′t/I′c≡Pot/Poc.
Ca2+ release [13]
SR vesicles (100 μg/ml), prepared from sheep heart [28], were added to a solution containing 100 mM KH2PO4 (pH 7), 4 mM MgCl2, 1 mM Na2ATP, 0.5 mM antipyrylazo III, 5 mM phospho(enol)pyruvate and 25 μg/ml pyruvate kinase. Extravesicular Ca2+ concentration was monitored at 710 nm. Vesicles were loaded with Ca2+ by adding four aliquots of CaCl2, each initially increasing the extravesicular Ca2+ concentration by 7.5 μM, with 3 min between each addition of Ca2+. Thapsigargin (200 nM) was added to block the Ca2+ ATPase, and finally 20 μM Ca2+ was used to initiate Ca2+-induced Ca2+ release. Ca2+ release with thapsigargin was subtracted from the initial rates of Ca2+-induced Ca2+ release. Experiments were performed with either vehicle alone (control) or vehicle plus peptide or recombinant loop, added before thapsigargin.
NMR spectroscopy
Peptides Cc and Cs were dissolved in 10% 2H2O/90% H2O to a final concentration of approx. 2 mM at pH 5.0. The samples for CDCL and SDCL were prepared in 50 mM potassium phosphate buffer (pH 6.5) containing 10% 2H2O, with a protein concentration of 1 mM in the presence of 200 mM KCl. Data was recorded at 25 or 5 °C on a Varian Inova 600 MHz spectrometer equipped with a pentaprobe. In order to obtain resonance assignments, one- and two-dimensional NMR spectroscopy was performed as described previously [21,25]. 15N/1H spectra were acquired using a sensitivity-enhanced 15N-HSQC (heteronuclear single-quantum coherence) pulse sequence [29]. Structural models for the Cc and Cs peptides were constructed using the program Biopolymer (Accelrys).
Circular dichroism
All peptide samples were diluted to 25 μM for CD measurements in 20% trifluoroethanol and 80% water, and the pH values were adjusted to 5.0. CD spectra were collected at 5 and 25 °C on a Jobin Yvon CD6 Dichrograph using a cell path length of 1 mm. Ten spectra were collected per sample, averaged and then subjected to a smoothing function.
Statistics
The significance of differences between values was tested using a Students t test, either one- or two-tailed, for independent or paired data as appropriate, or by the non-parametric ‘Sign’ test [30]. Differences were considered significant when P≤0.05.
RESULTS
The C peptides and recombinant II–III loop were added to the cis solution bathing the cytoplasmic side of native RyR2 channels. The channels were identified as RyRs by their Cs+ conductance of approx. 400 pS and their block by 30 μM Ruthenium Red.
Activation by peptide Cc
The C peptides maximally stimulate RyR1s at activating cytoplasmic Ca2+ concentrations of 10−4 M or 10−5 M [18,20], and were examined here with 10−4 M or 10−5 M cis Ca2+ and 2 mM MgATP to mimic in vivo conditions. Single-channel parameters under these control conditions (with and without MgATP) are given in Table 1. When MgATP was added, there was a trend towards a decline in Po with MgATP at 10−5 M cis Ca2+, or an increase at 10−4 M cis Ca2+, as reported previously [31]. Since there was a wide range of control channel activities (reflected in the standard errors in Table 1), average data below is presented as relative changes in channel activity to give equivalent weighting to the effects of the DHPR fragments on high- and low-activity channels.
Table 1. Effect of MgATP in the cytoplasmic (cis) solution on RyR activity.
Single-channel parameters under control conditions and in the presence of 2 mM MgATP and 10−5 M cis Ca2+ at +40 or −40 mV. Data are given (means±S.E.M.) for open probability (Po), mean open time (To) and mean closed time (Tc). Asterisks (*) indicate a significant difference between the initial reference value and the parameter value with MgATP. Data for 10−4 M cis Ca2+ is from [13] and is included here for comparison.
| +40 mV | −40 mV | ||||
|---|---|---|---|---|---|
| cis [Ca2+] | 10−5 M (n=5) | 10−4 M (n=10) | 10−5 M (n=5) | 10−4 M (n=10) | |
| Po | Initial | 0.293±0.149 | 0.25±0.08 | 0.256±0.141 | 0.17±0.08 |
| +MgATP | 0.201±0.084 | 0.40±0.11* | 0.169±0.016 | 0.38±0.09* | |
| To(ms) | Initial | 49.4±44.5 | 26.2±22.4 | 42.7±40.6 | 22.8±20.3 |
| +MgATP | 76.8±36.1* | 45.6±20.1* | 49.7±28.2 | 26.9±15.3* | |
| Tc (ms) | Initial | 198.9±107.1 | 42±56.2 | 82.9±63.3 | 161±57 |
| +MgATP | 132.4±56 | 76±30.5 | 124.2±92.1 | 67±47* | |
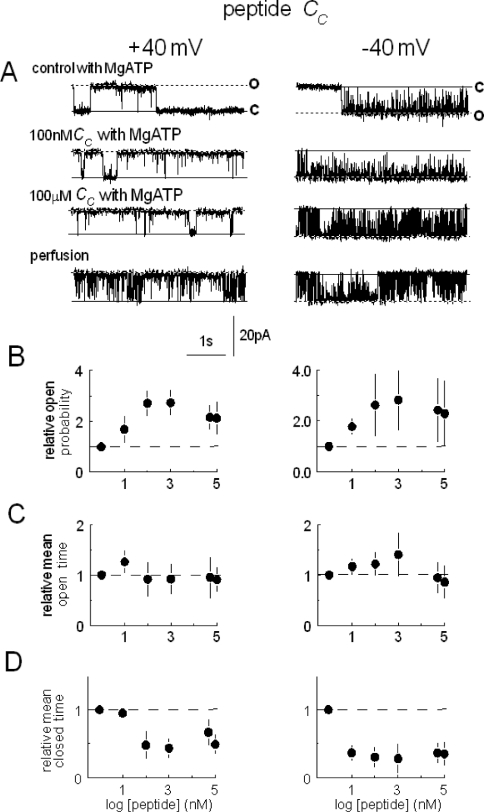
Peptide Cc increased relative open probability approx. 2–3-fold by inducing a >10-fold reduction in the average Tcs at both positive and negative potentials (Figure 1). When the cis chamber was perfused with 10 vol. of cis solution, the 100 μM peptide was diluted to approx. 30 nM, and ATP to approx. 600 nM [13]. Po after perfusion fell (Figure 1) because of the dilution of peptide and ATP; however, it remained greater than the initial activity in the absence of MgATP in all channels (results not shown), as expected if (i) the peptide remained at an activating concentration, and (ii) activation did not depend on the presence 2 mM MgATP. The activation by Cc was reversible after perfusion of lower concentrations of peptide (100 nM) from the cis chamber (n=4; Figure 2A).
Figure 1. Peptide Cc activates RyR2 channels by abbreviating channel closures.
