Abstract
Cyclin A is regulated primarily through transcription control during the mammalian cell cycle. A dual mechanism of cyclin A transcriptional repression involves, on the one hand, promoter-bound inhibitory complexes of E2F transcription factors and RB (retinoblastoma) family proteins, and on the other, chromatin-directed histone deacetylase activity that is recruited to the cyclin A promoter early in the cell cycle in association with these RB proteins. This dual regulation maintains transcriptional silence of the cyclin A locus until its transcription is required in S-phase. At that time, RB family members dissociate from E2F proteins and nucleosomal restructuring of the locus takes place, to permit transcriptional activation and resultant S-phase progression to proceed. We have identified a double bromo-domain-containing protein Brd2, which exhibits apparent ‘scaffold’ or transcriptional adapter functions and mediates recruitment of both E2F transcription factors and chromatin-remodelling activity to the cyclin A promoter. We have shown previously that Brd2-containing nuclear, multiprotein complexes contain E2F-1 and -2. In the present study, we show that, in S-phase, they also contain histone H4-directed acetylase activity. Overexpression of Brd2 in fibroblasts accelerates the cell cycle through increased expression of cyclin A and its associated cyclin-dependent kinase activity. Chromatin immunoprecipitation studies show that Brd2 is physically present at the cyclin A promoter and its overexpression promotes increased histone H4 acetylation at the promoter as it becomes transcriptionally active, suggesting a new model for the dual regulation of cyclin A.
Keywords: bromodomain, cell cycle, chromatin, cyclin A, histone acetylase, transcription
Abbreviations: BrdU, bromodeoxyuridine; CBP, CREB-binding protein; cdk, cyclin-dependent kinase; ChIP, chromatin immunoprecipitation; CMV, cytomegalovirus; CREB, cAMP-response-element-binding protein; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; FBS, fetal bovine serum; HA, haemagglutinin; HAT, histone acetyltransferase; HDAC, histone deacetylase; PSG, penicillin, streptomycin and glutamine; RB, retinoblastoma; NP40, Nonidet P40; PKA, protein kinase A; WT, wild-type
INTRODUCTION
The transcriptional control of certain key genes that dictate the mammalian cell cycle has been studied with respect to (i) direct recruitment of sequence-specific DNA-binding transcription activator proteins and (ii) protein complexes that are not DNA sequence-specific and modify nucleosomes or remodel chromatin structure. The cyclin A locus, in particular, which exerts primary control over the progression of DNA synthesis in S-phase, is transcriptionally repressed by HDAC (histone deacetylase) activity until the advent of S-phase, when the gene is derepressed and transcribed [1–3]. The liberation of E2F-1, -2 and -3 from inhibitory complexes with RB (retinoblastoma) protein and its family members, late in G1 phase, is timed to make E2F proteins available to participate in the transcriptional activation of cyclin A [2,4–6] and other genes that are required in S-phase. These two functions of transcription factor-driven promoter transactivation and chromatin modification, at the proper time and promoter, are required for normal S-phase progression. Disturbances in the RB–E2F regulatory axis [7–9] and chromatin structure [10] have received attention recently as hallmarks of cell-cycle destabilization and oncogenic transformation in a wide variety of tissues.
