Abstract
Ala-114, together with Asp-113, Tyr-115 and Gln-151, form the pocket that accommodates the 3′-OH of the incoming dNTP in the HIV-1 RT (reverse transcriptase). Four mutant RTs having serine, glycine, threonine or valine instead of Ala-114 were obtained by site-directed mutagenesis. While mutants A114S and A114G retained significant DNA polymerase activity, A114T and A114V showed very low catalytic efficiency in nucleotide incorporation assays, due to their high apparent Km values for dNTP. Discrimination between AZTTP (3′-azido-3′-deoxythymidine triphosphate) and dTTP was not significantly affected by mutations A114S and A114G in assays carried out with heteropolymeric template/primers. However, both mutants showed decreased susceptibility to AZTTP when poly(rA)/(dT)16 was used as substrate. Steady-state kinetic analysis of the incorporation of ddNTPs compared with dNTPs showed that substituting glycine for Ala-114 produced a 5–6-fold increase in the RT's ability to discriminate against ddNTPs (including the physiologically relevant metabolites of zalcitabine and didanosine), a result that was confirmed in primer-extension assays. In contrast, A114S and A114V showed wild-type ddNTP/dNTP discrimination efficiencies. Discrimination against ribonucleotides was not affected by mutations at position 114. Misinsertion and mispair extension fidelity assays as well as determinations of G→A mutation frequencies using a lacZ complementation assay showed that, unlike Tyr-115 or Gln-151 mutants, the fidelity of HIV-1 RT was not largely affected by substitutions of Ala-114. The role of the side-chain of Ala-114 in ddNTP/dNTP discrimination appears to be determined by its participation in van der Waals interactions with the ribose moiety of the incoming nucleotide.
Keywords: ddNTPs, drug resistance, fidelity, HIV, polymerase, reverse transcriptase
Abbreviations: AZT, azidothymidine; AZTTP, 3′-azido-3′-deoxythymidine triphosphate; DTT, dithiothreitol; RT, reverse transcriptase; ssDNA, single-stranded DNA
INTRODUCTION
HIV-1 RT (reverse transcriptase) is a multifunctional heterodimeric enzyme with RNA- and DNA-dependent DNA polymerase and RNase H activities, and is responsible for the conversion of the viral genomic RNA into double-stranded pre-integrative DNA (for a review, see [1]). The process of reverse transcription is error-prone, and contributes to the high degree of genetic variability of HIV (for recent reviews, see [2,3]). One of the consequences of the high mutation rates has been the emergence of drug-resistant variants, which constitutes a major obstacle for treatment of HIV infection ([4] and references cited therein). The HIV-1 RT heterodimer is composed of two subunits of 66 and 51 kDa [5], which are designated as p66 and p51 respectively.
Crystal structures of HIV-1 RT reveal that both subunits each contain four common subdomains, termed the ‘fingers’, ‘palm’, ‘thumb’ and ‘connection’ [6]. The 66 kDa polypeptide has an extra C-terminal domain spanning the last 120 residues, which provides an RNase H (endonuclease) activity. The overall folding of the subdomains is similar in p66 and p51, but the spatial arrangements of these subdomains differ markedly. The p66 subunit has a large nucleic-acid-binding cleft formed by the three subdomains known as ‘fingers’, ‘palm’ and ‘thumb’ [6,7]. The active site of the enzyme resides within the 66 kDa subunit that contains the catalytic aspartic acid residues (Asp-110, Asp-185 and Asp-186) [8–12]. The crystal structure of a ternary complex formed by the RT, a DNA–DNA template-primer and dTTP has shown that residues such as Lys-65, Arg-72, Asp-113, Ala-114, Tyr-115 and Gln-151 are involved in interactions with the incoming dNTP [13].
Asp-113, Ala-114, Tyr-115 and Gln-151 are all highly conserved residues in retroviral RTs (see sequence alignments in [2]), and form the pocket that accommodates the ribose moiety of the nucleoside triphosphate. Amino acid substitutions at these positions are usually deleterious for RT function. Examples are D113A and D113E [14], Q151A and Q151K [15–17], and Tyr-115 mutants, such as Y115A and Y115G, among others [18,19]. Tyr-115 acts as an ‘steric gate’, preventing the incorporation of ribonucleotides [20–22]. Mutants lacking an aromatic side chain at this position (i.e. Y115V, Y115A or Y115G) were shown to incorporate rNTPs with 103–105-fold higher efficiency than the wild-type enzyme [22]. Both Tyr-115 and Gln-151 appear to influence fidelity of DNA synthesis. Thus mutant Y115A showed a 5–10-fold higher misinsertion and mispair extension fidelity in comparison with the wild-type enzyme, in assays carried out with DNA templates [19,22]. This amino acid substitution also produced a 2.3-fold increase in the viral mutant frequency, as measured in cell culture during one round of HIV-1 replication [23]. On the other hand, the viral mutant frequency was reduced 5-fold when asparagine was substituted for Gln-151 [23], supporting the results obtained in cell-free reactions that showed the increased fidelity of mutant Q151N [17,24,25]. This mutant is also moderately resistant to ddNTPs [24]. A natural mutation at codon 151 (Q151M) showing wild-type fidelity is known to confer resistance to all dideoxynucleoside analogues used for treatment of HIV infection [26–28].
For Ala-114 mutants, available information is mostly limited to activity measurements using partially purified enzymes and homopolymeric template-primers [i.e. poly(rA)/oligo(dT) or poly(rC)/oligo(dG)]. In these assays, mutant A114V showed very low activity [29], while mutants A114G and A114S showed polymerase activities ranging from 22 to >80%, relative to the wild-type RT [8,29]. Studies with purified recombinant A114S RT showed that this mutation had a very small effect on ddTTP sensitivity [30], but conferred resistance to AZTTP (3′-azido-3′-deoxythymidine triphosphate) and phosphonoformate in assays carried out with homopolymeric template-primers [8,30]. Paradoxically, mutant viruses harbouring mutation A114S were viable and showed approx. 3-fold increased sensitivity to AZT (azidothymidine) when tested in cell culture [8]. In the present study, we have obtained a series of mutant RTs with substitutions at position 114 (i.e. A114S, A114G, A114T and A114V) in order to define its role in nucleotide recognition and fidelity of DNA synthesis.