Channel opening (in this and subsequent Figures) is upwards at +40 mV (left-hand panels) and downwards at −40 mV (right-hand panels), from the closed level c to the maximum single channel current o. In (A), control activity is shown with 2 mM MgATP and 100 μM Ca2+ in the cis chamber (first trace), after subsequent addition of 100 nM (second trace) or 100 μM peptide (third trace), and shortly after cis perfusion with a solution containing 100 μM Ca2+ and lacking MgATP and peptide (fourth trace). Note that the peptide concentration following perfusion is approx. 30 nM and the channel remains activated. Average relative Po is shown in (B), relative Tos in (C) and relative Tcs in (D) at +40 mV (left-hand panels) and −40 mV (right-hand panels). Means are for five experiments. In (C) to (D), the first symbol and broken line shows data obtained under control conditions.
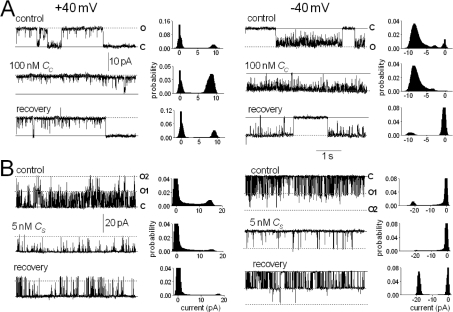
Figure 2. Reversible activation of RyR2 channels by peptide Cc and reversible inhibition by peptide Cs.
Data are shown at +40 mV (left-hand panels) and −40 mV (right-hand panels). In each panel, traces show 5 s of activity under each condition, and the associated histogram shows all-points histograms from the 30 s record. Under all conditions, 2 mM MgATP was present. The upper record shows control activity, the middle record shows activity in the presence of 100 nM peptide Cc (A) or 5 nM peptide Cs (B), and the third record shows activity between 30 and 60 s after perfusion of peptide from the cis chamber. Note that the bilayer in (B) contained two channels and that two open levels (O1 and O2) are indicated where appropriate.
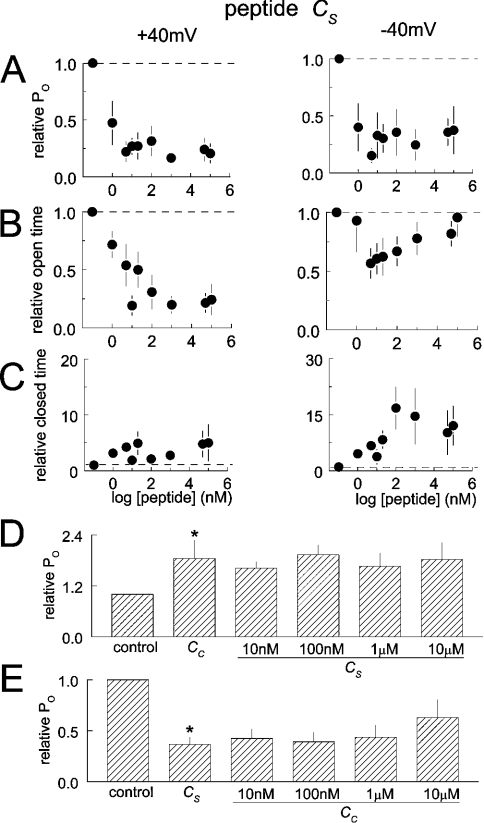
Inhibition by peptide Cs
Unlike Cc, peptide Cs inhibited RyR2 channels reversibly (Figure 2B). Po fell with Cs as low as 1 nM, at positive and negative potentials, and remained low with peptide concentrations up to 100 μM (Figure 3). The fall in activity was associated with complex changes in channel gating and was almost entirely due to a reduction in Tos at +40 mV. In contrast, although activity at −40 mV fell at low peptide concentrations because of a decrease in Tos, the Tos recovered at higher peptide concentrations, and low activity was maintained by an increase in closed durations. The recovery of open durations may reflect a lower affinity activation by the peptide. Because of the sequence (see the Experimental section) and structural (see below) similarities between peptides Cc and Cs, it was possible that they were exerting different actions on RyR2 by binding to the same site. To test this possibility, we added 10 μM Cs to the cis solution, followed by peptide Cs in increasing concentrations from 10 nM to 10 μM. The usual activation was seen with Cc, but Cs no longer inhibited the channels (Figure 3D). In the reverse experiment, initial exposure to Cs caused the usual inhibition. Subsequent exposure to low concentrations of Cc did not cause activation, but there was a trend towards reduced inhibition at higher concentrations of Cc (Figure 3E). These results provide strong evidence that the two C peptides act at the same or at closely related sites.
Figure 3. Changes in channel gating with peptide Cs.
(A), (B) and (C) show average relative Po (n=10), To and Tc (n=7 single-channel experiments) respectively, at +40 mV (left-hand panels) and −40 mV (right-hand panels). In (A) and (B), the first symbol and broken line show data obtained under control conditions. Note that the decline in Po is due to a fall in the duration of channel opening at +40 mV, but is due to more complex changes in the Tos and Tcs at −40 mV. (D) Average relative Po (data at +40 mV and −40 mV combined) from six experiments after exposure to 1 or 10 μM Cc, followed by exposure to 10 nM, 100 nM, 1 μM and 10 μM Cs. Note that Cs does not inhibit channels if added in the presence of Cc. (E) Average relative Po (data at +40 mV and −40 mV combined) from five experiments after exposure to 10 μM Cs, followed by exposure to 10 nM, 100 nM, 1 μM and 10 μM Cc. Note the relief of Cs inhibition at higher Cc concentrations. Asterisks indicate significant differences from the previous conditions.
The effects of the C peptides on RyR2 are specific effects, since we have shown that another acidic II–III loop peptide, as well as the scrambled peptide A sequence, do not alter the activity of these channels [13]. In addition, unrelated peptides, luteinizinghormone-releasing hormone (A. F. Dulhunty and S. M. Curtis, unpublished work) and inhibitory peptides for protein kinase A, protein kinase C and Ca2+/calmodulin kinase II [32], all fail to alter RyR activity.
The recombinant II–III loop
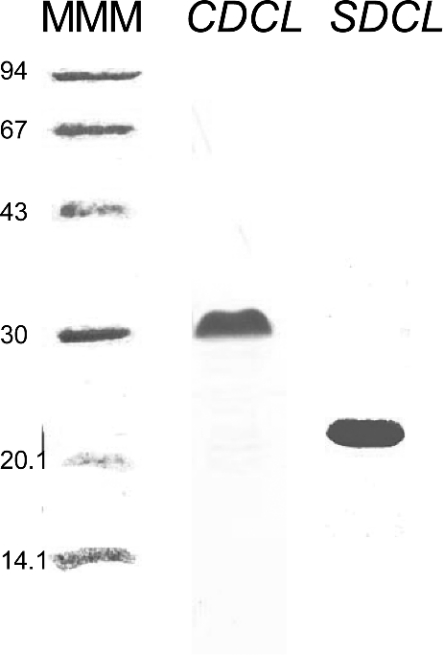
The recombinant peptides CDCL and SDCL corresponding to the II–III loop of the α1 subunit of the cardiac and skeletal DHPR respectively, and containing both the A and C sequences, were expressed and purified (see the Experimental section), and were found to run on SDS/PAGE at the same molecular masses as those shown in [9], i.e. 30 kDa for CDCL and 21 kDa for SDCL (Figure 4), which were higher than the calculated masses of 14.134 kDa and 15.230 kDa respectively.