This dual regulation is probably co-ordinated by transcriptional co-activator, adapter or ‘scaffold’ proteins that recruit the necessary biochemical factors [11]. Different types of massive, multicomponent protein complexes establish physical and functional links between sequence-specific DNA-binding proteins that are transcriptional activators, such as E2Fs, and the general transcription machinery, including RNA polymerase II and core promoter-bound factors [12]. These include, but are not limited to, the SAGA complex, which includes Gcn5, Ada1, Spt7 and Spt20 subunits and catalyses histone acetylation [13–15]; the Swi/Snf complex, an 11-subunit protein machine that increases chromatin accessibility through ATP hydrolysis [16–19]; and the Mediator complex [20–22]. A protein module called the bromodomain [23–26] has been noticed in connection with this co-activator role. This 110-amino-acid motif binds to acetyl-lysine groups that are present either in transcription factors such as Tat [27] or p53 [28] or in nucleosomal histones [29–31]. Structures have been solved for CBP [CREB (cAMP-response-element-binding protein)-binding protein] [28], TAFII250 [29], Gcn5 [30,32] and p/CAF [33] bromodomains. The bromodomain motif is present in authentic HATs (histone acetyltransferases), such as CBP/p300 [34,35], Gcn5 [36] and TAFII250 [37], as well as in acidic transcriptional activators such as Spt7 [38] (which along with Gcn5 participates in the SAGA complex [39]) and helicase superfamily members such as Snf2 and brahma [40] (which participate in the Swi/Snf complex). The bromodomain, which on its own does not encode HAT activity or other catalytic functions, finds its epitome in polybromo, which contains five bromodomains and appears to mediate interactions within co-activator complexes [41]. Some bromodomain-containing proteins, such as Rsc4, use additional domains to initiate contacts between chromatin and multiprotein chromatin-remodelling complexes, such as RSC [42]. These ‘scaffolding’ functions contribute to the epigenetic regulation of transcription through a mechanism called the histone code [43], wherein combinations of specific modifications of histones, such as acetylation, mediate the recruitment of diverse chromatin-remodelling machines [44].
Brahma contacts RB [45], which illuminates another aspect of chromatin status, namely transcriptional repression. RB and its family members p107 and p130 bind to E2F proteins and, as direct repressors, block E2F transcription activation function [2,6–9,46]. Indirectly, RB, p107 and p130 can also repress promoters through recruitment of HDAC activity [1,3,10,47–50], in concert with brahma function. The involvement of bromodomain-containing factors in both modes of chromatin remodelling, establishment of transcriptionally active euchromatin or transcriptionally silent heterochromatin, was first appreciated in yeast, wherein swi/snf mutations were observed to turn on as many genes as were turned off [51,52]. The transcriptional control of E2F-regulated cell-cycle genes is essential for proper progression through each stage of the cell cycle. Whereas transcriptional activation of one set of genes is necessary to enter a stage of the cell cycle, repression of certain other genes associated with the previous stage is necessary to exit from that stage [50]. The transcriptional machines that control these complex and timed processes of activation and repression at specific promoters are the focus of intensive investigation in recent works.
We have been investigating the structure and function of a double bromodomain protein called Brd2. We used a screen for novel, mitogen-responsive nuclear factors to purify an 85 kDa kinase from HeLa cell nuclear extracts, which was identified by microsequence analysis as RING3, and later renamed as Brd2 [53–55]. In studies to characterize Brd2, we used an antibody against recombinant Brd2 protein to immunoprecipitate the original activity from nuclear extracts and established that the cDNA encoded the appropriate activity in transfected cells. Furthermore, site-specific mutation of the putative active site Lys-574 to alanine destroyed kinase activity, which collectively supported the conclusion that the 85 kDa kinase was Brd2 [55]. Indirect immunofluorescence with the anti-Brd2 antibody confirmed nuclear localization of Brd2 protein in exponentially growing HeLa cells [55]. Brd2 nuclear localization is inducible in serum-starved fibroblasts by mitogenic stimulation and relies on a simple nuclear localization motif [56]. As expected, nuclear localization mutants fail to activate transcription [56]. Brd2 overexpression in NIH/3T3 cells causes foci to form synergistically with oncogenic ras [57], and lymphoid-restricted constitutive expression of Brd2 in transgenic mice increases cyclin A transcription and causes B cell leukaemia and lymphoma [58]. Taken together, these features suggest that this nuclear-localized and mitogen-inducible activity has a functional link to cell-cycle control and oncogenic transformation.