MATERIALS AND METHODS
Substrates
Stock solutions of dNTPs, rNTPs and ddNTPs (100 mM), poly(rA)∼450, oligo(dT)16, and [γ-32P]ATP were obtained from Amersham Biosciences. AZTTP was purchased from Moravek Biochemicals. Oligonucleotides PG5-25 (5′-CCAGAATGCTGGTAGGGCTATACAT-3′), 25PGG (5′-TGGTAGGGCTATACATTCTTATTAT-3′), 20PG5cg (5′-TGGTAGGGCTATACATTCGT-3′), 3TRP (5′-TGTGGCTTGCCAATACTCTGTC-3′), pT (5′-GGATTTTAGACAGGAACGGT-3′) and oligo(dT)16 were labelled at their 5′-termini with [γ-32P]ATP and T4 polynucleotide kinase (Promega), as described previously [19]. The phosphorylated primers were then annealed to their corresponding templates: D2-47 (5′-GGGATTAAATAAAATAGTAAGAATGTATAGCCCTACCAGCATTCTGG-3′) for PG5-25, D38G (5′-AAAATTAAATAAGATAATAAGAATGTATAGCCCTACCA-3′) for 25PGG, D2cg (5′-GGGATTAAATAAAATAGTACGAATGTATAGCCCTACCA-3′) for 20PG5cg, M_54 (5′-CCCATACAAAAGGAAACATGGGAAACATGGTGGACAGAGTATTGGCAAGCCACA-3′) for 3TRP, M13mp2 ssDNA (single-stranded DNA) for pT, and poly(rA)∼450 for oligo(dT)16. The templates and their corresponding primers were annealed in 150 mM NaCl and 150 mM magnesium acetate as described previously [19].
Mutagenesis, expression and purification of recombinant RTs
Site-directed mutagenesis was carried out with the Quik-Change Site-Directed Mutagenesis Kit (Stratagene), using plasmid pRT6 [31] as template and the mutagenic primers: 5′-GGATGTGGGTGATTCATATTTTTCAGTTCCC-3′ and 5′-GGGAACTGAAAAATATGAATCACCCACATC-3′ for A114S, 5′-GGATGTGGGTGATGGATATTTTTCAGTTCCC-3′ and 5′-GGGAACTGAAAAATATCCATCACCCACATC-3′ for A114G, 5′-GGATGTGGGTGATACATATTTTTCAGTTCCC-3′ and 5′-GGGAACTGAAAAATATGTATCACCCACATC-3′ for A114T, and 5′-GGATGTGGGTGATGTATATTTTTCAGTTCCC-3′ and 5′-GGGAACTGAAAAATATACATCACCCACATC-3′ for A114V (relevant mutations underlined), by following the manufacturer's instructions. After mutagenesis, the RT-coding regions were sequenced, and inserts containing the appropriate mutations were cloned in the p51 expression vector pT51H, by following previously described procedures [18,32]. Purification of wild-type and mutant RTs was carried out after independent expression of their subunits [18]. All RTs were purified as p66–p51 heterodimers. The 51 kDa polypeptide was obtained with an extension of 14 amino acid residues at its N-terminal end, which includes six consecutive histidine residues to facilitate its purification by metal chelate affinity chromatography.
Primer-extension assays
Template-primers D2-47/PG5-25, M_54/3TRP, M13mp2 ssDNA/pT and poly(rA)/oligo(dT)16 were used to determine the DNA polymerase activity of mutant RTs in the presence of different mixtures of nucleotides. Primers were labelled with 32P at their 5′ termini, and annealed to their corresponding templates as indicated above. A 15 μl volume of a solution containing 12–60 nM enzyme and 60 nM template-primer in 100 mM Hepes (pH 7.0) buffer, 30 mM magnesium acetate, 30 mM NaCl, 130 mM potassium acetate, 1 mM DTT (dithiothreitol), and 5% poly(ethylene glycol) 6000 were incubated at 37 °C for 10 min. Primer extensions were initiated by adding 15 μl of a mixture containing equimolar concentrations of all dNTPs (usually 50 μM to 1 mM, depending on the assay), in 130 mM potassium acetate, 1 mM DTT and 5% poly(ethylene glycol) 6000. Reactions were incubated for 0–60 min. Aliquots of 5 μl were withdrawn at different times, and reactions were then stopped by adding 5 μl of 10 mM EDTA in 90% formamide containing 3 mg/ml xylene cyanol FF and 3 mg/ml Bromophenol Blue. DNA synthesis products were separated on 6, 8 or 20% polyacrylamide–urea gels and visualized by phosphorimaging with a BAS 1500 scanner (Fuji). Extensions of primers with four rNTPs were carried out with D2-47/PG5-25, as described previously [22]. The template-primer M13mp2 ssDNA/pT was used in ddNTP/dNTP discrimination assays. Primer extensions were performed as described above, but elongation reactions were carried out for 60 min in the presence of a 50 μM concentration of each dNTP, and different ddNTPs were supplied at a concentration of 50 μM or 500 μM, depending on the assay.
Single-nucleotide extension assays
Nucleotide-incorporation assays were performed in 25 μl of 50 mM Hepes (pH 7.0) buffer, containing 15 mM magnesium acetate, 15 mM NaCl, 130 mM potassium acetate, 1 mM DTT and 5% poly(ethylene glycol) 6000 [33]. The template-primer concentration was 30 nM for D2-47/PG5-25 and D38G/25PGG, and 20 nM for D2cg/20PG5cg. The active enzyme concentration was approx. 1.5–8 nM. Reactions were initiated by incubating the enzyme with the corresponding annealed template-primer in the absence of nucleoside triphosphates (10 min at 37 °C), followed by addition of appropriate nucleotides at various concentrations: typically, in the range 32 nM–2.5 mM for dNTPs, 1 μM–4 mM for ddNTPs, and 100 μM–7.5 mM for rNTPs. Aliquots of 4–6 μl were withdrawn at different times and mixed with 6 μl of the EDTA/formamide solution described above to obtain the corresponding product against time plots, at each substrate concentration. Incubation times were usually in the range 10–90 s for dNTPs and ddNTPs, and up to 45 min for rNTPs. The extension products resulting from the incorporation of nucleotides at the 3′ end of the primer were resolved by electrophoresis in 20% polyacrylamide–urea gels, and primer extension was measured using a BAS 1500 scanner. Elongation measurements were used to determine the initial velocity for each concentration of nucleotide substrate. The amount of active enzyme in the assay was calculated from the intercept of the corresponding plots of product against time, obtained with saturating concentrations of the correct dNTP. The kcat and apparent Km values were determined after fitting the data to the Michaelis–Menten equation using the Gauss–Newton iteration method. Each of the experiments was performed independently at least three times.
Fidelity assays
Misinsertion and mispair extension fidelity assays were performed essentially as described previously [34,35], using a standing-start protocol. End-labelling of primers, template-primer annealing and polymerization reactions were performed in the conditions described for single-nucleotide extension assays with template-primers D2-47/PG5-25 and D2cg/20PG5cg. The concentrations of incorrect dNTPs used to determine catalytic parameters were in the range 250 μM–7.5 mM, and elongation reactions were incubated at 37 °C for up to 20–30 min. For mispair extension fidelity assays, three additional primers were used: PG5-25C, PG5-25G and PG5-25A. All of them are identical with PG5-25, but have C, G or A respectively at their 3′ end. Template-primer concentrations were kept at 30 nM in all assays. The dTTP concentrations used were in the range 20 μM–7.5 mM for elongation assays carried out with mismatched template-primers. Mispair extension reactions were incubated at 37 °C for up to 90 s for the elongation of A:C mispairs, and 10–20 min for A:A and A:G mispairs.