Figure 4. Expression of SDCL and CDCL.
Purified SDCL and CDCL on 12% denaturing polyacrylamide gels. Methods for expression and purification of the protein are given in the Experimental section. Molecular-mass markers are shown in the left-hand lane (MMM, with corresponding masses indicated in kDa), with 5 μg of CDCL in the centre lane (at approx. 30 kDa) and 5 μg of SDCL in the right-hand lane (at approx. 21 kDa).
CDCL and SDCL activate RyR2s
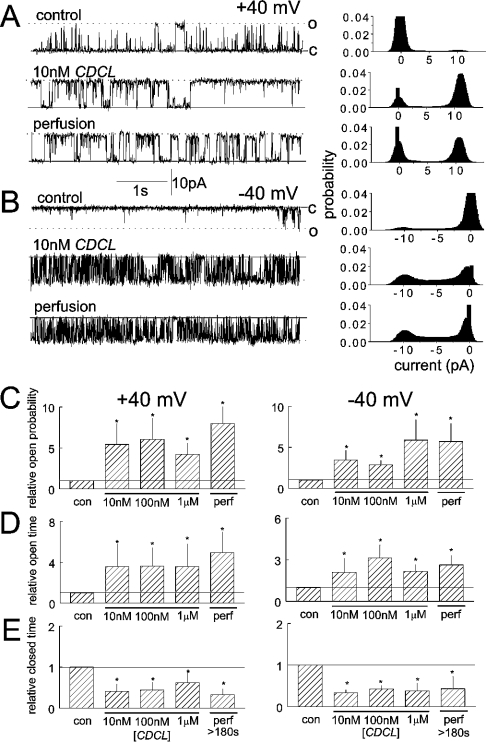
CDCL activated channels at concentrations ≥10 nM, at positive and negative potentials (Figure 5). The 5–6-fold increase in Po was due to a significant increase in channel To and abbreviation of closures at all concentrations. In contrast with the rapidly reversible effects of the C region peptides, the activation by CDCL was not reversible within the time frame of the bilayer experiment. Channel activity remained higher than control levels for up to 10 min after perfusion of CDCL from the cis chamber. This lack of reversibility is indicative of strong binding of the recombinant loop to the native RyR2 channel.
Figure 5. CDCL causes a high-affinity increase in RyR2 activity that is irreversible within the time frame of the experiment.
Brief (5 s) channel records (left-hand panels) and all-points histograms from a 30 s record (right-hand panels) are shown at +40 mV (A) and −40 mV (B). The upper record in each panel was obtained under control conditions with 10 μM cis Ca2+, the middle records in the presence of 10 nM CDCL and the final record >3 min after perfusion of CDCL from the cis chamber. The histograms in (C), (D) and (E) show average data for relative Po (C), relative To (D) and relative Tc (E) at +40 mV (left-hand panels) and −40 mV (right-hand panels). In each panel, data are shown under control conditions, with 10 nM, 100 nM and 1 μM CDCL and after perfusion of CDCL from the cis chamber. Asterisks indicate significant differences from control.
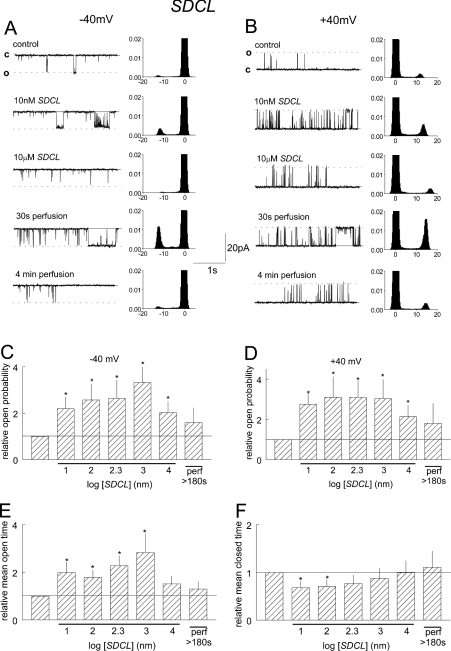
SDCL also activated cardiac channels at concentrations ≥10 nM in a voltage-independent manner (Figure 6). This was due to a 2–3-fold increase in the channel Tos together with a small decline in the Tcs which was greatest with 10 and 100 nM, and appeared to be less at higher loop concentrations. The increase in closed durations when SDCL was increased from 1 to 10 μM was accompanied by reduction in Tcs. Activity increased further when SDCL was perfused from the cis chamber, but then returned to control levels after approx. 3 min. These changes in activity are indicative of a high-affinity, slowly reversible, activation associated with SDCL binding plus an additional lower-affinity rapidly reversible inhibitory process.
Figure 6. SDCL causes a high-affinity, slowly reversible, increase in RyR2 activity.
(A) and (B) include 5 s current recordings (left-hand panels) and all-points histograms from 30 s of channel activity (right-hand panels) at +40 and −40 mV respectively. Data are shown under control conditions with 10 μM cis Ca2+, after addition of 10 nM, then 10 μM SDCL, and then 30 s and 4 min after perfusion of SDCL from the cis chamber. (C) and (D) show relative Po at +40 mV and −40 mV. Data for average relative Tc (E) and relative Tc (F) is the combined average of values obtained at +40 mV and −40 mV, since the results were very similar at both potentials. Average values are shown under control conditions, in the presence of 10 nM, 100 nM, 200 nM, 1 μM and 10 μM SDCL, and after perfusion of 10 μM SDCL from the cis chamber. Asterisks indicate significant differences from control.