In particular, we have hypothesized that the Brd2-driven, deregulated transcription of cyclin A in Eμ-BRD2 transgenic B cells may provide a mechanism for the oncogenic properties of constitutively expressed Brd2 [58]. In tissue culture experiments with transfected overexpression and reporter constructs, we found that Brd2 synergizes with oncogenic ras or certain ras effectors to transactivate a promoter-luciferase construct of cyclin A [57]. Overexpressed Brd2 transactivates several other E2F-regulated promoters of cell-cycle genes in synergy with oncogenic ras; furthermore, functional cis-acting E2F-binding sites are required for Brd2-dependent transcriptional function [57]. In addition, co-expression of WT (wild-type) RB protein, which is well known to halt E2F-dependent cell-cycle progression [6], ablates this Brd2-dependent transactivation [57]. Our observation that E2F-1 and -2 are present in Brd2 multiprotein complexes purified from nuclear extracts suggested that Brd2 might participate as a transcriptional co-activator in the dual regulation of cyclin A transcription described above.
The ras proteins are key mediators of the early cellular response to mitogens. Evidence suggests that ras-mediated mitogenic signalling promotes cell proliferation through the E2F/RB axis that mobilizes cyclin/cdk (cyclin-dependent kinase) activity in the G1 phase of the cell cycle. Both E2F and cyclin E/cdk2 are important downstream effectors of ras [59]. Ras activity is essential for the normal induction of cyclin A promoter activity [60] and ras inactivation blocks the normal serum-dependent accumulation of E2F, preventing the increase of cyclin A mRNA levels in vascular smooth-muscle cells [60]. Furthermore, E2F binding to the cyclin A promoter is necessary for serum responsiveness of cyclin A expression in NIH/3T3 fibroblasts [57]. These observations are the basis for the interest in ras-activated signal transduction pathways that link environmental mitogenic stimuli to cell-cycle regulators [61], particularly the less-well-understood coupling of ras and serum mitogenic signalling to cyclin A transcriptional regulation. The role of promoter histone acetylation and chromatin remodelling in such signalling-coupled processes has not been sufficiently studied.
Brd2 is a member of the BET family of proteins [62], defined by tandem bromodomains that are highly related to each other and an ET domain. Brd2 does not bind naked DNA directly (G. V. Denis, unpublished work), but the Brd2 bromodomains have been shown in vivo to bind acetylated histone H4 selectively [63]. Brd2 is homologous with the Drosophila homoeotic regulator fsh (female sterile homoeotic), which has chromatin modification functions [62,64], and is also related to mammalian TAFII250 [53], an authentic HAT [37] and scaffold for TFIID [65] that preferentially binds to acetylated histone H3 [63]. In yeast, Bdf1 [66,67], a homologue of Brd2, and Taf145 together serve a functional role similar to TAFII250 in mammals [68]; Bdf1 has been suggested to play a role in chromatin restructuring [31] and binds histones H3 and H4 in vitro [69], with a preference for acetylated histones [31]. The BET protein Brd4, also called MCAP, is essential to cell growth control and probably plays a role at the G2→M phase transition [70–72] in association with acetylated chromatin, particularly through binding to acetylated histone H4 [73]. Therefore BET proteins may be implicated in transcriptional control through histone modifications and chromatin remodelling. On the basis of these considerations, we hypothesized that, in addition to E2F proteins, the Brd2 multiprotein complex harbours HAT activity and that Brd2 bromodomain function is required for the observed Brd2-driven deregulation of cyclin A transcription. We also sought a mechanism to explain the ability of both WT Brd2 and a kinase-null point mutant to cause B cell leukaemia and lymphoma when constitutively expressed in the B lymphoid lineage of transgenic mice. The experiments we describe herein attempt to integrate what is already known about mitogenic signal transduction, histone acetylation and chromatin remodelling and control of the cell cycle, using transcriptional regulation of cyclin A as the focal point.