M13mp2 lacZ complementation assay to detect G→A mutations generated in the presence of biased dNTP pools
The frequencies of G→A transitions in RNA-dependent DNA polymerase reactions catalysed by wild-type and mutant HIV-1 RTs were determined as described previously, using a genetic screen based on a blue/white β-galactosidase complementation assay [36]. Briefly, a 141-nucleotide RNA template containing a 66 bp HIV-1 pol fragment that includes the sequence …AAGGAAACAUGGGAAACAUGGUGG… (Trp codons underlined) was reverse-transcribed in 50 mM Hepes (pH 7.0) containing 15 mM NaCl, 15 mM magnesium acetate, 130 mM potassium acetate and 10 mM DTT, either in the presence of a 50 μM concentration of each dNTP, or in the presence of a dNTP cocktail containing 0.1 μM dCTP, 440 μM dTTP, 40 μM dATP and 20 μM dGTP. The cDNA obtained was amplified by PCR with appropriate oligonucleotides, and purified PCR products encoding three tryptophan codons were cloned in-frame of the lacZα fragment of a modified replicative form of bacteriophage M13mp18. The ligation products were transformed into Escherichia coli XL-1 Blue MRF' cells. Transformed cells were plated on to minimal agar plates containing 8% X-Gal (5-bromo-4-chloroindol-3-yl β-D-galactopyranoside) and 20% IPTG (isopropyl β-D-thiogalactoside). G:T mispairs formed during RNA-dependent DNA synthesis result in G→A mutations that when occurring at any of the tryptophan codons render stop codons (colourless plaques). A few blue plaques (as control) and all colourless and light-blue plaques were analysed, and mutations were identified by DNA sequencing.
RESULTS
The crystal structure of the ternary complex of HIV-1 RT•DNA–DNA•dTTP [13] showed that Ala-114 is located in the bottom of the dNTP-binding site, which is mostly occupied by Tyr-115 (see Figure S1 at http://www.BiochemJ.org/bj/387/bj3870221add.htm). The side chain of Ala-114 is close to the 3′-OH group of the incoming dNTP. The distance between atom Cβ of Ala-114 and the 3′-OH of the ribose is 3.37 Å (1 Å=0.1 nm). In addition, atom Cβ of Ala-114 is also close to the peptidic -NH- group of Tyr-115 (2.91 Å) and to the catalytic carboxy group of Asp-185 (the distance between the Cβ of Ala-114 and the Oδ2 of Asp-185 is 3.23 Å). As a consequence, increasing the volume of the side chain at position 114 is likely to have a deleterious effect on the polymerase activity of the RT. Primer-extension measurements carried out with three different DNA–DNA template-primers [i.e. D2-47/PG5-25 (47/25-mer), M_54/3TRP (54/22-mer) and M13mp2 ssDNA/pT] in the presence of dNTP concentrations ranging from 50 μM to 1 mM showed that mutants A114G and A114S displayed a polymerase activity >90% of that shown by the wild-type enzyme. In contrast, A114V and A114T showed primer-extension efficiencies of 15–20% and 10–15% respectively, relative to the wild-type enzyme (results not shown). Steady-state kinetics of polymerization by wild-type and mutant RTs was investigated by using a 47-/25-mer heteropolymeric template-primer and dTTP (Table 1). The catalytic efficiency of mutant A114S was approx. 20% of the wild-type enzyme, while mutants A114V and A114T had very low catalytic efficiencies (<0.5% of the wild-type RT). All mutants showed significant increases in their corresponding Km values, which in the case of A114V and A114T were >200-fold higher than the wild-type enzyme. In contrast, the kcat values were not largely affected by replacements at Ala-114, although it should be noted that under steady-state conditions, the kcat values are probably coincident with the dissociation rates of the RT•DNA–DNA complex.
Table 1. Steady-state kinetic parameters of the wild-type (WT) and mutant RTs for the incorporation of dTTP, using template-primer D2-47/PG5-25.
| Enzyme | kcat (min−1) | Km (μM) | kcat/Km (μM−1·min−1) |
|---|---|---|---|
| WT RT | 3.54±1.35 | 0.32±0.10 | 11.08 |
| A114S | 4.63±1.69 | 2.02±0.95 | 2.29 |
| A114G | 1.40±0.29 | 4.07±2.30 | 0.34 |
| A114V | 4.73±1.52 | 71±15.5 | 0.067 |
| A114T | 5.28±1.19 | >100 | <0.05 |
Inhibition of dTMP incorporation by AZTTP
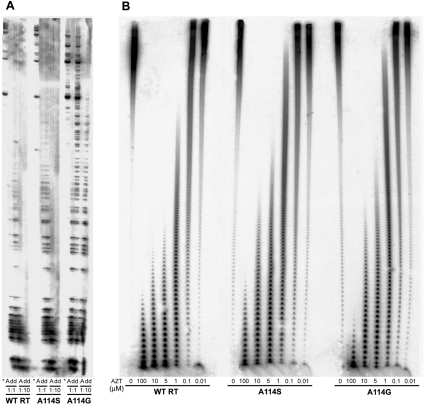
Previous reports showed that A114S confers resistance to AZTTP in cell-free assays [8,30], although viruses containing this mutation within their RT were sensitive to the inhibitor [8]. The selectivity values calculated as the catalytic efficiencies of incorporation of AZTTP compared with dTTP were less than 3-fold higher for mutants A114S and A114G than for the wild-type RT, in assays carried out with D2-47/PG5-25 (Table 2). In all cases, discrimination between AZTTP and dTTP was largely dependent on the apparent Km values, which were approx. 2–8 times higher for the inhibitor than for the natural substrate. This slightly increased resistance to AZTTP shown by both mutant RTs in comparison with the wild-type RT was almost undetectable in primer-extension assays using an M13-derived DNA template (Figure 1A), suggesting that those mutations had no significant effects on AZT resistance. However, in assays carried out with poly(rA)/oligo(dT)16, A114S and A114G were both less sensitive to AZTTP inhibition than the wild-type enzyme (Figure 1B), in agreement with data reported by other authors [8,30].