Ca2+ release from cardiac SR
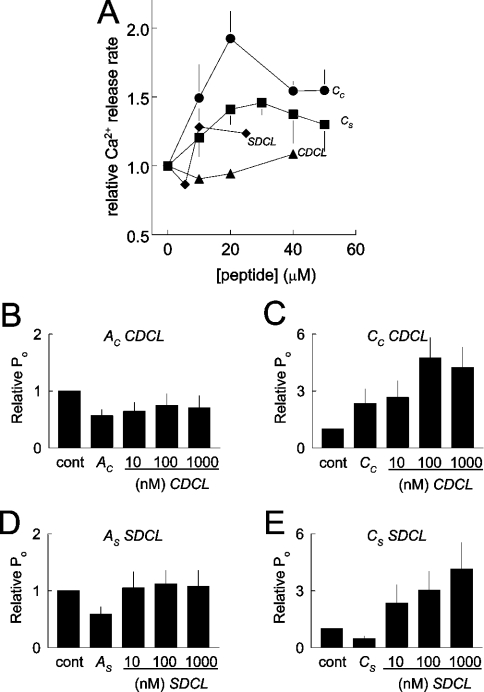
Measurements were made in the presence of thapsigargin (see the Experimental section), so that the Ca2+,Mg2+-ATPase was blocked, and changes in release rate could be attributed to Ca2+ efflux through the RyR. Cc, Cs and SDCL all enhanced Ca2+-induced Ca2+ release (Figure 7A). CDCL did not cause a significant change in the release, although there was a trend towards increased release with 40 μM protein. Even higher concentrations of CDCL may have significantly increased release, but could not be used because of low stock concentrations and a limit on volumes that could be added to the cuvette. In contrast with Ca2+-induced Ca2+ release, neither of the peptides or the recombinant loops altered resting Ca2+ release (results not shown).
Figure 7. CDCL and SDCL increase Ca2+-induced Ca2+ release from cardiac SR vesicles and compete strongly with the A peptides and partly with C peptides when activating RyR2 channels in bilayers.
(A) Relative rates of Ca2+-induced Ca2+ release are shown as a function of peptide concentration. Average data are shown for peptide Cs (●; n=5); Cc (■; n=5); SDCL (◆; n=2) and CDCL (▲; n=2). (B)–(E) Competition between the recombinant II–III loops and A or C peptide for activation of RyR2. In each experiment, 1 μM peptide was added for 4 min, and then the channel was exposed to 10 nM, 100 nM or 1 μM of II–III loop for 4 min at each concentration. Channels were treated with Ac and CDCL (B) (n=5), Cc and CDCL (C) (n=4), As and SDCL (D) (n=4), Cs and SDCL (E) (n=4).
As noted previously, modification of Ca2+ release required higher concentrations of DHPR fragments than effects on RyR channels in bilayers [9,22,33], possibly due to different conditions imposed by the different techniques.
Does the II–III loop bind to RyR2 via its A or C region?
To determine whether the A or C region of the loop was involved in the interaction between CDCL or SDCL and RyR2, the ability of the recombinant loops to activate the channel were assessed in the presence of either the A or the C peptide (Figures 7B–7E). The results indicate that the A region is strongly involved and suggest a weaker contribution from the C region. Addition of 1 μM Ac or As caused the usual reduction in RyR2 activity seen with activating cis Ca2+ concentration (in contrast with activation when cis Ca2+ concentration is lower) [13], but there were only small increases in activity with subsequent addition of CDCL or SDCL. In contrast, both CDCL and SDCL activated the channels following addition of Cc or Cs respectively, with usual levels of activation seen with 1 μM loop. However, activation by 10 nM of the loop was significantly less in the presence of the C peptides than in their absence (Figures 5 and 6). This is particularly apparent when CDCL was added with Cc (Figure 7C).
Structure of the C peptides, CDCL and SDCL
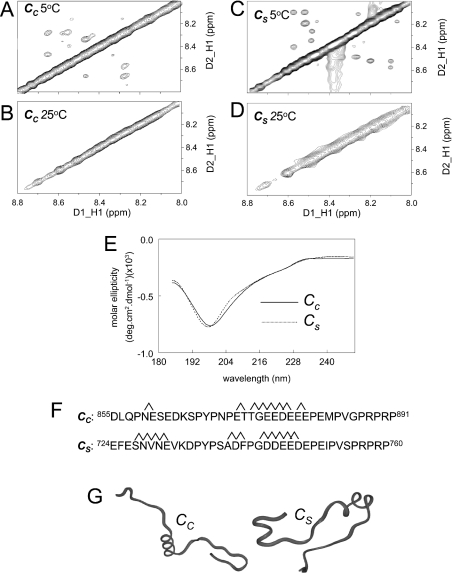
NMR TOCSY and NOESY experiments were performed to assign individual proton resonances (assignments not shown). The experiments were carried out at 5 °C (since weakly structured proteins are likely to be more stable at lower temperatures) as well as at 25 °C. Peptide Cs has been shown previously to have a random coil structure at room temperature [18]. The lack of cross-peaks in the NOESY spectra for Cc (Figure 8B) suggest that the cardiac C peptide is also random coil at 25 °C. In contrast, the cross-peaks in the NMR spectra at 5 °C indicated that both Cs and Cc have some helical content (Figures 8A and 8C). Assignment of these residues indicate that there are common elements of helical structure in both C peptides, encompassing residues 17–25 (Figure 8F), which have been depicted in Figure 8(G). The CD spectra are consistent with the fact that both peptides are mostly random coil in structure, with little difference apparent in their spectral profile (Figure 8E). Helical peptides display minima at 208 and 222 nm. The fact that both peptides show a minima at wavelengths less than 208 nm and a weak inflection near 222 nm suggests that the helical content is weak.
Figure 8. Comparison of structures of Cc and Cs.
(A)–(D) The amide–amide region of the Cc and Cs NOESY spectra respectively in 10% 2H2O/90% H2O, at 5 °C (A and C) or 25 °C (B and D). The off-diagonal cross-peaks in the 5 °C spectrum are indicative of helical structure, whereas the lack of cross-peaks at 25 °C indicate random coil structures. (E) CD spectra for Cc and Cs at 25 °C show a minima at approx. 200 nm, suggesting a mixture of random coil and helical structure. Assignment of the connectivities contributing to the NOE (nuclear Overhauser effect) cross-peaks at 25 °C indicated helical structure between residues as indicated in (F), allowing prediction of the structures in (G). In Cc, there is a helical turn involving residues 5 and 6, helical structure between residues 17 and 26. In Cs, there is helical structure between residues 4 and 8, and between residues 16 and 25.