EXPERIMENTAL
Molecular cloning
HA (haemagglutinin)-tagged constructs of WT and KA mutant Brd2 are described in [55,56]. All constructs used herein contained engineered consensus Kozak sequences and encoded an in-frame, N-terminal HA epitope. The ΔBD mutant was constructed by internal deletion of the bromodomains, with a reverse mutagenic primer 5′-GCGCGCGGTACCAAGTTGCAAAGCAGGCACCGAAGCCAT-3′ (which introduced a KpnI site after Gln-8 in the primary sequence) and a forward mutagenic primer 5′-GCGCGCGGTACCCCAATATCCAAGCCCAAGAGGAAAAGAGAG-3′ (which introduced a KpnI site after Gly-485). KpnI digestion and internal ligation achieved in-frame deletion and preserved the nuclear localization sequence. The ΔE2F mutant was constructed by translation termination of the protein just after the nuclear localization sequence at Lys-510 with the mutagenic primer 5′-CGCGCGGAATTCTTAGCCTCGATGCTTCTCTGCCTTCCGTTTCTTCTTTTTAGAGACTGGTAAAGGCCCTGGTTCTAGTGGTTC-3′. The ΔBD/KA double mutant was constructed by PCR mutagenesis of the published KA mutant [56] with the ΔBD deletion primers mentioned above. Recombinant human E2F-1 was cloned into the RSET system (Invitrogen, Carlsbad, CA, U.S.A.) for expression of His6-tagged recombinant protein in the BL21(DE3)(pLysE) strain of Escherichia coli (Invitrogen). Protein was purified by affinity chromatography on an Ni+ agarose column (Qiagen, Valencia, CA, U.S.A.). Epitope-tagged Brd2 constructs were cloned into the EcoRI and BamHI sites of the CMV (cytomegalovirus)-directed expression plasmid pAC-CMV for high-level expression of cDNA. Restriction mapping, sequencing and Southern blotting were used to verify all constructs. Recombinant viruses were generated in 293T cells with co-transfection of pAC-CMV plasmids [74] and pJM17 [75], encoding full-length adenovirus 5 genome without E3, and purified from cell lysates [74]. Control adenovirus was generated from a pAC-CMV plasmid without a cDNA insert.
Cell culture and transfection
Low passage NIH/3T3 cells (A.T.C.C., Manassas, VA, U.S.A.) were grown in DMEM (Dulbecco's modified Eagle's medium; Invitrogen) supplemented with 10% normal calf serum and PSG (penicillin, streptomycin and glutamine), maintained at less than 75% confluence and transfected in triplicate in 60 mm dishes at 50% confluence with plasmids by the Lipofectin® method (Invitrogen), then harvested at 100% confluence after 24–36 h. Transfections contained 3.0 μg of DNA/dish, comprised of 1.0 μg of Brd2 construct, 0.5 μg of cyclin A promoter luciferase reporter and 0.5 μg of Ki-ras expression vector, and were balanced with empty pcDNA(I) vector (Invitrogen). Normalization of luciferase activity was initially performed with reference to units of expressed β-galactosidase from a co-transfected vector and, after high reproducibility had been established, performed against protein concentration in the cell lysates. Extracts were prepared in reporter lysis buffer and luciferase activity was determined with an assay kit (Promega, Madison, WI, U.S.A.) and a luminometer. 293T cells were maintained in DMEM supplemented with 10% (v/v) FBS (fetal bovine serum; Invitrogen) and PSG and were maintained at 75% confluence for propagation of adenoviruses. Rat1 cells (A.T.C.C.) were cultured in DMEM supplemented with 10% FBS and PSG, and infected with purified adenoviruses after 36 h serum starvation in 10 cm dishes at 90% confluence.
