Abstract
The ADAM (a disintegrin and metalloprotease) family of proteins possess both proteolytic and adhesive domains. We have established previously that the disintegrin domain of ADAM28, an ADAM expressed by human lymphocytes, is recognized by the integrin α4β1. The present study characterizes the integrin binding properties of the disintegrin-like domains of human ADAM7, ADAM28 and ADAM33 with the integrins α4β1, α4β7 and α9β1. Cell-adhesion assays demonstrated that, similar to ADAM28, the ADAM7 disintegrin domain supported α4β1-dependent Jurkat cell adhesion, whereas the ADAM33 disintegrin domain did not. The lymphocyte integrin α4β7 was also found to recognize both disintegrin domains of ADAM7 and ADAM28, but not of ADAM33. This is the first demonstration that mammalian disintegrins are capable of interacting with α4β7. All three disintegrin domains supported α9β1-dependent cell adhesion. Recognition by both α4β1 and α4β7 of ADAM7 and ADAM28 was activation-dependent, requiring either the presence of Mn2+ or an activating monoclonal antibody for cell attachment. Charge-to-alanine mutagenesis experiments revealed that the same residues within an individual ADAM disintegrin domain function in recognizing multiple integrins. However, the residues within a specific region of each ADAM disintegrin-like domain required for integrin binding were distinct. These results establish that ADAM7 and ADAM28 are recognized by the leucocyte integrins α4β1, α4β7 and α9β1. ADAM33 exclusively supported only α9β1-dependent adhesion.
Keywords: adisintegrin and metalloprotease (ADAM), cell adhesion, disintegrin, integrin, lymphocyte, mutagenesis
Abbreviations: ADAM, adisintegrin and metalloprotease; CHO cells, Chinese-hamster ovary cells; Dis–Fc, recombinant disintegrin–Fc fusion protein; mAb, monoclonal antibody; MAdCAM, mucosal addressin adhesion molecule
INTRODUCTION
ADAM (a disintegrin and metalloprotease) proteins are members of the metzincins that also include the related matrix metalloproteases. The defining feature of the ADAMs is a conserved modular domain organization consisting of a prodomain, a metalloprotease domain, a disintegrin-like domain, a cysteine-rich domain, an EGF (epidermal growth factor)-like domain, a transmembrane domain and a cytoplasmic domain [1,2]. The ADAMs comprise nearly 40 proteins with members being identified in a variety of mammalian species as well as lower eukaryotes such as Caenorhabditis elegans, Xenopus and Drosophila [3].
Although the metalloprotease domain of several ADAMs functions as a cell-surface sheddase [4,5], the functional relevance of the other ADAM domains remains unclear. The disintegrin-like domain of the ADAMs exhibits extensive homology with the small non-enzymic peptide constituents of snake venom that are potent integrin antagonists [6,7]. Multiple studies have reported integrin recognition of ADAM disintegrin-like domains. The mechanism of integrin recognition of snake venom disintegrins has been attributed to tripeptide sequences positioned on a structurally defined loop [8–10]. A similar region within ADAM disintegrin-like domains also exists. Although some charged residues within the putative disintegrin loop of the ADAMs are critical for integrin recognition [11–14], many of the residues that comprise the integrin recognition site are located outside of the putative loop [15].
Integrins are a cell–cell and cell–matrix receptor superfamily composed of non-covalently linked heterodimers. Integrins have broad and significant biological roles in processes such as embryonic development, haematopoiesis, wound repair and immune response [16]. Recognition of multiple ADAM disintegrins by the same integrin family member has been demonstrated for the recognition of ADAM2, ADAM3 and ADAM9 by the integrin α6β1 [13,14,17,18] and the recognition of ADAM2, ADAM3 and ADAM28 by α4β1 [19,20], whereas α9β1 was shown to recognize ADAM2, ADAM3, ADAM12 and ADAM15 [19,21], leading to the hypothesis that α9β1 interacts with all prototypical ADAMs [12]. Promiscuity is not restricted to the receptor, but is additionally a property of the ADAM ligands. ADAM9 is recognized by α6β1 and αvβ5 [18,22] and ADAM15 binds α9β1, αvβ3 or α5β1 [21,23,24]. A recent study suggests that multiple integrins expressed on the same cell facilitate cell adhesion to a single ADAM disintegrin domain [19]. This complex pattern of integrin-ADAM recognition is reminiscent of the well-established integrin–extracellular matrix and integrin–cellular counter receptor interactions.
Members of the integrin family can be segregated into five groups based on the sequence homology of α subunits and the similarity of ligand specificities [25]. The smallest grouping is comprised of the α4 and α9 integrins: α4β1, α4β7 and α9β1. These two integrin subunits are unique among all integrin α subunits in that they lack inserted A-domains and are not post-translationally processed into heavy and light chains [26]. The integrins α4β1, α4β7 and α9β1 share similar ligand specificity and functionality. The integrin α4β1 functions in lymphocyte infiltration of tissue through VCAM-1 (vascular cell adhesion molecule-1) [27]. The integrin α4β7 interacts with MAdCAM-1 (mucosal addressin adhesion molecule-1) for proper homing of lymphocytes to lymphoid structures such as Peyer's patches to establish and maintain gut-associated immunity [28–30]. The integrin α9β1 has been demonstrated to be expressed by neutrophils and to mediate adhesion events that facilitate transendothelial migration [31,32].
As the biological role of the ADAMs becomes more evident, distinct functional classes of these ADAMs are recognizable. Two dominant classes are proteins involved in fertilization, which include ADAM1, ADAM2 and ADAM3 [33,34], and ADAM10, ADAM17, ADAM19 (MADDAM), ADAM28 and ADAM33 that have been implicated in immune functions. ADAM10 processes the chemokine fractalkine, which promotes monocytic chemotaxis [35]; ADAM17 cleaves tumour necrosis factor-α and L-selectin [36–38]; MADDAM is crucial for dendritic cell differentiation [39]; ADAM28 is localized to mature B-cells (R. D. Bowditch, unpublished work), cleaves a CD23 peptide [40] and binds α4β1 [20]; and ADAM33 has been genetically linked to asthma [41].
In the present study, we explore the possibility that the integrins α4β1, α4β7 and α9β1 recognize multiple mammalian disintegrin domains. We assessed the adhesive properties of the newly identified ADAM7 due to its homology and chromosomal proximity to ADAM28. We also evaluated the potential integrin recognition of ADAM33. On the basis of the current knowledge of these ADAM disintegrin domains and their respective integrin ligand properties established in this study, we propose that these ADAMs are functional ligands for α4 and α9 subclasses of the integrin family.