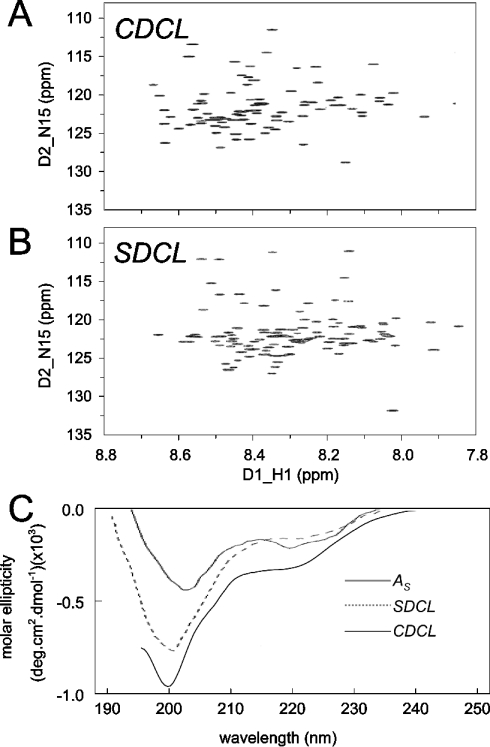
Examination of the 15N/1H-HSQC NMR spectra for CDCL and SDCL indicates that the spectral profile of both recombinant proteins are similar. Although the three-dimensional structure of either of these proteins has not been determined, the chemical-shift distribution and line widths of the cross-peaks are good indicators of secondary structure and the oligomeric state respectively. Based on preliminary NMR data obtained for SDCL and the spectral similarities with CDCL, we can assert that the two proteins are monomeric and adopt a helical/random coil structure. NMR spectra for CDCL and SDCL show that both recombinant proteins contain similar amounts of α-helical content (Figures 9A and 9B). Local differences are due to differences in amino acid composition, as well as minor structural variations. These differences will be elucidated in future determination of the solution structures. The secondary-structure composition was confirmed with the CD analysis, which indicated that CDCL and SDCL have a random coil/helical content, but substantially less helix than that in the approx. 80% helical As peptide (Figure 9C). At this level of analysis, there did not appear to be a major difference between the secondary structures of the CDCL and the SDCL.
Figure 9. Comparison of the structural profiles of CDCL and SDCL.
(A) and (B) 15N/1H-HSQC spectra for CDCL and SDCL. The similar spectral distribution of cross-peaks suggest a similar degree of structure in the two recombinant proteins. (C) CD spectra for the 80% helical As peptide and for CDCL and SDCL. The spectra for the recombinant proteins show minima at approx. 200 nm, indicating less helical content than As, which has a minimum at 204 nm. The more pronounced dip at approx. 222 nm in CDCL is indicative of greater helical content in that protein.
DISCUSSION
Novel results show that the 36-residue peptides corresponding to the C region of the DHPR II–III loop, as well as the ∼120-residue recombinant loops, interact with native RyR2 channels. Functional changes reflecting binding to the RyR2 complex were seen with skeletal and cardiac DHPR sequences. NMR and CD analysis showed that both the cardiac Cc and the Cs peptide had a partial α-helical structure. The results (i) are consistent with observations that other parts of the skeletal and cardiac DHPR can bind to RyR2 and (ii) support the hypothesis that both the skeletal and cardiac DHPRs have the potential for physical/conformational coupling to RyR2.
Interactions between DHPRs and RyRs
An isoform-independent ability of DHPRs to interact with RyRs is indicated by the cross-reactions between skeletal or cardiac DHPR fragments and skeletal or cardiac RyRs ([13,14,20], C. S. Haarmann, A. F. Dulhunty and D. R. Laver, unpublished results, and the present study). Some myocyte studies indicate physical interactions between the cardiac DHPR and RyR and voltage-activated Ca2+ release through RyR2 in the cardiac tissue has been reported [5–7,34,35], although the contribution of this mechanism to EC coupling is questioned [1,2]. There are studies in other tissues indicating isoform-independent interactions between DHPRs and RyR. For example, cardiac DHPRs (CaV1.2), as well as CaV1.3, but not skeletal DHPRs (CaV1.1), in brain co-immunoprecipitate RyR1 [36]. In spinal cord white matter, CaV1.2 co-immunoprecipitates RyR1, while CaV1.3 precipitates RyR2, and functional evidence is presented suggesting an external Ca2+-independent coupling process like that in skeletal muscle [37]. These observations have led to suggestions of functional depolarization-dependent coupling between the cardiac DHPR and RyR1 in the central nervous system. With hindsight, the isoform-independent interactions between cardiac and skeletal DHPRs and RyRs are not surprising given the homology between the isoforms of each protein [38,39]. An isoform-independent ability to interact is attractive from an evolutionary perspective, since a primitive system could diverge into either the external Ca2+-dependent or Ca2+-independent (i.e. cardiac or skeletal type) EC coupling in muscle and in other tissues with adjustments to the ratio of DHPRs to RyRs, targeting signals or RyR gating mechanisms.
Previous studies of Ca2+ release from cardiac SR may not have detected functional interactions with C region peptides or recombinant loops [10,19], because they were not performed under conditions which reveal the effects, i.e. Ca2+-induced Ca2+ release in the presence of thapsigargin. Previous bilayer studies may have failed to detect interactions between DHPR fragments and RyR2s because the channels were CHAPS-purified [9,12], and may have been modified during purification or separated from an essential co-protein. The possibility of a requirement for a co-protein is suggested because the skeletal DHPR A fragment interacts with RyR1 only in the presence of FKBP12 [3,22,40].
The C peptides and II–III loops can either activate or inhibit RyR2s. Enhanced Ca2+ release and channel activation were seen with peptide Cc and SDCL. On the other hand, Ca2+ release was increased with Cs, while channel activity was inhibited, indicating that the bilayer conditions may favour inhibition, while Ca2+ release conditions favour activation. It is notable that an increase in Tos at high Cs concentrations indicated a low-affinity activation with this peptide. A dual effect of the C peptides is also seen with RyR1s, where activation or inhibition of channels is seen under different conditions [18]. Evidence for dual effects of the II–III loop fragments is also seen with (i) SDCL, which activates RyR2 at low concentrations, but tends to inhibit at high concentrations, and (ii) CDCL, which tends to reduce Ca2+ release at low concentrations, but enhances release at high concentrations. The A peptides either activate RyR2s with nanomolar concentrations of cis Ca2+ or inhibit with 10–100 μM cis Ca2+ [13].
Binding sites for the DHPR fragments
It is not clear whether the DHPR fragments bind to the RyR2 or to proteins that remain associated with the cytoplasmic side of the channel, such as FK506-binding proteins, Ca2+/calmodulin kinase II (on RyR1) or protein kinase A (on RyR2) [32,41–43]. However, the binding sites for the DHPR fragments are most likely to be on RyR2, since binding to the protein is seen in two-hybrid [4] and surface plasmon resonance studies [3]. Nevertheless, we cannot exclude the possibility that some effects are mediated by binding to an associated protein such as sorcin, which binds to both the RyR and DHPR in cardiac and in skeletal muscle, and is thought to facilitate communication between the channels [44]. Similarly, on the luminal side of the SR, calsequestrin modulates RyR activity by binding to the anchoring proteins triadin and junctin [45,46].
Do Cc and Cs bind to the same sites on RyR2s? Peptides Cc and Cs have similar C-terminal, but different N-terminal, sequences. Although the peptides are thought to bind to the same sites on RyR1 because their actions on the channel are similar [18,20,47]), their different actions on RyR2 do not necessarily indicate different binding sites. Indeed, the competition experiments (Figure 3) provide strong evidence for the peptides binding to a similar site. It is not uncommon for compounds binding to a single site to have different actions. Calmodulin can either activate or inhibit RyRs, depending on its association with Ca2+ (its Ca2+-dependent conformation), even though Ca2+/calmodulin and apo-calmodulin bind to the same, or overlapping, sites [48,49]. Thus the C peptides may bind to a common site, with the functional effects of the binding depending on the sequence/structural differences between the peptides.