Northern-blot and immunoblot analyses
RNA was purified from 107 adenovirus-infected Rat1 cells, using the guanidine/sodium lauryl sarcosine method, resolved in formaldehyde gels, blotted on to nitrocellulose and probed with a radiolabelled cDNA probe for Brd2 in Express Hyb solution (Clontech BD Biosciences, San Jose, CA, U.S.A.). Cell extracts were prepared as described in [55,57] for immunoblotting. Briefly, total cell extracts were prepared on ice by detergent lysis of PBS-washed monolayers in ice-cold buffer A [50 mM disodium glycerol 2-phosphate, 20 mM Tris/HCl (pH 7.5), 10 mM KCl, 1 mM MgCl2, 1 mM Na3VO4, 1 mM DTT (dithiothreitol), 0.2 mM EDTA and 0.1% NP40 (Nonidet P40), supplemented with PMSF, pepstatin, aprotinin and leupeptin; Calbiochem, EMD Biosciences, San Diego, CA, U.S.A.] to produce a cytosolic extract. Nuclei and cell fragments were subsequently extracted for 30 min with ice-cold buffer B [420 mM NaCl, 50 mM disodium glycerol 2-phosphate, 20 mM Tris/HCl (pH 7.5), 10% (v/v) glycerol, 1 mM Na3VO4, 1 mM DTT, 0.2 mM EDTA and supplemented with the same protease inhibitors]. Cytosolic and nuclear supernatants were pooled and insoluble materials were removed by additional centrifugation at 4 °C. Proteins were resolved by SDS/PAGE (12% gel), blotted on to PVDF membranes (Bio-Rad Laboratories, Richmond, CA, U.S.A.), blocked with 5% (w/v) non-fat dry milk in TBST [100 mM NaCl, 10 mM Tris/HCl (pH 8.0), 5 mM 2-mercaptoethanol, 1 mM EDTA and 0.1% Tween 20] and probed with 1:1500 (v/v) mouse monoclonal antibody to the HA epitope, clone HA.11 (Covance, Berkeley, CA, U.S.A.), or 1:2000 (v/v) rabbit polyclonal antibodies, either to cyclin A (1:250, H-432), cyclin D1 (1:500, H-295; both from Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), cyclin E (1:200) or cyclophilin B (1:2000, both from AbCam, Cambridge, MA, U.S.A.), which were detected with 1:7500 (v/v) anti-rabbit or anti-mouse secondary antibody directed against immunoglobulin heavy and light chains, conjugated with horseradish peroxidase (Roche, Indianapolis, IN, U.S.A.). Secondary antibody was visualized with a chemiluminescent reagent system (PerkinElmer Life Sciences, Boston, MA, U.S.A.) and an X-Omat Blue XB-1 film (Eastman Kodak, Rochester, NY, U.S.A.).
Immunoprecipitation and in vitro phosphorylation
The amount of recombinant protein in 107 infected Rat1 cell extracts was determined by anti-HA immunoblotting and the amount of extract used was normalized to the amount of each derivative. For immunoprecipitation, extracts were diluted with ice-cold buffer A, precleared with Protein A/G–agarose (Santa Cruz Biotechnology) and incubated overnight at 4 °C with gentle agitation with 20 μg of anti-HA epitope antibody. Immunocomplexes were collected and washed three times with ice-cold RIPA buffer [200 mM NaCl, 50 mM Tris/HCl (pH 7.5), 50 mM disodium glycerol-2-phosphate, 1 mM DTT, 1 mM EDTA, 1% NP40 and 0.1% SDS] and once with buffer A by microcentrifugation in the cold. In-gel autophosphorylation in E2F-1-containing polyacrylamide gels was performed by denaturation of proteins in guanidine and renaturation overnight, followed by autophosphorylation [55]. Phosphorylation in solution was performed at 30 °C for 15 min with 50 μg of recombinant E2F-1 substrate in 0.5 ml of buffer A supplemented with 100 mM NaCl, 5 mM MgCl2, 0.5% Tween 20, 1 mM DTT, 0.5 mM EDTA, 50 μM disodium ATP and 10 μCi of [γ-32P]ATP (3000 Ci/mmol; New England Nuclear, Boston, MA, U.S.A.), PMSF, leupeptin, pepstatin and aprotinin, but without Na3VO4. The catalytic subunit of PKA (protein kinase A) and all other reagents unless otherwise specified were from Sigma (St. Louis, MO, U.S.A.). Phosphorylated proteins were precipitated by ice-cold 10% (w/v) trichloroacetic acid and washed with ice-cold acetone, then solubilized with SDS sample buffer for PAGE on 12% gels. Gels were stained with Coomassie Brilliant Blue, destained in acetic acid and methanol, dried and subjected to autoradiography.