EXPERIMENTAL
Materials
The Jurkat cell line was obtained from the A.T.C.C. (Manassas, VA, U.S.A.). The RPMI 8866 cell line was a gift from Dr J. C. Wilkins (University of Manitoba, Manitoba, Canada). Both lymphoma cell lines were maintained in RPMI 1640, containing 10 mM Hepes, 1 mM sodium pyruvate, 10% (v/v) fetal calf serum, 1% L-glutamine and 1% penicillin–streptomycin. The parental K562 cell line was purchased from the A.T.C.C., and the α4-K562 cell line was a gift from Dr R. Isberg (Tufts University School of Medicine, Boston, MA, U.S.A.). The α9-transfected CHO cells (Chinese-hamster ovary cells) were described previously [32]. The anti-integrin mAbs (monoclonal antibodies) P1H4 (anti-α4), 44H6 (anti-α4), 6S6 (anti-β1), YFC118.3 (anti-β2) and Y9A2 (anti-α9β1) were purchased from Chemicon International (Temecula, CA, U.S.A.). The anti-β7 mAb FIB 27 was obtained from BD PharMingen (San Diego, CA, U.S.A.). The anti-β1 mAb AIIB2 was kindly provided by Dr C. Damsky (University of California, San Francisco). The anti-β1-activating mAb QE2E5 [42] was affinity-purified from supernatants harvested from a hybridoma cell line provided by Dr R. Faull (Royal Adelaide Hospital, Adelaide, South Australia, Australia). The anti-β7-activating mAb 2G3 [43] and recombinant soluble MAdCAM–Ig were generously supplied by Dr T. Yednock (Elan Pharmaceuticals, San Francisco, CA, U.S.A.). The CS-1 peptide (EILDVPST) was purchased from the American Peptide Company (Sunnyvale, CA, U.S.A.).
Recombinant disintegrin fusion proteins
Methods for the generation of the ADAM28 Dis–Fc (recombinant disintegrin–Fc fusion protein) construct were described previously [15,20]. DNA fragments of ADAM7 (1121–1784 bp) and ADAM33 (1202–1615 bp) containing the full-length disintegrin domain coding regions were obtained by nested PCR of a human cDNA lymph node and liver library (ClonTech, Palo Alto, CA, U.S.A.) respectively. By performing PCR overlap extension reactions, the full-length sequence encoding the disintegrin domains of ADAM7 (residues Tyr397-Gly497) and ADAM33 (residues Ala418-Gly510) was annealed in frame with a 5′-GP67 secretion signal sequence and a 3′ human IgG3 Fc-fusion tag. The human IgG3 Fc heavy chain lacked the hinge region to eliminate Fc-mediated dimerization of the recombinant proteins. PCR products were subcloned into the TOPO pIB/V5-His vector (Invitrogen, Carlsbad, CA, U.S.A.). ADAM12 Dis–Fc was generated as discussed for ADAM7 and ADAM33 by using a nested PCR approach on a human cDNA liver library. The recombinant protein contained the entire ADAM12 disintegrin domain spanning residues Asn414-Gly519. Charge-to-alanine substitution mutants were produced with the Quik Change site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.). Synthetic oligonucleotides were obtained from the Molecular Biology Resource Facility at The University of Oklahoma Health Science Center or Sigma-Genosys (Woodlands, TX, U.S.A.). After sequence verification, recombinant Dis–Fc proteins were produced by stable transfection of High 5 cells with CellFectin (Invitrogen). Conditioned media were harvested, poly(ethylene glycol)-concentrated and applied on to a Protein G–Sepharose column (Amersham Biosciences, Uppsala, Sweden). Recombinant protein was eluted by the addition of 100 mM citric acid (pH 3.0), and column fractions were immediately neutralized with 1 M Tris/HCl (pH 9.0). All recombinant disintegrin proteins used in the present study migrated as a single band when evaluated by SDS/PAGE under reduced and non-reduced conditions, suggesting that the purified recombinant disintegrin domains did not possess any gross structural perturbations resulting in aberrant disulphide bond formation.
Cell-adhesion assay
The assay utilized was modified from that previously described for demonstrating cell adhesion to immobilized fibronectin [44]. For Jurkat and RPMI 8866 cell-adhesion experiments, recombinant disintegrin domains were immobilized on Immulon-2 96-well plates (Dynatech Laboratories, Chantilly, VA, U.S.A.) by incubating desired concentrations of recombinant protein in 0.1 M NaHCO3 (pH 8.4) overnight at 4 °C in a total volume of 100 μl. Wells were subsequently blocked with 2% (w/v) BSA in 0.1 M NaHCO3 (pH 8.4) at room temperature (22 °C) for 1 h. Cells were washed three times with Hepes-Tyrode's buffer (5 mM Hepes, 150 mM NaCl, 12 mM NaHCO3, 2.6 mM KCl, 5 mM D-glucose, 0.2 mg/ml BSA, 0.5 mM MgCl2 and 1 mM CaCl2). Where indicated, the buffer additionally contained 1 mM MnCl2. After washes, cells were adjusted to 2×106 cells/ml in Hepes-Tyrode's containing 10 μg/ml of the appropriate activating mAb: QE2E5 (anti-β1) or 2G3 (anti-β7). CHO cell-adhesion assays were as described for Jurkat cells except that cells were resuspended to 1×106 cells/ml in Hepes-Tyrode's buffer that lacked exogenous integrin activators. Aliquots of 100 μl of the cell suspension were incubated with immobilized substrates for 1 h at 37 °C in an atmosphere of 5% CO2. For inhibition experiments, inhibitors were suspended in Hepes-Tyrode's buffer at twice the final desired concentration (50 μl/well) and subsequently diluted with cells (50 μl/well) at double the final concentration. Non-adherent cells were displaced with three consecutive washes of Hepes-Tyrode's buffer. Adherent cells were disclosed by colorimetric assessment of endogenous acid phosphatase activity by the addition of 100 μl/well of 1% Triton X-100, 50 mM sodium acetate (pH 5.0) and 6 mg/ml p-nitrophenyl phosphatase and incubating at 37 °C for 2 h. After the addition of 50 μl/well of 1 M NaOH, absorbance at 405 nm was determined. Calculation of the number of adherent cells/well was derived from a standard curve of absorbance values generated from a known number of cells.