CDCL and SDCL are likely to bind to the same site(s) on RyR2. Both recombinant proteins induced high-affinity, slowly reversible, activation of RyR2s, which differed from the rapidly reversible effects of the C peptides. CDCL and SDCL could bind to the RyR2 channel complex via either their C region or their A region, since both A and C peptides bind to the complex. The A region of the skeletal II–III loop is important in II–III loop binding to the skeletal RyR [50,51]. We also found that binding of the A peptide reduces activation of RyR2 by the recombinant II–III loops substantially, indicating that SDCL and CDCL may bind to RyR2 at the A binding site. The affinity of RyR2 for SDCL and CDCL was apparently decreased in the presence of the C peptides, indicating either that the II–III loops also bind to the C site on RyR2, or that the negatively charged C peptide binds to the A region of the loops and prevents their interaction with RyR2. This latter possibility seems unlikely, since peptides As and Cs do not bind to each other [18]. Thus we propose a model in which CDCL/SDCL bind to RyR2 at both the A binding site and the C binding site. If the A site is occluded by peptide A binding, then binding to the C site causes weak activation. If the C site is occupied by peptide C, activation through binding to the A site occurs at higher loop concentrations.
Structure of the C peptides and recombinant II–III loops
We have shown previously that, at room temperature, the Cs peptide is random coil [18] and is one of a group of intrinsically unstructured, functionally important proteins [52]. However, weak hydrogen bonding interactions and weak secondary-structure elements can be detected if NMR experiments are performed at a lower temperature. When this was done for Cs and Cc peptides in the present study, weak helical components were evident in both peptides, although only a small number of the total amino acid residues were involved in the formation of the nascent helix. Overall, peptide Cs has marginally more helical content than Cc, but the helix in the conserved central negatively charged region was stronger in Cc. Given the sequence similarity, it is no real surprise that the region has a similar structure in the two peptides. The appearance of structure at low temperature raises the possibility that secondary-structural elements may be stabilized when the DHPR binds to the RyR in vivo. The slight secondary-structure differences in the C region do not alter the ability of Cc or CDCL to interact with either RyR2 (see above) or RyR1 [20]. Preliminary work performed on the full skeletal II–III loop suggests that the C region is also mostly random coil in structure (M. G. Casarotto and A. F. Dulhunty, unpublished work), as do published structure prediction studies [14,53]. In contrast with our data, the predictions suggest that there is significantly more helical content in the cardiac C region than in the skeletal C region [14,53].
Physiological significance of peptide effects on RyR2
Skeletal EC coupling in expression systems is specific for the skeletal isoforms of both the DHPR II–III loop and the RyR. In contrast, the physical interaction between the proteins is not isoform-specific (see above). Thus the loss of skeletal EC coupling when the cardiac C region replaces the skeletal C region in the DHPR cannot be attributed to an inability of the DHPR to interact with the RyR when the C region contains either the cardiac sequence or a helical structure [53]. This suggests that the specificity in EC coupling is for expression of appropriate numbers of DHPRs or for their targeting into tetrads in exact opposition to the RyR. There are many fewer DHPRs than RyRs in cardiac muscle [23], and the DHPRs are not normally located opposite RyRs [24].
It is clear that direct DHPR–RyR coupling does not contribute significantly to EC coupling in the heart [1,2]. However, the ability of the two proteins to interact raises the possibilities that (i) physical interaction could contribute to some signal transduction processes in the heart and (ii) isoform-independent physical interactions between the two proteins could occur in other tissues (such as brain) if the proteins are appropriately targeted.
In conclusion, we show for the first time that both the C region of the DHPR II–III loop and the recombinant full-length DHPR II–III loop are capable of isoform-independent functional interactions with RyR2. A novel structural analysis shows that the C region of the CDCL and SDCL peptides is a mixture of α-helix and random coil. Furthermore, we show that the overall helical content of the full DHPR II–III loop is comparable in the recombinant CDCL and SDCL proteins. The results add to an increasing body of evidence that differences between EC coupling in cardiac and skeletal muscle is related more to factors regulating expression levels and targeting of the DHPR and RyR than to intrinsic differences between the ability of the proteins to physically interact with each other.
Acknowledgments
We are grateful to Suzy Pace and to Joan Stivala for assistance with preparation and characterization of SR vesicles, to Sarah Watson for assistance with some of the single-channel studies, and to Louise Cengia for assistance with measurements of Ca2+ release. We thank Professor Gerhard Meissner for providing us with cDNA for SDCL and CDCL. The project was supported by a grant from the Australian National Health and Medical Research Council # 224235.
References
- 1.Piacentino V., 3rd, Dipla K., Gaughan J. P., Houser S. R. Voltage-dependent Ca2+ release from the SR of feline ventricular myocytes is explained by Ca2+-induced Ca2+ release. J. Physiol. 2000;523:533–548. doi: 10.1111/j.1469-7793.2000.t01-1-00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wier W. G., Balke C. W. Ca2+ release mechanisms, Ca2+ sparks, and local control of excitation–contraction coupling in normal heart muscle. Circ. Res. 1999;85:770–776. doi: 10.1161/01.res.85.9.770. [DOI] [PubMed] [Google Scholar]
- 3.O'Reilly F. M., Ronjat M. Direct interaction of the skeletal dihydropyridine receptor α1 subunit with skeletal and cardiac ryanodine receptors. Biophys. J. 1999;76:A466. [Google Scholar]
- 4.Proenza C., O'Brien J., Nakai J., Mukherjee S., Allen P. D., Beam K. G. Identification of a region of RyR1 that participates in allosteric coupling with the α1S (CaV1.1) II–III loop. J. Biol. Chem. 2002;277:6530–6535. doi: 10.1074/jbc.M106471200. [DOI] [PubMed] [Google Scholar]