Co-immunoprecipitation was performed with three washes of cyclin A immunocomplexes in ice-cold buffer A (supplemented with 50 mM NaCl, 5 mM disodium ATP, 2 mM MgCl2, 1 mM EDTA and 0.1% Tween 20, but without KCl or NP40) and cyclin A-associated kinase activity was assayed with a cyclin-dependent kinase assay kit (Upstate Biotechnology, Lake Placid, NY, U.S.A.) and [γ-32P]ATP (New England Nuclear).
Cell-cycle kinetics
Rat1 cell monolayers at 90% confluence in 10 cm dishes were starved for 36 h in DMEM containing 0.2% FBS and PSG, then exposed for 6 h to fresh media containing 0.5% FBS, PSG, 0.4 μg/ml methyl-[5-(2-thienylcarbonyl)-1H-benzimidazol-2-yl]-carbamate (nocodazole; Calbiochem, EMD Biosciences) and 1011 plaque-forming units of recombinant adenovirus, which encoded HA-epitope-tagged Brd2 or empty vector control. Cells were then stimulated with FBS to a final concentration of 3.5% to initiate cell-cycle entry, which marked the zero time point. Cells were harvested by trypsinization at different times after serum stimulation, washed with serum-free DMEM and fixed overnight in 70% (v/v) ethanol. The fixed cells were washed with PBS and stained with 18 μg/ml propidium iodide in the presence of RNase A (American Bioanalytical, Natick, MA, U.S.A.) and then analysed with a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ, U.S.A.) for the determination of DNA content. Alternatively, cells were pulse-labelled with 10 μM BrdU (bromodeoxyuridine; Becton Dickinson) for the final 60 min before harvest, fixed with paraformaldehyde, permeabilized with saponin, treated with DNase I and then stained for incorporated BrdU with FITC-conjugated anti-BrdU antibody and for DNA content with 7-aminoactinomycin D (Becton Dickinson). Analysis of 20000 events was performed with Cell Quest and Windows Multiple Document Interface version 2.8 (Becton Dickinson) and Summit version 3.1 software (Cytomation, Fort Collins, CO, U.S.A.) for flow cytometry.
ChIP (chromatin immunoprecipitation)
Rat1 cells were infected as described above, stimulated with 3.5% FBS; then, 8 or 18 h later (for G1 phase or mid-S-phase experiments respectively), cells were incubated with 1% formaldehyde for 10 min to cross-link recombinant proteins. Then, after the reaction was quenched with 125 mM glycine (final concentration), cells were scraped into ice-cold PBS supplemented with protease inhibitors, microcentrifuged at 4 °C and resuspended in ice-cold ChIP lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris/HCl, pH 8.0), supplemented with protease inhibitors. After 10 min on ice, the mixture was diluted 2.5-fold with ice-cold ChIP dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris/HCl (pH 8.0) and 16.7 mM NaCl], supplemented with protease inhibitors. Chromatin was then sheared with a 550 Sonic Dismembrator probe sonicator (Fisher Scientific, Morris Plains, NJ, U.S.A.) and samples were maintained on ice. In preliminary experiments, we determined that three pulses of 10 s each at 18% power gave optimal shearing of DNA (a median size of 500 bp fragments). The sonicated material was microcentrifuged at 16000 g for 10 min at 4 °C to remove debris and 5% (v/v) of the supernatant was saved as ‘input’chromatin. The chromatin solution was precleared with BSA and salmon sperm DNA/Protein A–agarose (Upstate Biotechnology) and then chromatin was immunoprecipitated by overnight incubation at 4 °C with specific antibodies, either mouse monoclonal antibody against the HA epitope (Covance) or high-affinity, biotinylated rat monoclonal antibody (3F10) against acetylated histone H4 (Roche). Immunocomplexes were collected with Protein A–agarose or streptavidin–agarose (Upstate Biotechnology) respectively in the presence of salmon sperm DNA (Promega) and BSA, washed once with each of buffer 1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl and 20 mM Tris/HCl, pH 8.0), buffer 2 (same as buffer 1, but in 500 mM NaCl), buffer 3 (0.25 M LiCl, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris/HCl, pH 