Table 2. Kinetic parameters for the incorporation of AZTTP and ddTTP by wild-type (WT) and mutant RTs, using template-primer D2-47/PG5-25.
| Enzyme | Nucleotide | kcat (min−1) | Km (μM) | kcat/Km (μM−1·min−1) | Selectivity* |
|---|---|---|---|---|---|
| WT RT | AZTTP | 3.63±0.49 | 2.42±0.47 | 1.509 | 0.135 |
| ddTTP | 4.92±0.78 | 8.56±1.34 | 0.575 | 5.2×10−2 | |
| A114S | AZTTP | 5.19±1.51 | 11.5±2.12 | 0.451 | 0.197 (1.4) |
| ddTTP | 0.83±0.55 | 4.56±2.18 | 0.182 | 7.9×10−2 (1.5) | |
| A114G | AZTTP | 1.12±0.14 | 9.1±1.1 | 0.123 | 0.361 (2.7) |
| ddTTP | 1.13±0.22 | 385±79 | 2.9×10−3 | 8.5×10−3 (0.16) | |
| A114V | ddTTP | 2.05±0.12 | 412±56 | 5.0×10−3 | 7.5×10−2 (1.4) |
* Selectivity=[kcat (inhibitor)/Km (inhibitor)]/[kcat (dTTP)/Km (dTTP)]. Values between parentheses represent the ratio of the selectivity of each mutant enzyme to that shown by the wild-type RT.
Figure 1. AZTTP inhibition of wild-type (WT) and mutant RTs.
(A) Primer-extension assays carried out in the presence of 30 nM M13mp2 ssDNA/pT and all four dNTPs (each at 50 μM) are indicated with an asterisk. Reaction mixtures marked with an ‘A’ or ‘dd’ also contained AZTTP or ddTTP at 50 μM (1:1 ratio) or 500 μM (1:10 ratio), depending on the assay. (B) Elongation reactions carried out with 30 nM poly(rA)/oligo(dT)16 and 10 μM dTTP, with varying concentrations of AZTTP. The RT concentrations in the assays shown in both (A) and (B) were estimated to be approx. 15 nM, and samples were incubated at 37 °C for 60 min.
Discrimination between dNTPs and ddNTPs or rNTPs by Ala-114 mutants
The proximity of the 3′-OH of the ribose of the incoming dNTP to the side chain at position 114 suggests that ddNTP/dNTP discrimination could be affected by substitutions of Ala-114. The determination of kinetic parameters for the incorporation of ddTTP using D2-47/PG5-25 showed similar selectivity values for wild-type RT and mutants A114S and A114V (Table 2). The catalytic efficiency (kcat/Km) for the incorporation of ddTTP by the wild-type RT was approx. 5.2% in comparison with dTTP. However, this value was reduced to 0.85% for mutant A114G. The difference between the wild-type RT and mutant A114G in ddTTP/dTTP selectivity is the result of a large increase in the Km value for ddTTP observed with A114G. The kcat for the incorporation of dTTP is similar to that shown by ddTTP in both enzymes.
The reduced susceptibility of A114G to ddNTPs was also observed in assays that monitored the incorporation of dCTP and ddCTP, as well as with dATP and ddATP. Both ddCTP and ddATP are the physiologically relevant derivatives of approved RT inhibitors, such as zalcitabine and didanosine. The incorporation of ddCTP compared with dCTP by mutant A114G was approx. 6-fold less efficient than for the wild-type enzyme and mutant A114S (Table 3), and similar effects were observed when measuring incorporation of ddATP compared with dATP (Table 4). For all tested enzymes, the selectivity values obtained with both ddNTPs relative to their corresponding dNTPs were similar to those obtained with ddTTP and dTTP, and mutations had similar effects on the catalytic efficiencies of dNTP incorporation. However, substituting glycine for Ala-114 produced significant reductions in the relative kcat values of incorporation of ddCTP compared with dCTP, and ddATP compared with dATP, indicating that, unlike in the case of ddTTP, selectivity differences resulted from the combined effect of the kcat and Km values.
Table 3. Kinetic parameters for the incorporation of dCTP and ddCTP by wild-type (WT) and mutant RTs, using template-primer D38G/25PGG.
| Enzyme | Nucleotide | kcat (min−1) | Km (μM) | kcat/Km (μM−1·min−1) | Selectivity* |
|---|---|---|---|---|---|
| WT RT | dCTP | 2.99±0.25 | 0.100±0.026 | 29.74 | |
| ddCTP | 1.33±0.11 | 0.960±0.171 | 1.38 | 4.65×10−2 | |
| A114S | dCTP | 5.50±1.71 | 0.575±0.260 | 9.56 | |
| ddCTP | 0.79±0.17 | 0.797±0.309 | 0.99 | 1.04×10−1 (2.2) | |
| A114G | dCTP | 2.45±0.70 | 1.34±0.60 | 1.83 | |
| ddCTP | 0.40±0.01 | 26.5±3.4 | 1.49×10−2 | 8.14×10−3 (0.17) |
* Selectivity=[kcat (ddCTP)/Km (ddCTP)]/[kcat (dCTP)/Km (dCTP)]. Values between parentheses represent the ratio of the selectivity of each mutant enzyme to that shown by the wild-type RT.
Table 4. Kinetic parameters for the incorporation of dATP, ddATP and rATP by wild-type (WT) and mutant RTs, using template-primer D2cg/20PG5cg.
| Enzyme | Nucleotide | kcat (min−1) | Km (μM) | kcat/Km (μM−1·min−1) | Selectivity* |
|---|---|---|---|---|---|
| WT RT | dATP | 3.25±1.00 | 0.047±0.016 | 69.59 | |
| ddATP | 2.33±0.14 | 0.636±0.220 | 3.67 | 5.3×10−2 | |
| rATP | 1.03±0.32 | 620±184 | 1.7×10−3 | 2.4×10−5 | |
| A114S | dATP | 10.31±4.23 | 1.22±0.32 | 8.45 | |
| ddATP | 2.22±0.52 | 5.48±1.28 | 0.41 | 4.8×10−2 (0.9) | |
| rATP | 0.46±0.12 | 830±156 | 5.5×10−4 | 6.5×10−5 (2.7) | |
| A114G | dATP | 5.39±1.71 | 4.85±2.46 | 1.11 | |
| ddATP | 1.60±0.50 | 129±42 | 1.2×10−2 | 1.1×10−2 (0.2) | |
| rATP | 0.045±0.015 | 577±159 | 7.9×10−5 | 7.1×10−5 (2.9) |
* Selectivity=[kcat (incorrect)/Km (incorrect)]/[kcat (correct)/Km (correct)], where incorrect nucleotides were ddATP or rATP, while the correct nucleotide is dATP. Values between parentheses represent the ratio of the selectivity of each mutant enzyme to that shown by the wild-type RT.