- 5.Hryshko L. V., Kobayashi T., Bose D. Possible inhibition of canine ventricular sarcoplasmic reticulum by BAY K 8644. Am. J. Physiol. 1989;257:H407–H414. doi: 10.1152/ajpheart.1989.257.2.H407. [DOI] [PubMed] [Google Scholar]
- 6.Katoh H., Schlotthauer K., Bers D. M. Transmission of information from cardiac dihydropyridine receptor to ryanodine receptor: evidence from BayK 8644 effects on resting Ca2+ sparks. Circ. Res. 2000;87:106–111. doi: 10.1161/01.res.87.2.106. [DOI] [PubMed] [Google Scholar]
- 7.Cohen N. M., Lederer W. J. Changes in the calcium current of rat heart ventricular myocytes during development. J. Physiol. 1988;406:115–146. doi: 10.1113/jphysiol.1988.sp017372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Bers D. M. A cardiac dihydropyridine receptor II–III loop peptide inhibits resting Ca2+ sparks in ferret ventricular myocytes. J. Physiol. 2001;537:17–26. doi: 10.1111/j.1469-7793.2001.0017k.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu X., Xu L., Meissner G. Activation of the skeletal muscle calcium release channel by a cytoplasmic loop of the dihydropyridine receptor. J. Biol. Chem. 1994;269:6511–6516. [PubMed] [Google Scholar]
- 10.El-Hayek R., Antoniu B., Wang J., Hamilton S. L., Ikemoto N. Identification of calcium release-triggering and blocking regions of the II–III loop of the skeletal muscle dihydropyridine receptor. J. Biol. Chem. 1995;270:22116–22118. doi: 10.1074/jbc.270.38.22116. [DOI] [PubMed] [Google Scholar]
- 11.Gurrola G. B., Arevalo C., Sreekumar R., Lokuta A. J., Walker J. W., Valdivia H. H. Activation of ryanodine receptors by imperatoxin A and a peptide segment of the II–III loop of the dihydropyridine receptor. J. Biol. Chem. 1999;274:7879–7886. doi: 10.1074/jbc.274.12.7879. [DOI] [PubMed] [Google Scholar]
- 12.Stange M., Tripathy A., Meissner G. Two domains in dihydropyridine receptor activate the skeletal muscle Ca2+ release channel. Biophys. J. 2001;81:1419–1429. doi: 10.1016/S0006-3495(01)75797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulhunty A. F., Curtis S. M., Cengia L., Sakowska M., Casarotto M. G. Peptide fragments of the dihydropyridine receptor can modulate cardiac ryanodine receptor channel activity and sarcoplasmic reticulum Ca2+ release. Biochem. J. 2004;379:161–172. doi: 10.1042/BJ20031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bannister M. L., Williams A. J., Sitsapesan R. Removal of clustered positive charge from dihydropyridine receptor II–III loop peptide augments activation of ryanodine receptors. Biochem. Biophys. Res. Commun. 2004;314:667–674. doi: 10.1016/j.bbrc.2003.12.128. [DOI] [PubMed] [Google Scholar]
- 15.Tanabe T., Beam K. G., Powell J. A., Numa S. Restoration of excitation–contraction coupling and slow calcium current in dysgenic muscle by dihydropyridine receptor complementary DNA. Nature (London) 1988;336:134–139. doi: 10.1038/336134a0. [DOI] [PubMed] [Google Scholar]
- 16.Wilkens C. M., Kasielke N., Flucher B. E., Beam K. G., Grabner M. Excitation–contraction coupling is unaffected by drastic alteration of the sequence surrounding residues L720–L764 of the α1S II–III loop. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5892–5897. doi: 10.1073/pnas.101618098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahern C. A., Bhattacharya D., Mortenson L., Coronado R. A component of excitation–contraction coupling triggered in the absence of the T671–L690 and L720–Q765 regions of the II–III loop of the dihydropyridine receptor α1s pore subunit. Biophys. J. 2001;81:3294–3307. doi: 10.1016/S0006-3495(01)75963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haarmann C. S., Green D., Casarotto M. G., Laver D. R., Dulhunty A. F. The random coil ‘C’ fragment of the dihydropyridine receptor II–III loop can activate or inhibit native skeletal ryanodine receptors. Biochem. J. 2003;372:305–316. doi: 10.1042/BJ20021763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto T., Rodriguez J., Ikemoto N. Ca2+-dependent dual functions of peptide C. The peptide corresponding to the Glu724–Pro760 region (the so-called determinant of excitation-contraction coupling) of the dihydropyridine receptor α1 subunit II–III loop. J. Biol. Chem. 2002;277:993–1001. doi: 10.1074/jbc.M105837200. [DOI] [PubMed] [Google Scholar]
- 20.Dulhunty A. F., Haarmann C., Green D., Laver D. R., Board P. G., Casarotto M. G. Interactions between dihydropyridine receptors and ryanodine receptors in striated muscle. Prog. Biophys. Mol. Biol. 2002;79:45–75. doi: 10.1016/s0079-6107(02)00013-5. [DOI] [PubMed] [Google Scholar]
- 21.Casarotto M. G., Gibson F., Pace S. M., Curtis S. M., Mulcair M., Dulhunty A. F. A structural requirement for activation of skeletal ryanodine receptors by peptides of the dihydropyridine receptor II–III loop. J. Biol. Chem. 2000;275:11631–11637. doi: 10.1074/jbc.275.16.11631. [DOI] [PubMed] [Google Scholar]
- 22.Dulhunty A. F., Laver D. R., Gallant E. M., Casarotto M. G., Pace S. M., Curtis S. Activation and inhibition of skeletal RyR channels by a part of the skeletal DHPR II–III loop: effects of DHPR Ser687 and FKBP12. Biophys. J. 1999;77:189–203. doi: 10.1016/S0006-3495(99)76881-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bers D. M., Stiffel V. M. Ratio of ryanodine to dihydropyridine receptors in cardiac and skeletal muscle and implications for E–C coupling. Am. J. Physiol. 1993;264:C1587–C1593. doi: 10.1152/ajpcell.1993.264.6.C1587. [DOI] [PubMed] [Google Scholar]
- 24.Franzini-Armstrong C., Protasi F., Ramesh V. Comparative ultrastructure of Ca2+ release units in skeletal and cardiac muscle. Ann. N.Y. Acad. Sci. 1998;853:20–30. doi: 10.1111/j.1749-6632.1998.tb08253.x. [DOI] [PubMed] [Google Scholar]
- 25.Casarotto M. G., Green D., Pace S. M., Curtis S. M., Dulhunty A. F. Structural determinants for activation or inhibition of ryanodine receptors by basic residues in the dihydropyridine receptor II–III loop. Biophys. J. 2001;80:2715–2726. doi: 10.1016/S0006-3495(01)76240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catanzariti A. M., Soboleva T. A., Jans D. A., Board P. G., Baker R. T. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porath J., Olin B. Immobilized metal ion affinity adsorption and immobilized metal ion affinity chromatography of biomaterials: serum protein affinities for gelimmobilized iron and nickel ions. Biochemistry. 1983;22:1621–1630. doi: 10.1021/bi00276a015. [DOI] [PubMed] [Google Scholar]