8.0) and then twice with 10 mM Tris/HCl, pH 8.0 and 1 mM EDTA. Non-specific control immunoprecipitations were performed with rabbit IgG–agarose under the same conditions. Chromatin immunoprecipitated products were eluted with 0.1 M NaHCO3 and 1% SDS, cross-links were reversed at 65 °C for 6 h, RNA was digested with RNase A, protein was digested with proteinase K and DNA was extracted with phenol (American Bioanalytical) and chloroform (1:1, v/v). The purified DNA was precipitated from ethanol with NaCl and glycogen (Roche), dissolved in water and quantified with PicoGreen (Molecular Probes, Eugene, OR, U.S.A.). Input and immunoprecipitated DNAs were quantified with SYBR Green reagents in a Mx3000P real-time PCR system (Stratagene, La Jolla, CA, U.S.A.) with primers to the Rattus norvegicus gene cyclin A2, 5′-TGAAGGCCGGGAAGGTGC-3′ and 5′-CTCATCGTTTATAGGAAGGTCC-3′, to give an 89-bp amplicon that begins at nt 116 of the open reading frame, which we determined in preliminary experiments to give optimal amplification of the template.
Histone acetylation
Rabbit anti-Brd2 co-immunocomplexes were assayed for the associated histone acetylases [76] by incubation at 30 °C for 30 min in a 30 μl reaction, either with 10 μg of mixed calf thymus histones or with 10 μg of each core histone (Roche) and 0.25 μCi of [14C]acetyl-CoA (59 mCi/mmol; Amersham Biosciences, Piscataway, NJ, U.S.A.) in 50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 10 mM sodium butyrate, 0.5 mM DTT, 0.2 mM PMSF and 0.1 mm EDTA. The reaction was quenched by centrifugation at 4 °C. Histones in the supernatant were precipitated with trichloroacetic acid and recovered on GF/C filters (Whatman, Clifton, NJ, U.S.A.). Incorporation of radioactivity was determined with a scintillation counter or by autoradiography. Anti-HA immunocomplexes were collected from lysates of adenovirus-infected cells and washed under co-immunoprecipitation conditions (three washes with ice-cold buffer A, supplemented with 50 mM NaCl, 5 mM disodium ATP, 2 mM MgCl2, 1 mM EDTA and 0.1% Tween 20) and used to acetylate histones as above. The histones were recovered with trichloroacetic acid precipitation, dissolved in urea and resolved in 20% polyacrylamide gel that had been polymerized in the presence of 10 mM Triton X-100. Histone separation was accomplished with electrophoresis in a urea/acetic acid system and visualization with Coomassie Blue stain or En3Hance (New England Nuclear) and autoradiography.
RESULTS
Ectopic expression of Brd2 transactivates the cyclin A promoter
Evidence that Brd2 synergizes with ras and certain ras effectors to transactivate the cyclin A promoter, among other E2F-dependent promoters of cell-cycle-regulatory genes [57], motivated the experiments described herein to measure Brd2-dependent cell-cycle progression. Published results indicate that Brd2 possesses functional domains responsible for association with chromatin (its double bromodomains), an ET domain, a nuclear localization signal, a kinase domain and a C-terminal domain, which accounts for Brd2 association with nuclear multiprotein complexes that contain E2F proteins. We constructed mutants to explore the domains that account for Brd2-dependent transactivation function in the cyclin A model system. We generated one mutant that harbours a deletion of the N-terminal bromodomains (ΔBD) and another that harbours a deletion of the C-terminal E2F association domain and kinase domain (ΔE2F), as schematically shown in Figure 1(A). All constructs retained the nuclear localization motif that we defined [56] and were tagged at the N-terminus with the HA epitope, which does not interfere with transcriptional activation or nuclear localization [56]. These mutants are shown in comparison with the full-length WT protein and a full-length mutated protein that contains a point mutation of the putative catalytic Lys-574 to alanine (KA), which we previously showed eliminates intrinsic kinase activity [55].