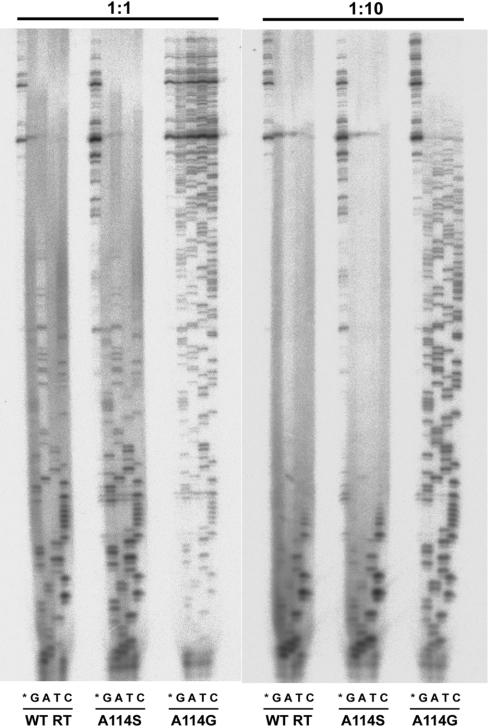
Further evidence for the ddNTP resistance phenotype displayed by A114G was obtained in primer-elongation assays carried out with nucleotide mixtures containing all dNTPs, and one ddNTP. As shown in Figure 2, band patterns were similar for A114S and the wild-type RT in reactions carried out using a 1:1 or a 1:10 dNTP/ddNTP ratio. However, much larger extensions were observed with A114G, thereby indicating a rather inefficient incorporation of ddNTP by this mutant.
Figure 2. Gel assay for ddNTP/dNTP discrimination.
Comparison of wild-type (WT) HIV-1 RT and mutants A114S and A114G at two different concentration ratios of dNTP compared with ddNTP. Reactions were carried out in the presence of 30 nM M13mp2 ssDNA/pT, and all four dNTPs each at a 50 μM concentration. Reaction mixtures marked with ‘G’, ‘A’, ‘T’ or ‘C’ also contained the corresponding ddNTP at a concentration of 50 μM (1:1 ratio) or 500 μM (1:10 ratio). No ddNTPs were added in reactions marked with an asterisk.
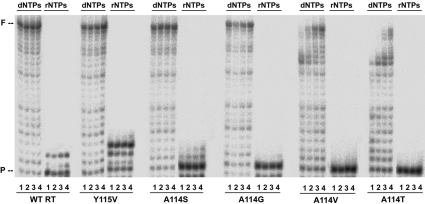
Tyr-115 was previously identified as a ‘steric gate’ discriminating against rNTPs [20–22]. Therefore we tested if the neighbouring residue, Ala-114, had any effect on rNTP/dNTP selectivity. Selectivity values obtained from steady-state kinetic parameters for the incorporation of rATP or dATP on template-primer D2cg/20PG5cg (38-/20-mer) were in the range (2.4–7.1)×10−5 for wild-type RT and mutants A114S and A114G (Table 4). Although selectivity values were somewhat higher for both mutants than for the wild-type enzyme, differences were not statistically significant. Furthermore, primer extensions in the presence of all four rNTPs were very inefficient for all Ala-114 mutants, except for Y115V (Figure 3).
Figure 3. Extension of primer PG5-25 using dNTPs or rNTPs as nucleotide substrates by wild-type (WT) RT, and mutants Y115V, A114S, A114G, A114V and A114T.
Reactions were carried out in the presence of 30 nM D2-47/PG5-25. All nucleotides were supplied at a 1 mM concentration each, and the amount of active enzyme in the assay was approx. 15–30 nM. Lanes 1–4 correspond to the analysis of aliquots taken after 15, 30, 60 and 120 min respectively. ‘P’ indicates the position of the 25-mer primer, and ‘F’ stands for the full-length product of 47 nucleotides.
Fidelity of DNA synthesis
Misinsertion and mispair extension fidelity assays were used to estimate the fidelity of wild-type RT and mutants A114G and A114S, using a heteropolymeric DNA–DNA template-primer (D2-47/PG5-25). Misinsertion fidelity assays involved kinetic measurements for the incorporation of a correct (T) or an incorrect (C, G or A) nucleotide at the 3′ end of the primer. All tested RTs showed similar misinsertion ratios for A:C and A:G mispairs, ranging from 6.7×10−5 to 1.6×10−4, and from 2.8×10−4 to 6.1×10−4 respectively (Table 5). Discrimination between correct and incorrect dNTPs was mostly due to the large differences in the Km values obtained with correct and incorrect nucleotides, although the contribution of the Km was limited for mutants A114S and A114G. The kinetics of mispair extension were studied for correctly matched base pairs (A:T) and for mismatches A:C, A:G and A:A. In all cases, we measured the incorporation of a correct T opposite A at the 3′ end of the primer. The results are shown in Table 6. Wild-type RT shows a mispair extension ratio of 2.8×10−3 with the template-primer having an A:C mispair, and lower mispair extension ratios (in the order of 10−5) for A:A and A:G mispair termini. Differences between the wild-type RT and mutants A114G and A114S were relatively small, except in the case of the extension of A:G mispairs by A114G, which was 8-fold more efficient than for the wild-type enzyme. The small differences found between wild-type and mutants A114S and A114G were also consistent with the results of primer extension experiments carried out in the presence of three dNTPs. Primer-extension patterns were similar for all three enzymes (results not shown).
Table 5. Misinsertion fidelity of wild-type (WT) and mutant RTs, as obtained using template-primer D2-47/PG5-25.
ND, not determined.
| Enzyme | Nucleotide | kcat (min−1) | Km (μM) | kcat/Km (μM−1·min−1) | Misinsertion ratio (fins)* |
|---|---|---|---|---|---|
| WT RT | dCTP | 1.64±0.51 | 1693±876 | 9.7×10−4 | 8.8×10−5 |
| dGTP | 1.64±0.56 | 537±75 | 3.1×10−3 | 2.8×10−4 | |
| dATP | 0.018±0.002 | ND | ND | ND | |
| A114S | dCTP | 0.50±0.15 | 1408±265 | 3.6×10−4 | 1.6×10−4 (1.8) |
| dGTP | 0.17±0.06 | 121±37 | 1.4×10−3 | 6.1×10−4 (2.2) | |
| dATP | <5.7×10−3 | ND | ND | ND | |
| A114G | dCTP | 0.077±0.009 | 3359±553 | 2.3×10−5 | 6.7×10−5 (0.8) |
| dGTP | 0.18±0.08 | 910±353 | 2.0×10−4 | 5.8×10−4 (2.1) | |
| dATP | <1.1×10−3 | ND | ND | ND |
* fins=[kcat (incorrect)/Km (incorrect)]/[kcat (correct)/Km (correct)], where incorrect nucleotides were dCTP, dGTP or dATP, while the correct nucleotide is dTTP (kinetic parameters for the incorporation of this nucleotide are given in Table 1). Values between parentheses represent the ratio of the selectivity of each mutant enzyme to that shown by the wild-type RT.