Flow cytometry
RPMI 8866 cells were washed twice with PBS containing 0.1% BSA. Cells (5×105) were incubated in a total volume of 200 μl with the appropriate primary anti-integrin antibody (10 μg/ml) for 30 min at 4 °C. The cells were washed twice with PBS/BSA and incubated with either 10 μg/ml goat anti-mouse IgG-specific FITC-conjugated antibody (Caltag Laboratories, Burlingame, CA, U.S.A.) or with goat anti-rat IgG-specific FITC-conjugated antibody (Chemicon International, Temecula, CA, U.S.A.). The cells were incubated with the secondary antibody for 30 min at 4 °C. Cells were washed twice with PBS/BSA and immediately analysed on an FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA, U.S.A.).
RESULTS
The disintegrin domain of ADAM7 is an α4β1 ligand
We have established previously that the disintegrin domain of ADAM28, a disintegrin expressed by mature B-lymphocytes (R. D. Bowditch, unpublished work), is recognized by the integrin α4β1 [20]. In the present study, we examined the potential of the homologous ADAM7 and ADAM33 disintegrin domains to facilitate α4β1-dependent cell adhesion (Figure 1a).
Figure 1. The disintegrin domain of ADAM7 facilitates integrin α4β1-dependent cell adhesion.
(a) Alignment of the primary amino acid sequences of the disintegrin domains of human ADAM7, ADAM28 and ADAM33. Identities of ADAM28 with ADAM7 and ADAM33 disintegrin domains are shaded, with the respective homology of ADAM7 and ADAM33 to ADAM28 displayed at the far left. The predicted integrin α9β1 recognition motif is indicated by thick underlining and contains the putative disintegrin loops. Arrows indicate charged residues selected for alanine substitution in the ADAM7 and ADAM28 disintegrin domains. (b) Microtitre wells were coated with the indicated concentrations of ADAM7 (■), ADAM28 (○) or ADAM33 Dis–Fc (▲) recombinant proteins. ADAM7 and ADAM33 Dis–Fc contained the complete disintegrin domains Tyr397-Gly497 and Ala418-Gly510 respectively and ADAM28 Dis–Fc is as described previously [15,20]. The T-lymphoma cell line Jurkat (2×105 cells/well) was added to wells in Hepes/Tyrode's buffer containing 1 mM CaCl2, 0.5 mM MgCl2 and 10 μg/ml QE2E5 mAb for exogenous activation of the β1 integrins. The number of adherent cells per well was quantified based on a standard curve generated by assessing the endogenous acid phosphatase activity as described in the Experimental section. Adherent cells/well=adherent cells(recombinant protein)−adherent cells(BSA). Results shown are the means±S.D. for a representative experiment performed in triplicate. (c) Microtitre wells were coated with 10 μg/ml of purified, recombinant ADAM28 (white bars) or ADAM7 Dis–Fc (black bars). Jurkat cells were added to wells as described previously in a buffer containing 1 mM MnCl2. Cells were incubated with the recombinant ligands in the presence of 5 μg/ml of various integrin subunit mAbs or with 100 μg/ml of the α4β1 ligand mimetic CS-1 peptide. Results are expressed as the average percentage inhibition±S.D. for a representative experiment performed in triplicate. Percentage inhibition={1−[adherent cells(inhibitor)−adherent cells(BSA)]/[adherent cells(no inhibitor)−adherent cells(BSA)]}×100. (d) α4-K562 (●) or K562 (○) cells were added to microtitre wells (2×105 cells/well) coated with various concentrations of ADAM7 Dis–Fc in the presence of 10 μg/ml QE2E5. Adherent cells/well=adherent cells(recombinant protein)−adherent cells(BSA). Results are expressed as the mean adhesion±S.D. for a representative experiment performed in triplicate.
The recombinant disintegrin domains of ADAM7 (ADAM7 Dis–Fc) and ADAM33 (ADAM33 Dis–Fc) were initially tested for their ability to mediate Jurkat cell adhesion (Figure 1b). As reported previously, the disintegrin domain of ADAM28 facilitated Jurkat cell adhesion in a dose-dependent and saturable manner when isolated (ADAM28 Dis–Fc; Figure 1b) or in the context of other domains (Dis-Cys-EGF-Fc) [20]. A similar but more apparent trend was observed with the ADAM7 Dis–Fc (Figure 1b). Whereas wells coated with 5 μg/ml of the ADAM28 disintegrin domain exhibited a maximal 47.5±3.7% adhesion of the total input cells, the same concentration of recombinant ADAM7 disintegrin resulted in 88.5±3.0% adhesion. Considerably lower concentrations of the disintegrin domain of ADAM7 were required to generate measurable levels of Jurkat cell adhesion when compared with ADAM28. As shown in Figure 1(b), no detectable cell adhesion was attained with ADAM33 Dis–Fc at the highest concentration tested (5 μg/ml).
To establish that Jurkat cell adhesion to the ADAM7 Dis–Fc was α4β1-dependent, function-blocking anti-human mAbs to selected α and β integrin subunits were used as inhibitors in the adhesion assay. Similar to the recombinant ADAM28 disintegrin domain, Jurkat cell adhesion to the ADAM7 Dis–Fc was inhibited by the anti-α4 and anti-β1 function-blocking mAbs, indicating that the observed adhesion required α4β1 (Figure 1c). The non-inhibitory anti-α4 (mAb), 44H6, did not substantially block Jurkat cell adhesion to ADAM7 Dis–Fc. Integrin α4β1 dependence was supported by the potent inhibition of Jurkat cell adhesion to ADAM7 Dis–Fc by the fibronectin CS-1-derived peptide (EILDVPST; Figure 1c). Stable transfection of the human α4-subunit cDNA into the K562 erythroleukaemic cell line conferred the ability to adhere to ADAM7 Dis–Fc, establishing that adhesion was directly dependent on the presence of the α4β1 integrin (Figure 1d). The α4-K562 cells exhibited similar adhesion trends as the Jurkat cell line when the adhesion levels of ADAM28 and ADAM7 Dis–Fc were compared (results not shown).
The disintegrin domains of ADAM7 and ADAM28 support α4β7-dependent cell adhesion
Since the integrins α4β7 and α4β1 exhibit similar ligand recognition specificities and share a common α-subunit, recognition of the ADAM28, ADAM7 and ADAM33 disintegrin domains by α4β7 was investigated. For these experiments, the B-cell-derived RPMI 8866 cell line was utilized. This cell line is classified as an α4 high, β7 high, β1 low expressing cell line, which was verified by flow cytometry (Figure 2a).