- 28.Laver D. R., Roden L. D., Ahern G. P., Eager K. R., Junankar P. R., Dulhunty A. F. Cytoplasmic Ca2+ inhibits the ryanodine receptor from cardiac muscle. J. Membr. Biol. 1995;147:7–22. doi: 10.1007/BF00235394. [DOI] [PubMed] [Google Scholar]
- 29.Kay L. E., Keifer P., Sarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 30.King B. M., Bear G. Some (almost) assumption-free tests. In: Minium E. W., editor. Statistical Reasoning in Psychology and Education. New York: Wiley; 1993. pp. 474–496. [Google Scholar]
- 31.Xu L., Mann G., Meissner G. Regulation of cardiac Ca2+ release channel (ryanodine receptor) by Ca2+, H+, Mg2+, and adenine nucleotides under normal and simulated ischemic conditions. Circ. Res. 1996;79:1100–1109. doi: 10.1161/01.res.79.6.1100. [DOI] [PubMed] [Google Scholar]
- 32.Dulhunty A. F., Laver D., Curtis S. M., Pace S., Haarmann C., Gallant E. M. Characteristics of irreversible ATP activation suggest that native skeletal ryanodine receptors can be phosphorylated via an endogenous CaMKII. Biophys. J. 2001;81:3240–3252. doi: 10.1016/S0006-3495(01)75959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dulhunty A. F., Curtis S. M., Watson S., Cengia L., Casarotto M. G. Multiple actions of imperatoxin A on ryanodine receptors: interactions with the II–III loop “A” fragment. J. Biol. Chem. 2004;279:11853–11862. doi: 10.1074/jbc.M310466200. [DOI] [PubMed] [Google Scholar]
- 34.Hobai I. A., Howarth F. C., Pabbathi V. K., Dalton G. R., Hancox J. C., Zhu J. Q., Howlett S. E., Ferrier G. R., Levi A. J. “Voltage-activated Ca release” in rabbit, rat and guinea-pig cardiac myocytes, and modulation by internal cAMP. Pflugers Arch. 1997;435:164–173. doi: 10.1007/s004240050496. [DOI] [PubMed] [Google Scholar]
- 35.Howlett S. E., Zhu J. Q., Ferrier G. R. Contribution of a voltage-sensitive calcium release mechanism to contraction in cardiac ventricular myocytes. Am. J. Physiol. 1998;274:H155–H170. doi: 10.1152/ajpheart.1998.274.1.H155. [DOI] [PubMed] [Google Scholar]
- 36.Mouton J., Marty I., Villaz M., Feltz A., Maulet Y. Molecular interaction of dihydropyridine receptors with type-1 ryanodine receptors in rat brain. Biochem. J. 2001;354:597–603. doi: 10.1042/0264-6021:3540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouardouz M., Nikolaeva M. A., Coderre E., Zamponi G. W., McRory J. E., Trapp B. D., Yin X., Wang W., Woulfe J., Stys P. K. Depolarization-induced Ca2+ release in ischemic spinal cord white matter involves L-type Ca2+ channel activation of ryanodine receptors. Neuron. 2003;40:53–63. doi: 10.1016/j.neuron.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Mikami A., Imoto K., Tanabe T., Niidome T., Mori Y., Takeshima H., Narumiya S., Numa S. Primary structure and functional expression of the cardiac dihydropyridine-sensitive calcium channel. Nature (London) 1989;340:230–233. doi: 10.1038/340230a0. [DOI] [PubMed] [Google Scholar]
- 39.Otsu K., Willard H. F., Khanna V. K., Zorzato F., Green N. M., MacLennan D. H. Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J. Biol. Chem. 1990;265:13472–13483. [PubMed] [Google Scholar]
- 40.Lamb G. D., El-Hayek R., Ikemoto N., Stephenson D. G. Effects of dihydropyridine receptor II–III loop peptides on Ca2+ release in skinned skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 2001;279:C891–C905. doi: 10.1152/ajpcell.2000.279.4.C891. [DOI] [PubMed] [Google Scholar]
- 41.Ahern G. P., Junankar P. R., Dulhunty A. F. Single channel activity of the ryanodine receptor calcium release channel is modulated by FK-506. FEBS Lett. 1994;352:369–374. doi: 10.1016/0014-5793(94)01001-3. [DOI] [PubMed] [Google Scholar]
- 42.Marx S. O., Gaburjakova J., Gaburjakova M., Henrikson C., Ondrias K., Marks A. R. Coupled gating between cardiac calcium release channels (ryanodine receptors) Circ. Res. 2001;88:1151–1158. doi: 10.1161/hh1101.091268. [DOI] [PubMed] [Google Scholar]
- 43.Marx S. O., Reiken S., Hisamatsu Y., Jayaraman T., Burkhoff D., Rosemblit N., Marks A. R. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101:365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- 44.Meyers M. B., Puri T. S., Chien A. J., Gao T., Hsu P. H., Hosey M. M., Fishman G. I. Sorcin associates with the pore-forming subunit of voltage-dependent L-type Ca2+ channels. J. Biol. Chem. 1998;273:18930–18935. doi: 10.1074/jbc.273.30.18930. [DOI] [PubMed] [Google Scholar]
- 45.Beard N. A., Sakowska M. M., Dulhunty A. F., Laver D. R. Calsequestrin is an inhibitor of skeletal muscle ryanodine receptor calcium release channels. Biophys. J. 2002;82:310–320. doi: 10.1016/S0006-3495(02)75396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gyorke I., Hester N., Jones L. R., Gyorke S. The role of calsequestrin, triadin, and junctin in conferring cardiac ryanodine receptor responsiveness to luminal calcium. Biophys. J. 2004;86:2121–2128. doi: 10.1016/S0006-3495(04)74271-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haarmann C. S., Dulhunty A. F., Laver D. R. Regulation of skeletal ryanodine receptors by dihydropyridine receptor II–III loop peptides: relief of Mg2+ inhibition. Biochem. J. 2004 doi: 10.1042/BJ20040786. 10.1042/BJ20040786. [DOI] [PMC free article] [PubMed]
- 48.Tang W., Sencer S., Hamilton S. L. Calmodulin modulation of proteins involved in excitation–contraction coupling. Front. Biosci. 2002;7:d1583–d1589. doi: 10.2741/tang. [DOI] [PubMed] [Google Scholar]
- 49.Samso M., Wagenknecht T. Apocalmodulin and Ca2+–calmodulin bind to neighboring locations on the ryanodine receptor. J. Biol. Chem. 2002;277:1349–1353. doi: 10.1074/jbc.M109196200. [DOI] [PubMed] [Google Scholar]
- 50.Lu X., Xu L., Meissner G. Phosphorylation of dihydropyridine receptor II–III loop peptide regulates skeletal muscle calcium release channel function: evidence for an essential role of the β-OH group of Ser687. J. Biol. Chem. 1995;270:18459–18464. doi: 10.1074/jbc.270.31.18459. [DOI] [PubMed] [Google Scholar]
- 51.O'Reilly F. M., Robert M., Jona I., Szegedi C., Albrieux M., Geib S., De Waard M., Villaz M., Ronjat M. FKBP12 modulation of the binding of the skeletal ryanodine receptor onto the II–III loop of the dihydropyridine receptor. Biophys. J. 2002;82:145–155. doi: 10.1016/S0006-3495(02)75381-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 53.Kugler G., Weiss R. G., Flucher B. E., Grabner M. Structural requirements of the dihydropyridine receptor α1S II–III loop for skeletal-type excitation–contraction coupling. J. Biol. Chem. 2004;279:4721–4728. doi: 10.1074/jbc.M307538200. [DOI] [PubMed] [Google Scholar]