Table 6. Mispair extension fidelity of wild-type (WT) and mutant RTs, as obtained using template-primer D2-47/PG5-25.
| Enzyme | Base pair at the 3′ end* | kcat (min−1) | Km (μM) | kcat/Km (μM−1·min−1) | Mispair extension ratio (fext)† |
|---|---|---|---|---|---|
| WT RT | A:C | 1.30±0.14 | 41.3±5.6 | 3.2×10−2 | 2.8×10−3 |
| A:A | 0.15±0.04 | 266±107 | 5.7×10−4 | 5.1×10−5 | |
| A:G | 0.19±0.04 | 513±130 | 3.7×10−4 | 3.3×10−5 | |
| A114S | A:C | 1.64±0.53 | 280±134 | 5.9×10−3 | 2.6×10−3 (0.9) |
| A:A | 0.098±0.034 | 319±121 | 3.1×10−4 | 1.3×10−4 (2.6) | |
| A:G | 0.112±0.056 | 415±142 | 2.7×10−4 | 1.2×10−4 (3.5) | |
| A114G | A:C | 0.78±0.03 | 858±348 | 9.1×10−4 | 2.6×10−3 (0.9) |
| A:A | 0.032±0.007 | 824±228 | 3.9×10−5 | 1.1×10−4 (2.2) | |
| A:G | 0.022±0.006 | 230±53 | 9.6×10−5 | 2.8×10−4 (8.3) |
* The first base corresponds to the template and the second base to the primer.
† fext=[kcat (mismatched)/Km (mismatched)]/[kcat (matched)/Km (matched)], where kinetic parameters for the incorporation of dTTP on matched template-primers are given in Table 1. Numbers in parentheses represent the relative increase in the mispair extension ratio shown by the mutant RT relative to the wild-type enzyme. Our analysis assumes that RTs bind with roughly equal affinity to the matched and mismatched template-primer ends [22,35].
Further evidence of the small effect of residue 114 mutation on the fidelity of DNA synthesis was obtained using a lacZ complementation assay designed to measure the frequency of G→A mutations arising in RNA-dependent DNA polymerase reactions. When reverse transcription reactions were performed in the presence of equimolar concentrations of each dNTP, the G→A mutation frequency obtained with the wild-type RT was 3.43×10−4. This value was increased more than 10-fold when polymerase reactions were carried out in the presence of biased dNTP pools (Table 7). This effect was also observed with mutant RTs carrying the amino acid substitutions A114S, A114G and A114V. All RT mutants showed a reduced G→A mutation frequency compared with the wild-type RT at low [dCTP]/[dTTP], but differences were not statistically significant.
Table 7. Mutation frequencies accompanying reverse transcription in the presence of biased dNTP pools.
The number of mutant plaques that carried a G→A substitution in the target sequence and the total number of plaques were scored in four independent experiments. The frequency of G→A transitions (fG→A) was calculated as the number of G→A substitutions identified at tryptophan codons, divided by the number of target nucleotides (six, in the template used in the assay) and the total number of plaques analysed. WT, wild-type.
| Number of plaques analysed | |||||
|---|---|---|---|---|---|
| Enzyme | [dCTP] (μM)* | Total | Mutated | Number of G→A transitions | fG→A |
| WT RT | 50 | 486 | 1 | 1 | 3.43×10−4 |
| 0.1 | 555 | 13 | 13 | 3.90×10−3 | |
| A114S | 50 | 448 | 2 | 2 | 7.44×10−4 |
| 0.1 | 578 | 8 | 8 | 2.31×10−3 | |
| A114G | 50 | 402 | 0 | 0 | <4.14×10−4 |
| 0.1 | 701 | 6 | 6 | 1.43×10−3 | |
| A114V | 50 | 696 | 2 | 2 | 4.79×10−4 |
| 0.1 | 822 | 8 | 11 | 2.23×10−3 | |
* Nucleotide concentrations in the assays were either 50 μM of each dNTP, or 0.1 μM dCTP, 440 μM dTTP, 40 μM dATP and 20 μM dGTP.
DISCUSSION
All tested amino acid substitutions affecting Ala-114 led to significant increases in the Km values for dNTP incorporation and produced enzymes with diminished polymerase activity. Although previous reports showed that HIV-1 variants with the A114S mutation were viable [8], amino acid substitutions at this position are rare in clinical isolates. For instance, the Stanford HIV RT and Protease Sequence Database [37] contains only eight isolates (out of 11895) with mutations at Ala-114 [i.e. A114S, A114G, A114T, A114P (two isolates) and A114E (three isolates)]. In addition, A114P and A114E were found to be deleterious when introduced in the equivalent position of the RT-coding region of a murine leukaemia virus [38]. It should also be noted that sequences deposited in the database may contain sequencing errors and could represent non-infectious virus. For example, in one of the isolates containing the substitution A114G (GenBank® accession number AF347414) [39], we also found a stop codon within the RT-coding region, as well as another deleterious mutation at codon 115 (Y115K). The key role of Ala-114 in RT function is suggested further by its conservation in most retroviral RTs as well as in retrotransposons lacking LTRs (long terminal repeats) [40].
Ala-114 is part of the dNTP-binding site of HIV-1 RT [13]. Its side chain is oriented towards the ribose moiety of the incoming nucleotide. The methyl group of Ala-114 is less than 3.5 Å away from the 3′-OH group of the ribose and the carboxy group of the catalytic Asp-185. In addition, the 3′-OH forms a hydrogen bond with the -NH- group of Tyr-115, which is only 2.9 Å away from the Cβ of Ala-114. Therefore any increase in the volume of the side chain of Ala-114 is expected to alter the positioning of the dNTP. In agreement with this proposal, we found that mutants A114V and A114T had the lowest catalytic efficiencies in comparison with the wild-type enzyme due to their high Km values for dNTP incorporation. In the case of mutants A114G and A114S, we also observed an increase in the Km values for dNTP incorporation. However, these enzymes retained significant polymerase activity.
The side chain of Ala-114 lies close to the 3′-OH of the ribose which projects into a small cavity (known as the 3′ pocket) lined by the side chains of Asp-113, Ala-114, Tyr-115 and Gln-151 (see Figure S1 at http://www.BiochemJ.org/bj/387/bj3870221add.htm), suggesting that Ala-114 could play a role in the discrimination between chain-elongation inhibitors and dNTPs. Previous reports have shown that both A114G and A114S were resistant to AZT in cell-free assays, but ddNTP/dNTP discrimination was not studied in detail with any of those mutants [8,30]. In the case of the AZT resistance displayed by A114S, conflicting results were reported by the same authors, since they observed that viruses harbouring this mutation were sensitive to the inhibitor in cell culture [8]. Although we confirmed that both mutants were less sensitive to inhibition in assays carried out with homopolymeric template-primers, single-nucleotide incorporation assays revealed only subtle differences between mutant and wild-type enzymes in their ability to discriminate against AZTTP. In addition, none of these enzymes showed detectable ATP-dependent excision activity on primers terminated with AZT (T. Matamoros and L. Menéndez-Arias, unpublished work). Taken together, these data support the evidence obtained in cell culture, indicating that A114S does not confer resistance to AZT.