Figure 2. The disintegrin domains of ADAM28 and ADAM7 are recognized by the immunological integrin α4β7.
(a) Flow-cytometric analysis of integrin subunit expression on RPMI 8866 cells. RPMI 8866 cells were incubated with the primary mAbs 44H6 (anti-α4), Y9A2 (anti-α9β1), 6S6 (anti-β1), YCFII8.3 (anti-β2) and FIB27 (anti-β7). After washing, the bound primary antibody was disclosed with appropriate secondary FITC-conjugated antibody (unfilled curve) as described in the Experimental section. Experiments containing only secondary conjugated antibody served as negative controls (filled curve). (b) Adhesion assays were performed as described previously by adding RPMI 8866 cells (2×105 cells/well) to microtitre wells coated with various concentrations of purified, recombinant ADAM7 (■), ADAM28 (○) or ADAM33 Dis–Fc (▲). Cells were suspended in Hepes-Tyrode's buffer containing 1 mM CaCl2, 0.5 mM MgCl2 and 10 μg/ml of the anti-β7 activating mAb 2G3. Adherent cells/well=adherent cells(recombinant protein)−adherent cells(BSA). Results shown are the average±S.D. for a representative experiment performed in triplicate. (c) RPMI 8866 cells (2×105 cells/well) suspended in a buffer containing 1 mM MnCl2 were added to microtitre wells coated with 10 μg/ml of the indicated recombinant ADAM28 (white bars) or ADAM7 Dis–Fc (black bars) in the presence of 5 μg/ml of either function-blocking or non-function-blocking mAbs that recognize distinct integrin subunits. Results are expressed as the average percentage inhibition±S.D. for a representative experiment performed in triplicate. Percentage inhibition={1−[adherent cells(inhibitor)−adherent cells(BSA)]/[adherent cells(no inhibitor)−adherent cells(BSA)]}×100.
The RPMI 8866 cells attached in a dose-dependent and saturable manner to both the ADAM7 and ADAM28 Dis–Fc, whereas no significant adhesion was observed with ADAM33 Dis–Fc (Figure 2b). Integrin specificity was demonstrated by using function-blocking mAbs to the integrin subunits that would potentially be involved in this recognition (Figure 2c). Only function-blocking mAbs to the α4 and β7 integrin subunits significantly decreased RPMI 8866 cell adhesion to ADAM28 and ADAM7 Dis–Fc. Function-blocking mAbs to the β1 and β2 integrin subunits and the non-function-blocking anti-α4 mAb 44H6 had no significant inhibitory effects. Furthermore, the α4β7 recombinant ligand MAdCAM–Ig inhibited the adhesion of RPMI 8866 cells to both ADAM28 and ADAM7 Dis–Fc (Figure 3b). In contrast with the results obtained with Jurkat cells, ADAM28 Dis–Fc showed significantly higher levels of adhesion with RPMI 8866 cells when compared with the ADAM7 Dis–Fc, suggesting that the ADAM28 disintegrin domain has a higher avidity for α4β7 than the ADAM7 disintegrin domain.
Figure 3. Recognition of the disintegrin domains of ADAM7 and ADAM28 by α4β1 and α4β7 integrins is activation-dependent.
(a) Jurkat cells (2×105/well) suspended in Hepes-Tyrode's buffer containing either 1 mM MnCl2, 10 μg/ml of the anti-β1-activating antibody QE2E5 or only 1 mM CaCl2 and 0.5 mM MgCl2 were added to wells coated with 10 μg/ml each of ADAM7 (black bars) or ADAM28 Dis–Fc (white bars). (b) RPMI 8866 cell adhesion to 10 μg/ml each of ADAM7 (black bars) or ADAM28 (white bars) Dis–Fc or 2.5 μg/ml MAdCAM-Ig (hatched bars) was investigated in Hepes-Tyrode's buffer containing 1 mM MnCl2, 10 μg/ml of the anti-β7-activating antibody 2G3 or only 1 mM CaCl2 and 0.5 mM MgCl2. α4β7 specificity was demonstrated for ADAM7 and ADAM28 by including 10 μg/ml MAdCAM–Ig in solution with the 2G3-activating antibody. Results shown in (a, b) are the average±S.D. for a representative experiment conducted in triplicate. Adherent cells/well=adherent cells(recombinant protein)−adherent cells(BSA).
α4β1 and α4β7-dependent adhesion to ADAM7 and ADAM28 disintegrin domains requires integrin activation
In adhesion experiments with the two non-adherent cell lines described above, the activating mAbs QE2E5 (anti-β1) or 2G3 (anti-β7) were used. We have established previously that α4β1 recognition of the disintegrin domain of ADAM28 depends on integrin activation. Similarly, Jurkat cell adhesion to the disintegrin domain of ADAM7 required either the presence of the mAb QE2E5 or 1 mM MnCl2 (Figure 3a). It was also demonstrated that α4β7-mediated RPMI 8866 cell adhesion to the disintegrin domains of ADAM7 or ADAM28 required exogenous integrin activation (Figure 3b). This was distinct from the results obtained with immobilized MAdCAM-Ig.
Homologous disintegrin domains utilize distinct but overlapping residues for integrin recognition
Six different charge-to-alanine disintegrin-loop mutants of ADAM7 and ADAM28 Dis–Fc were generated for addressing integrin recognition mechanisms utilized by homologous mammalian disintegrin domains (Figure 1a). In the present study, the mutagenesis was focused within the putative disintegrin loop, a region previously implicated in adhesion to several mammalian disintegrins, which also encompasses the predicted α9β1 integrin recognition motif R(X6)DLPEF [12].
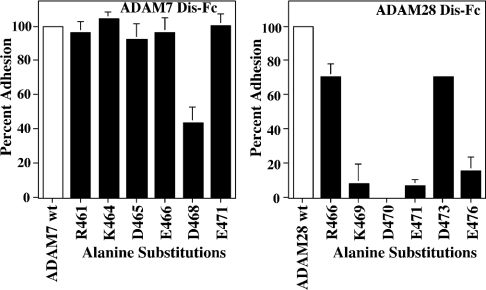
Six disintegrin-loop mutants of ADAM7 Dis–Fc were evaluated for their ability to support α4β1-dependent cell adhesion when compared with the native domain (Figure 4a). Only the conserved Arg461/Arg466 and Lys464/Lys469 residues of ADAM7 and ADAM28 respectively exhibited similar effects when alanine substitution mutants were compared. Even though Asp465, Glu466 and Glu471 of ADAM7 Dis–Fc were not critical for Jurkat cell adhesion, the homologous residues were required for maintaining the α4β1 ligand properties of the ADAM28 disintegrin domain [15]. As with many other mammalian disintegrins, the Asp468 of ADAM7 was crucial in facilitating integrin-dependent cell adhesion [12,13].