A114S and wild-type RT had similar ddNTP/dNTP discrimination efficiencies in single-nucleotide incorporation assays, as well as primer extensions carried out with different proportions of ddNTPs and dNTPs. However, we observed that the incorporation of ddNTP by A114G was very inefficient in comparison with the incorporation of dNTP, resulting in enzymes that showed a 5–6-fold reduced ddNTP/dNTP discrimination ability in comparison with the wild-type RT. This effect is caused by the large difference in Km values of ddNTP and dNTP incorporation obtained with A114G compared with the wild-type RT. Although a mechanistic interpretation of the data is problematical due to the difficulties in relating the steady-state kinetic parameters kcat and Km to steps in the mechanism of nucleotide addition, kinetic data suggest that removal of the methyl group of Ala-114 has a larger effect on ddNTP binding than on dNTP binding. The substitution of glycine for Ala-114 produces a small increase in the size of the 3′ pocket, which probably reduces van der Waals interactions between the ribose moiety of the incoming nucleotide and the side chain at position 114. This effect is probably more important for ddNTP than for dNTP binding, because of the contribution to dNTP binding of the hydrogen bond established between the 3′-OH of the ribose and the main chain -NH- group of Tyr-115 [13]. The presence of a bulky azido group in the 3′ position of the ribose, as occurs with AZTTP, minimizes the effect of increasing the size of the 3′ pocket, thereby explaining the relatively small differences in nucleotide selectivity found in AZTTP-incorporation assays.
Fidelity assays based on the determination of kinetic constants for the incorporation of nucleotides on template-primers, or in the identification of G→A mutations arising during reverse transcription in the presence of biased dNTP pools revealed that all mutants of Ala-114 were as faithful as the wild-type RT. The largest differences were observed for A:G mispairs using A114G, which showed a mispair extension ratio eight times higher than the wild-type enzyme. Nevertheless, A:G mispairs are very poor substrates of all RTs in comparison with A:A, and particularly with A:C mispairs, suggesting that, despite the differences, their overall contribution to fidelity should be relatively small. Although fidelity of HIV-1 RT mutants containing substitutions at position 114 has not been previously analysed, it has been reported that murine leukaemia virus carrying a A154S substitution in its RT-coding region (equivalent of A114S of HIV-1 RT) showed reduced a viral titre in comparison with the wild-type construct and produced a slight decrease in fidelity [38].
Examination of the available crystal structure of the ternary complex of Taq DNA polymerase bound to template-primer and a nucleotide [41] indicates that Ile-614 and Glu-615 are located under the sugar moiety of the incoming dNTP, and are the equivalent residues of Ala-114 and Tyr-115 in HIV-1 RT. Ile-614 and Glu-615 of Taq DNA polymerase are conserved in E. coli DNA polymerase I, where they are designated as Ile-709 and Glu-710. These residues are part of the highly conserved motif A, found in many polymerases [40,42]. Replacement of Glu-710 of E. coli DNA polymerase I (Klenow fragment) by alanine decreases discrimination against rNTPs 1000-fold [43], suggesting that this residue is functionally equivalent to Tyr-115 of HIV-1 RT. However, mutational analysis of Ile-614 of Taq DNA polymerase revealed that this position was highly mutable [44], in contrast with our findings with Ala-114 of HIV-1 RT. In addition, Taq DNA polymerase mutants with substitutions at Ile-614 had altered rNTP/dNTP discrimination efficiencies [45], and were less faithful than the wild-type enzyme [46]. The role of this residue in fidelity of DNA synthesis was confirmed for E. coli DNA polymerase I using mutants with substitutions involving Ile-709 [47]. Taken together, these results suggest that there is only limited functional equivalence between motifs A of HIV-1 RT and type I DNA polymerases.
In summary, unlike other residues within the 3′ pocket, such as Tyr-115 or Gln-151 which have been identified as part of the fidelity centre of HIV-1 RT, while affecting ddNTP/dNTP discrimination [17–22,24,25], our data support the role of Ala-114 as a key element of the dNTP-binding site, contributing to ddNTP/dNTP discrimination, but without a major influence on fidelity of DNA synthesis. In addition, substituting glycine for Ala-114 leads to an RT that shows cross-resistance to antiretroviral drugs, such as zalcitabine and didanosine.
Online data
Acknowledgments
This work was supported in part by Fondo de Investigación Sanitaria (FIS)-FEDER grant 01/0067-01 and by an institutional grant to the Centro de Biología Molecular “Severo Ochoa” from the Fundación Ramón Areces. Support from the Red Temática Cooperativa de Investigación en SIDA (FIS G03/173) is also acknowledged.
References
- 1.Telesnitsky A., Goff S. P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J. W., Hughes S. H., Varmus H. E., editors. Retroviruses. Plainview: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 2.Menéndez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:91–147. doi: 10.1016/s0079-6603(02)71042-8. [DOI] [PubMed] [Google Scholar]
- 3.Svaroskaia E. S., Cheslock S. R., Zhang W.-H., Hu W.-S., Pathak V. K. Retroviral mutation rates and reverse transcriptase fidelity. Front. Biosci. 2003;8:D117–D134. doi: 10.2741/957. [DOI] [PubMed] [Google Scholar]
- 4.Menéndez-Arias L. Targeting HIV: antiretroviral therapy and development of drug resistance. Trends Pharmacol. Sci. 2002;23:381–388. doi: 10.1016/s0165-6147(02)02054-0. [DOI] [PubMed] [Google Scholar]
- 5.Di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 6.Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 7.Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P., et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larder B. A., Kemp S. D., Purifoy D. J. M. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc. Natl. Acad. Sci. U.S.A. 1989;86:4803–4807. doi: 10.1073/pnas.86.13.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Grice S. F. J., Naas T., Wohlgensinger B., Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer P. L., Ferris A. L., Hughes S. H. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J. Virol. 1992;66:1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hostomsky Z., Hostomska Z., Fu T.-B., Taylor J. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J. Virol. 1992;66:3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaushik N., Rege N., Yadav P. N. S., Sarafianos S. G., Modak M. J., Pandey V. N. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry. 1996;35:11536–11546. doi: 10.1021/bi960364x. [DOI] [PubMed] [Google Scholar]
- 13.Huang H., Chopra R., Verdine G. L., Harrison S. C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 14.Harris D., Kaushik N., Pandey P. K., Yadav P. N. S., Pandey V. N. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J. Biol. Chem. 1998;273:33624–33634. doi: 10.1074/jbc.273.50.33624. [DOI] [PubMed] [Google Scholar]
- 15.Sarafianos S. G., Pandey V. N., Kaushik N., Modak M. J. Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Biochemistry. 1995;34:7207–7216. doi: 10.1021/bi00021a036. [DOI] [PubMed] [Google Scholar]
- 16.Kaushik N., Harris D., Rege N., Modak M. J., Yadav P. N. S., Pandey V. N. Role of glutamine-151 of human immunodeficiency virus type-1 reverse transcriptase in RNA-directed DNA synthesis. Biochemistry. 1997;36:14430–14438. doi: 10.1021/bi970645k. [DOI] [PubMed] [Google Scholar]
- 17.Weiss K. K., Isaacs S. J., Tran N. H., Adman E. T., Kim B. Molecular architecture of the mutagenic active site of human immunodeficiency virus type 1 reverse transcriptase: roles of the β8-αE loop in fidelity, processivity and substrate interactions. Biochemistry. 2000;39:10684–10694. doi: 10.1021/bi000788y. [DOI] [PubMed] [Google Scholar]
- 18.Martín-Hernández A. M., Domingo E., Menéndez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 1996;15:4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 19.Martín-Hernández A. M., Gutiérrez-Rivas M., Domingo E., Menéndez-Arias L. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res. 1997;25:1383–1389. doi: 10.1093/nar/25.7.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao G., Orlova M., Georgiadis M. M., Hendrickson W. A., Goff S. P. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl. Acad. Sci. U.S.A. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyer P. L., Sarafianos S. G., Arnold E., Hughes S. H. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3056–3061. doi: 10.1073/pnas.97.7.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cases-González C. E., Gutiérrez-Rivas M., Menéndez-Arias L. Coupling ribose selection to fidelity of DNA synthesis: the role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 2000;275:19759–19767. doi: 10.1074/jbc.M910361199. [DOI] [PubMed] [Google Scholar]
- 23.Mansky L. M., Le Rouzic E., Benichou S., Gajary L. C. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J. Virol. 2003;77:2071–2080. doi: 10.1128/JVI.77.3.2071-2080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaushik N., Talele T. T., Pandey P. K., Harris D., Yadav P. N. S., Pandey V. N. Role of glutamine 151 of human immunodeficiency virus type-1 reverse transcriptase in substrate selection as assessed by site-directed mutagenesis. Biochemistry. 2000;39:2912–2920. doi: 10.1021/bi991376w. [DOI] [PubMed] [Google Scholar]
- 25.Weiss K. K., Bambara R. A., Kim B. Mechanistic role of residue Gln151 in error prone DNA synthesis by human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT): pre-steady state kinetic study of the Q151N HIV-1 RT mutant with increased fidelity. J. Biol. Chem. 2002;277:22662–22669. doi: 10.1074/jbc.M200202200. [DOI] [PubMed] [Google Scholar]
- 26.Shirasaka T., Kavlick M. F., Ueno T., Gao W.-Y., Kojima E., Alcaide M. L., Chokekijchai S., Roy B. M., Arnold E., Yarchoan R., Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueno T., Shirasaka T., Mitsuya H. Enzymatic characterization of human immunodeficiency virus type 1 reverse transcriptase resistant to multiple 2′,3′-dideoxynucleoside 5′-triphosphates. J. Biol. Chem. 1995;270:23605–23611. doi: 10.1074/jbc.270.40.23605. [DOI] [PubMed] [Google Scholar]
- 28.Ueno T., Mitsuya H. Comparative enzymatic study of HIV-1 reverse transcriptase resistant to 2′,3′-dideoxynucleotide analogs using the single-nucleotide incorporation assay. Biochemistry. 1997;36:1092–1099. doi: 10.1021/bi962393d. [DOI] [PubMed] [Google Scholar]
- 29.Wrobel J. A., Chao S.-F., Conrad M. J., Merker J. D., Swanstrom R., Pielak G. J., Hutchison C. A., III A genetic approach for identifying critical residues in the fingers and palm subdomains of HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 1998;95:638–645. doi: 10.1073/pnas.95.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arion D., Sluis-Cremer N., Parniak M. A. Mechanism by which phosphonoformic acid resistance mutations restore 3′-azido-3′-deoxythymidine (AZT) sensitivity to AZT-resistant HIV-1 reverse transcriptase. J. Biol. Chem. 2000;275:9251–9255. doi: 10.1074/jbc.275.13.9251. [DOI] [PubMed] [Google Scholar]
- 31.Quiñones-Mateu M. E., Soriano V., Domingo E., Menéndez-Arias L. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology. 1997;236:364–373. doi: 10.1006/viro.1997.8748. [DOI] [PubMed] [Google Scholar]
- 32.Menéndez-Arias L. Studies on the effects of truncating α-helix E' of p66 human immunodeficiency virus type 1 reverse transcriptase on template-primer binding and fidelity of DNA synthesis. Biochemistry. 1998;37:16636–16644. doi: 10.1021/bi981830g. [DOI] [PubMed] [Google Scholar]
- 33.Gutiérrez-Rivas M., Menéndez-Arias L. A mutation in the primer grip region of HIV-1 reverse transcriptase that confers reduced fidelity of DNA synthesis. Nucleic Acids Res. 2001;29:4963–4972. doi: 10.1093/nar/29.24.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boosalis M. S., Petruska J., Goodman M. F. DNA polymerase insertion fidelity: gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 35.Mendelman L. V., Boosalis M. S., Petruska J., Goodman M. F. Nearest neighbor influences on DNA polymerase insertion fidelity. J. Biol. Chem. 1989;264:14415–14423. [PubMed] [Google Scholar]
- 36.Cases-González C. E., Menéndez-Arias L. Increased G→A transition frequencies displayed by primer grip mutants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 2004;78:1012–1019. doi: 10.1128/JVI.78.2.1012-1019.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhee S.-Y., Gonzales M. J., Kantor R., Betts B. J., Ravela J., Shafer R. W. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halvas E. K., Svarovskaia E. S., Pathak V. K. Role of murine leukemia virus reverse transcriptase deoxyribonucleoside triphosphate-binding site in retroviral replication and in vivo fidelity. J. Virol. 2000;74:10349–10358. doi: 10.1128/jvi.74.22.10349-10358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beerenwinkel N., Schmidt B., Walter H., Kaiser R., Lengauer T., Hoffmann D., Korn K., Selbig J. Diversity and complexity of HIV-1 drug resistance: a bioinformatics approach to predicting phenotype from genotype. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8271–8276. doi: 10.1073/pnas.112177799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Korolev S., Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 43.Astatke M., Ng K., Grindley N. D. F., Joyce C. M. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel P. H., Loeb L. A. DNA polymerase active site is highly mutable: evolutionary consequences. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5095–5100. doi: 10.1073/pnas.97.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel P. H., Loeb L. A. Multiple amino acid substitutions allow DNA polymerases to synthesize RNA. J. Biol. Chem. 2000;275:40266–40272. doi: 10.1074/jbc.M005757200. [DOI] [PubMed] [Google Scholar]
- 46.Patel P. H., Kawate H., Adman E., Ashbach M., Loeb L. A. A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity. J. Biol. Chem. 2001;276:5044–5051. doi: 10.1074/jbc.M008701200. [DOI] [PubMed] [Google Scholar]
- 47.Shinkai A., Loeb L. A. In vivo mutagenesis by Escherichia coli DNA polymerase I. J. Biol. Chem. 2001;276:46759–46764. doi: 10.1074/jbc.M104780200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.