Figure 4. Residues required for integrin α4β1- and α4β7-mediated cell adhesion to two homologous disintegrin domains.
(a) Microtitre wells were coated with 5 μg/ml of either native (white bar) or the indicated charge-to-alanine mutant of (black bars) ADAM7 or ADAM28 Dis–Fc. Jurkat cells were added as described previously in the presence of 10 μg/ml QE2E5. Adhesion assays were performed as described in the Experimental section, and results shown are the average±S.D. for three independent experiments each performed in triplicate. Percentage adhesion=[adherent cells(native or mutant rDis–Fc)−adherent cells(BSA)]/[adherent cells(native rDis–Fc)−adherent cells(BSA)]×100. (b) RPMI 8866 cell adhesion to 5 μg/ml of native ADAM7 or ADAM28 Dis–Fc (white bar) or charge-to-alanine mutants of ADAM7 or ADAM28 Dis–Fc (black bars) was evaluated in the presence of 10 μg/ml of 2G3. Results shown are the average±S.D. for three independent experiments each performed in triplicate. Percentage adhesion=[adherent cells(native or mutant rDis–Fc)−adherent cells(BSA)]/[adherent cells(native rDis–Fc)−adherent cells(BSA)]×100.
A similar trend was evident when the ADAM7 and ADAM28 Dis–Fc charge-to-alanine mutants were tested for their relative ability to mediate α4β7-dependent cell adhesion (Figure 4b). Similar to the α4β1 recognition of ADAM28 [15], alanine substitution of residues Lys469, Asp470, Glu471 and Glu476 significantly decreased α4β7-dependent cell adhesion to ADAM28 Dis–Fc. However, alanine substitution of the homologous residues in ADAM7 did not affect α4β7-dependent cell adhesion. As observed for α4β1-dependent adhesion, the Asp468 in ADAM7 Dis–Fc negatively affected α4β7-dependent adhesion (Figure 4a). Interestingly, the homologous Asp473–Ala substitution within ADAM28 Dis–Fc had no effect on either α4β1 or α4β7 interaction.
The disintegrin domains of ADAM7, ADAM28 and ADAM33 are recognized by the integrin α9β1
It has been hypothesized that α9β1 integrin is a receptor for prototypical ADAMs [12]. Transfected CHO cells expressing human α9 subunit adhered to recombinant ADAM28, ADAM7 and ADAM33 Dis–Fc proteins, whereas mock-transfected CHO cells did not (Figure 5). Adhesion of CHO cells to α9 was also inhibited with Y9A2, a function-blocking anti-α9β1 mAb (Figure 5). Results resembled levels attained with ADAM12 Dis–Fc, a disintegrin that has been previously identified as an α9β1 ligand [21].
Figure 5. The integrin α9β1 functions as a receptor for multiple mammalian disintegrin domains.
Adhesion of CHO cells (1×105 cells/well) expressing the human α9 integrin subunit (■) or mock-transfected cells (○) to various recombinant mammalian disintegrin domains was examined in a concentration-dependent manner. The α9β1 function-blocking mAb Y9A2 was included at 5 μg/ml at the highest ligand concentration (▲). Adhesion assays were performed as described previously in Hepes-Tyrode's buffer containing 1 mM CaCl2 and 0.5 mM MgCl2 with no exogenous integrin activators. ADAM12 Dis–Fc possessed the entire ADAM12 disintegrin domain spanning residues Asn414–Gly519. Results shown are the average±S.D. for a representative experiment performed in triplicate.
To define further the molecular mechanism involved in integrin recognition of mammalian disintegrins, we tested our ADAM7 and ADAM28 alanine substitution mutants for the ability to support α9β1-mediated cell adhesion (Figure 6). As with α4β1 and α4β7 recognition of ADAM28, substitution of Lys469, Asp470, Glu471 and Glu476 with alanine resulted in decreased cell adhesion when compared with the native sequence. Only replacement of Asp468 with alanine within ADAM7 Dis–Fc elicited an adhesive defect.
Figure 6. Mutagenesis analysis of ADAM7 and ADAM28 disintegrin domains as ligands for the integrin α9β1.
Microtitre wells were coated with 10 μg/ml of native ADAM7 or ADAM28 Dis–Fc (white bars) or an appropriate charge-to-alanine mutant (black bars). α9-CHO cells were added (1×105 cells/well) to wells and the extent of adhesion was determined as previously mentioned. Results shown are the average±S.D. for three independent experiments each performed in triplicate. Percentage adhesion=[adherent cells(native or mutant rDis–Fc)−adherent cells(BSA)]/[adherent cells(native rDis–Fc)−adherent cells(BSA)]×100.
DISCUSSION
Results of the present study established that the homologous disintegrin-like domains of human ADAM28 and ADAM7 are recognized by the leucocyte integrins α4β1, α4β7 and α9β1. As a model system for addressing the integrin ligand properties of ADAMs, disintegrin domains were recombinantly produced as Fc-fusion (Dis–Fc) proteins. This approach was based on previous studies demonstrating that the adhesive properties of isolated ADAM disintegrin domains are the same as domains expressed in the context of additional ecto-domains [18,20,21,23,24,45]. Cell adhesion to ADAM28 or ADAM7 Dis–Fc required exogenous activation of the receptor by inclusion of MnCl2 or an activating mAb. Charge-to-alanine mutagenesis studies demonstrated that the same residues within specific ADAM disintegrin domains are used for the recognition of multiple integrins. The residues required for integrin binding, at least within the putative disintegrin loop, of each ADAM disintegrin domain differ. The disintegrin domain of human ADAM33 differs from ADAM7 and ADAM28 in that it is recognized by α9β1, but not by α4β1 or α4β7.
It is apparent that particular ADAMs may be recognized by multiple integrins. Multi-receptor binding is common among integrin ligands [46]. Here, we have demonstrated that both the disintegrin domains of ADAM28 and ADAM7 are recognized by the lymphocyte integrins α4β1 and α4β7. In contrast with α4β1, the ADAM28 disintegrin domain consistently generated a higher level of α4β7-dependent cell adhesion when compared with the ADAM7 disintegrin domain, suggesting an integrin preference for these ADAMs. This is the first demonstration that mammalian disintegrins are capable of interacting with α4β7. Only two other non-RGD-containing ADAMs, ADAM9 and ADAM23, have been reported to interact with non-very-late-antigen integrin family members [22,45]. Taken together, these findings suggest a broader and more diverse involvement of the integrin superfamily in ADAM's function. EC3, a heterodimeric disintegrin derived from the venom of Echis carinatus, is the only snake venom disintegrin shown to be a potent inhibitor of α4β7 function [47]. However, there is no significant homology between EC3 and the disintegrin-like domains of ADAM28 or ADAM7 that would indicate a conserved mechanism for generating α4β7 recognition. Interestingly, the disintegrin-like domain of human ADAM33 contains an MGD tripeptide sequence that is reminiscent of the MLD motif found in the snake venom disintegrins EC3, VLO5 and EO5 which is required for the recognition of integrins α4β1 and α9β1 [48]. However, ADAM33 was found to be not recognized by the integrin α4β1. It should also be noted that the MGD sequence in human ADAM33 is not conserved in the mouse homologue of ADAM33.
It has been postulated recently that the integrin α9β1 interacts with most prototypical ADAMs, a characteristic attributed to a highly conserved disintegrin loop motif [12]. This prediction is supported by current evidence that the disintegrin domains of ADAM2 and ADAM3 interact with α9β1 [19]. We have expanded these results by demonstrating that ADAM7, ADAM28 and ADAM33 Dis–Fc are also recognized by α9β1. Even though the putative α9β1 recognition motif is common to all ADAMs recognized by α9β1, our evidence suggests that each ADAM exhibits distinct integrin ligand properties. As demonstrated in the present study, some ADAMs exhibit both α4β1 and α9β1 ligand properties, and others such as ADAM33 are recognized exclusively by α9β1.
The integrin binding surfaces of the homologous disintegrins of ADAM7 and ADAM28 are overlapping but distinct. Although similar regions of the domain interact with the integrin, the residues responsible within this region were unique for each ADAM. Even though the level of homology is not as extensive as with ADAM7 and ADAM28, similar results have been obtained with mutagenesis studies of the functionally linked ADAM2 and ADAM3 disintegrin domains [13,14,49]. When evaluating the binding properties of a particular ADAM, the same residues comprised the molecular contacts for multiple integrins, suggesting a highly conserved binding interface specific for each ADAM. Even though both α4 ligands require a conserved aspartic residue positioned in an extended CD loop for integrin binding, slight structural differences between VCAM and MAdCAM account for the unique binding activities of each ligand [50,51]. Perhaps, similar differences exist in the architecture of ADAM7 and ADAM28, accounting for the distinct avidities for α4β1 versus α4β7 and differences in residue requirement for integrin interaction.
A common theme evident from the present study is that all the integrins involved have proven roles in the migration of leucocytes from the vasculature to sites in the periphery. In the light of our current model that proposes that integrins may regulate ectodomain shedding by binding ADAMs [2], it is interesting to speculate about the functional consequences of the interaction of this group of ADAMs with the α4/α9 subset of the integrin family. The mature B-cell expression pattern of ADAM28 and the expression of α4β7 on a defined population of circulating memory B-cells [52] suggests that, in certain physiological settings, ADAM28 and α4β7 would be expressed on the same subset of lymphocytes. Association of ADAM28 through the disintegrin domain with α4β7 may target or sequester the sheddase, thus altering the repertoire of lymphocyte surface adhesion molecules or cytokines affecting lymphocyte activity.
The present study establishes that the disintegrin domains of ADAM28 and ADAM7 are ligands for a functionally related subset of integrins composed of α4β1, α4β7 and α9β1. By analysing six different charge-to-alanine disintegrin-loop mutants of these newly identified integrin ligands, we present evidence that suggests that the recognition of the same mammalian disintegrin by multiple integrins utilizes a highly conserved integrin recognition interface. Even though the residues that constitute the molecular contacts of various ADAMs are distinct, it appears that the integrin interface on each individual domain is spatially overlapping. Additionally, we have demonstrated that the disintegrin domain of ADAM33 is capable of mediating α9β1-dependent cell adhesion.
Acknowledgments
This work was supported by NIH grant no. AI47314.
References
- 1.White J. M. ADAMs: modulators of cell–cell and cell–matrix interactions. Curr. Opin. Cell Biol. 2003;15:598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Bridges L. C., Bowditch R. D. ADAM–integrin interactions: potential integrin regulated ectodomain shedding activity. Curr. Pharm. Des. 2005 doi: 10.2174/1381612053381747. in the press. [DOI] [PubMed] [Google Scholar]
- 3.Seals D. F., Courtneidge S. A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 4.Moss M. L., Lambert M. H. Shedding of membrane proteins by ADAM family proteases. Essays Biochem. 2002;38:141–153. doi: 10.1042/bse0380141. [DOI] [PubMed] [Google Scholar]
- 5.Schlondorff J., Blobel C. P. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J. Cell Sci. 1999;112:3603–3617. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- 6.Kamiguti A. S., Zuzel M., Theakston R. D. Snake venom metalloproteinases and disintegrins: interactions with cells. Braz. J. Med. Biol. Res. 1998;31:853–862. doi: 10.1590/s0100-879x1998000700001. [DOI] [PubMed] [Google Scholar]
- 7.Huang T. F. What have snakes taught us about integrins? Cell. Mol. Life Sci. 1998;54:527–540. doi: 10.1007/s000180050181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkinson R. A., Saudek V., Pelton J. T. Echistatin: the refined structure of a disintegrin in solution by 1H NMR and restrained molecular dynamics. Int. J. Pept. Protein Res. 1994;43:563–572. doi: 10.1111/j.1399-3011.1994.tb00558.x. [DOI] [PubMed] [Google Scholar]
- 9.Marcinkiewicz C., Vijay-Kumar S., McLane M. A., Niewiarowski S. Significance of RGD loop and C-terminal domain of echistatin for recognition of alphaIIb beta3 and alpha(v) beta3 integrins and expression of ligand-induced binding site. Blood. 1997;90:1565–1575. [PubMed] [Google Scholar]
- 10.Adler M., Lazarus R. A., Dennis M. S., Wagner G. Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist. Science. 1991;253:445–448. doi: 10.1126/science.1862345. [DOI] [PubMed] [Google Scholar]
- 11.Yuan R., Primakoff P., Myles D. G. A role for the disintegrin domain of cyritestin, a sperm surface protein belonging to the ADAM family, in mouse sperm-egg plasma membrane adhesion and fusion. J. Cell Biol. 1997;137:105–112. doi: 10.1083/jcb.137.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eto K., Huet C., Tarui T., Kupriyanov S., Liu H. Z., Puzon-McLaughlin W., Zhang X. P., Sheppard D., Engvall E., Takada Y. Functional classification of ADAMs based on a conserved motif for binding to integrin alpha 9beta1: implications for sperm-egg binding and other cell interactions. J. Biol. Chem. 2002;277:17804–17810. doi: 10.1074/jbc.M200086200. [DOI] [PubMed] [Google Scholar]
- 13.Bigler D., Takahashi Y., Chen M. S., Almeida E. A., Osbourne L., White J. M. Sequence-specific interaction between the disintegrin domain of mouse ADAM 2 (fertilin beta) and murine eggs. Role of the alpha(6) integrin subunit. J. Biol. Chem. 2000;275:11576–11584. doi: 10.1074/jbc.275.16.11576. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi Y., Bigler D., Ito Y., White J. M. Sequence-specific interaction between the disintegrin domain of mouse ADAM 3 and murine eggs: role of beta1 integrin-associated proteins CD9, CD81, and CD98. Mol. Biol. Cell. 2001;12:809–820. doi: 10.1091/mbc.12.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridges L. C., Hanson K. R., Tani P. H., Mather T., Bowditch R. D. Integrin alpha4beta1-dependent adhesion to ADAM 28 (MDC-L) requires an extended surface of the disintegrin domain. Biochemistry. 2003;42:3734–3741. doi: 10.1021/bi026871y. [DOI] [PubMed] [Google Scholar]
- 16.Hynes R. O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell (Cambridge, Mass.) 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 17.Almeida E. A., Huovila A. P., Sutherland A. E., Stephens L. E., Calarco P. G., Shaw L. M., Mercurio A. M., Sonnenberg A., Primakoff P., Myles D. G., et al. Mouse egg integrin alpha 6 beta 1 functions as a sperm receptor. Cell (Cambridge, Mass.) 1995;81:1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]
- 18.Nath D., Slocombe P. M., Webster A., Stephens P. E., Docherty A. J., Murphy G. Meltrin gamma(ADAM-9) mediates cellular adhesion through alpha(6)beta(1)integrin, leading to a marked induction of fibroblast cell motility. J. Cell Sci. 2000;113:2319–2328. doi: 10.1242/jcs.113.12.2319. [DOI] [PubMed] [Google Scholar]
- 19.Tomczuk M., Takahashi Y., Huang J., Murase S., Mistretta M., Klaffky E., Sutherland A., Bolling L., Coonrod S., Marcinkiewicz C., et al. Role of multiple beta1 integrins in cell adhesion to the disintegrin domains of ADAMs 2 and 3. Exp. Cell Res. 2003;290:68–81. doi: 10.1016/s0014-4827(03)00307-0. [DOI] [PubMed] [Google Scholar]
- 20.Bridges L. C., Tani P. H., Hanson K. R., Roberts C. M., Judkins M. B., Bowditch R. D. The lymphocyte metalloprotease MDC-L (ADAM 28) is a ligand for the integrin alpha4beta1. J. Biol. Chem. 2002;277:3784–3792. doi: 10.1074/jbc.M109538200. [DOI] [PubMed] [Google Scholar]
- 21.Eto K., Puzon-McLaughlin W., Sheppard D., Sehara-Fujisawa A., Zhang X. P., Takada Y. RGD-independent binding of integrin alpha9beta1 to the ADAM-12 and -15 disintegrin domains mediates cell-cell interaction. J. Biol. Chem. 2000;275:34922–34930. doi: 10.1074/jbc.M001953200. [DOI] [PubMed] [Google Scholar]
- 22.Zhou M., Graham R., Russell G., Croucher P. I. MDC-9 (ADAM-9/Meltrin gamma) functions as an adhesion molecule by binding the alpha(v)beta(5) integrin. Biochem. Biophys. Res. Commun. 2001;280:574–580. doi: 10.1006/bbrc.2000.4155. [DOI] [PubMed] [Google Scholar]
- 23.Nath D., Slocombe P. M., Stephens P. E., Warn A., Hutchinson G. R., Yamada K. M., Docherty A. J., Murphy G. Interaction of metargidin (ADAM-15) with alphavbeta3 and alpha5beta1 integrins on different haemopoietic cells. J. Cell Sci. 1999;112:579–587. doi: 10.1242/jcs.112.4.579. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X. P., Kamata T., Yokoyama K., Puzon-McLaughlin W., Takada Y. Specific interaction of the recombinant disintegrin-like domain of MDC-15 (metargidin, ADAM-15) with integrin alphavbeta3. J. Biol. Chem. 1998;273:7345–7350. doi: 10.1074/jbc.273.13.7345. [DOI] [PubMed] [Google Scholar]
- 25.Hynes R. O. Integrins: bidirectional, allosteric signaling machines. Cell (Cambridge, Mass.) 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 26.Palmer E. L., Ruegg C., Ferrando R., Pytela R., Sheppard D. Sequence and tissue distribution of the integrin alpha 9 subunit, a novel partner of beta 1 that is widely distributed in epithelia and muscle. J. Cell Biol. 1993;123:1289–1297. doi: 10.1083/jcb.123.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Springer T. A. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell (Cambridge, Mass.) 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 28.Berlin C., Berg E. L., Briskin M. J., Andrew D. P., Kilshaw P. J., Holzmann B., Weissman I. L., Hamann A., Butcher E. C. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell (Cambridge, Mass.) 1993;74:185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 29.Rott L. S., Briskin M. J., Andrew D. P., Berg E. L., Butcher E. C. A fundamental subdivision of circulating lymphocytes defined by adhesion to mucosal addressin cell adhesion molecule-1. Comparison with vascular cell adhesion molecule-1 and correlation with beta 7 integrins and memory differentiation. J. Immunol. 1996;156:3727–3736. [PubMed] [Google Scholar]
- 30.Mebius R. E., Streeter P. R., Michie S., Butcher E. C., Weissman I. L. A developmental switch in lymphocyte homing receptor and endothelial vascular addressin expression regulates lymphocyte homing and permits CD4+ CD3− cells to colonize lymph nodes. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11019–11024. doi: 10.1073/pnas.93.20.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang T., Yednock T., Issekutz A. C. Alpha9beta1 integrin is expressed on human neutrophils and contributes to neutrophil migration through human lung and synovial fibroblast barriers. J. Leukoc. Biol. 1999;66:809–816. doi: 10.1002/jlb.66.5.809. [DOI] [PubMed] [Google Scholar]
- 32.Taooka Y., Chen J., Yednock T., Sheppard D. The integrin alpha9beta1 mediates adhesion to activated endothelial cells and transendothelial neutrophil migration through interaction with vascular cell adhesion molecule-1. J. Cell Biol. 1999;145:413–420. doi: 10.1083/jcb.145.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans J. P. Fertilin beta and other ADAMs as integrin ligands: insights into cell adhesion and fertilization. BioEssays. 2001;23:628–639. doi: 10.1002/bies.1088. [DOI] [PubMed] [Google Scholar]
- 34.Evans J. P. Sperm disintegrins, egg integrins, and other cell adhesion molecules of mammalian gamete plasma membrane interactions. Front. Biosci. 1999;4:D114–D131. doi: 10.2741/evans. [DOI] [PubMed] [Google Scholar]
- 35.Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood. 2003;102:1186–1195. doi: 10.1182/blood-2002-12-3775. [DOI] [PubMed] [Google Scholar]
- 36.Black R. A., Rauch C. T., Kozlosky C. J., Peschon J. J., Slack J. L., Wolfson M. F., Castner B. J., Stocking K. L., Reddy P., et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature (London) 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 37.Moss M. L., Jin S. L., Milla M. E., Bickett D. M., Burkhart W., Carter H. L., Chen W. J., Clay W. C., Didsbury J. R., Hassler D., et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature (London) 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 38.Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 39.Fritsche J., Moser M., Faust S., Peuker A., Buttner R., Andreesen R., Kreutz M. Molecular cloning and characterization of a human metalloprotease disintegrin – a novel marker for dendritic cell differentiation. Blood. 2000;96:732–739. [PubMed] [Google Scholar]
- 40.Fourie A. M., Coles F., Moreno V., Karlsson L. Catalytic activity of ADAM8, ADAM15, and MDC-L (ADAM28) on synthetic peptide substrates and in ectodomain cleavage of CD23. J. Biol. Chem. 2003;278:30469–30477. doi: 10.1074/jbc.M213157200. [DOI] [PubMed] [Google Scholar]
- 41.Van Eerdewegh P., Little R. D., Dupuis J., Del Mastro R. G., Falls K., Simon J., Torrey D., Pandit S., McKenny J., Braunschweiger K., et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature (London) 2002;418:426–430. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 42.Faull R. J., Wang J., Leavesley D. I., Puzon W., Russ G. R., Vestweber D., Takada Y. A novel activating anti-beta1 integrin monoclonal antibody binds to the cysteine-rich repeats in the beta1 chain. J. Biol. Chem. 1996;271:25099–25106. doi: 10.1074/jbc.271.41.25099. [DOI] [PubMed] [Google Scholar]
- 43.Tidswell M., Pachynski R., Wu S. W., Qiu S. Q., Dunham E., Cochran N., Briskin M. J., Kilshaw P. J., Lazarovits A. I., Andrew D. P., et al. Structure-function analysis of the integrin beta 7 subunit: identification of domains involved in adhesion to MAdCAM-1. J. Immunol. 1997;159:1497–1505. [PubMed] [Google Scholar]
- 44.Faull R. J., Kovach N. L., Harlan J. M., Ginsberg M. H. Affinity modulation of integrin alpha 5 beta 1: regulation of the functional response by soluble fibronectin. J. Cell Biol. 1993;121:155–162. doi: 10.1083/jcb.121.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cal S., Freije J. M., Lopez J. M., Takada Y., Lopez-Otin C. ADAM 23/MDC3, a human disintegrin that promotes cell adhesion via interaction with the alphavbeta3 integrin through an RGD-independent mechanism. Mol. Biol. Cell. 2000;11:1457–1469. doi: 10.1091/mbc.11.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada K. M. Adhesive recognition sequences. J. Biol. Chem. 1991;266:12809–12812. [PubMed] [Google Scholar]
- 47.Marcinkiewicz C., Calvete J. J., Marcinkiewicz M. M., Raida M., Vijay-Kumar S., Huang Z., Lobb R. R., Niewiarowski S. EC3, a novel heterodimeric disintegrin from Echis carinatus venom, inhibits alpha4 and alpha5 integrins in an RGD-independent manner. J. Biol. Chem. 1999;274:12468–12473. doi: 10.1074/jbc.274.18.12468. [DOI] [PubMed] [Google Scholar]
- 48.Bazan-Socha S., Kisiel D. G., Young B., Theakston R. D., Calvete J. J., Sheppard D., Marcinkiewicz C. Structural requirements of MLD-containing disintegrins for functional interaction with alpha 4 beta 1 and alpha 9 beta1 integrins. Biochemistry. 2004;43:1639–1647. doi: 10.1021/bi035853t. [DOI] [PubMed] [Google Scholar]
- 49.Zhu X., Bansal N. P., Evans J. P. Identification of key functional amino acids of the mouse fertilin beta (ADAM2) disintegrin loop for cell-cell adhesion during fertilization. J. Biol. Chem. 2000;275:7677–7683. doi: 10.1074/jbc.275.11.7677. [DOI] [PubMed] [Google Scholar]
- 50.Tan K., Casasnovas J. M., Liu J. H., Briskin M. J., Springer T. A., Wang J. H. The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition. Structure. 1998;6:793–801. doi: 10.1016/s0969-2126(98)00080-x. [DOI] [PubMed] [Google Scholar]
- 51.Newham P., Craig S. E., Seddon G. N., Schofield N. R., Rees A., Edwards R. M., Jones E. Y., Humphries M. J. Alpha4 integrin binding interfaces on VCAM-1 and MAdCAM-1. Integrin binding footprints identify accessory binding sites that play a role in integrin specificity. J. Biol. Chem. 1997;272:19429–19440. doi: 10.1074/jbc.272.31.19429. [DOI] [PubMed] [Google Scholar]
- 52.Rott L. S., Briskin M. J., Butcher E. C. Expression of alpha4beta7 and E-selectin ligand by circulating memory B cells: implications for targeted trafficking to mucosal and systemic sites. J. Leukoc. Biol. 2000;68:807–814. [PubMed] [Google Scholar]