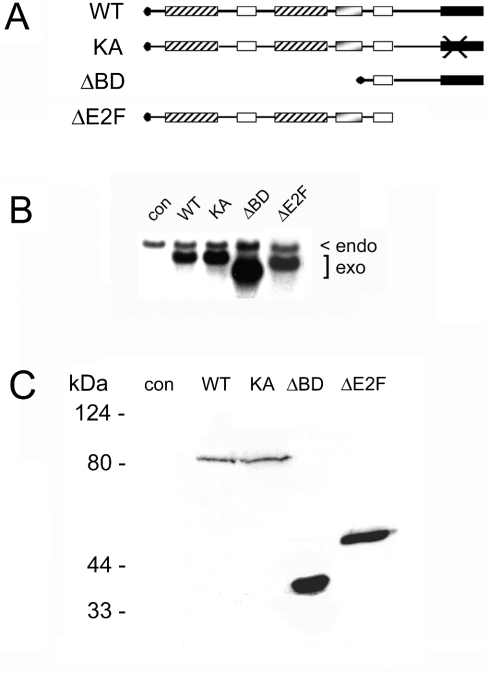
Figure 1. Brd2 derivatives and expression.
(A) Brd2 domains and deletions. Schematic shows the N-terminal HA epitope tag (●), dual, mutually related bromodomains (cross-hatched boxes), positively charged domains (open boxes), negatively charged domain (shaded box) and the C-terminal kinase domain that mediates interaction with E2F protein-containing complexes (solid box). The second positively charged domain contains the nuclear localization sequence. Point mutation of Lys-574 to alanine is indicated by a large X. (B) Northern-blot analysis of adenovirus expression of Brd2. Rat1 cells were infected with control (con) or Brd2-expressing adenovirus constructs (WT, KA, ΔBD, ΔE2F) and stimulated with serum. The Brd2 probe detects endogenous Brd2 mRNA (endo) and exogenous, adenovirus-directed expression of mRNA (exo). (C) Immunoblot analysis of expression of constructs. Extracts of infected cells were probed with mouse anti-HA monoclonal antibody and detected with rabbit anti-mouse secondary antibody conjugated to horseradish peroxidase.
We chose Rat1 cells for cell-cycle studies because they are relatively normal fibroblasts and are easily transfected or infected with adenovirus 5 [74]. We used adenovirus 5 to deliver CMV promoter-driven Brd2 or its derivatives. Quiescent, serum-starved Rat1 cells were infected with Brd2-expressing adenovirus or control adenovirus and stimulated with serum to initiate cell-cycle entry. Vector-encoded mRNA expression began almost immediately after infection and was significantly stimulated after the addition of serum. Under these conditions, >99% of the cells in the population became infected with adenovirus and expressed an adenovirus-encoded green fluorescent protein (results not shown), so that statistically each group of infected cells could be treated as a population for the purposes of cell-cycle analysis. Northern-blot analysis of Brd2 mRNA expression is shown in Figure 1(B). Expression is shown after 8 h of serum stimulation for endogenous (endo) and exogenous, adenovirus-encoded (exo) Brd2 mRNAs of the correct size: 2.26 kb for WT and KA, 0.89 kb for ΔBD and 1.55 kb for ΔE2F. The cells were infected either with empty vector adenovirus (con) or with adenovirus that encoded full-length WT Brd2 (WT), the full-length K574A (Lys574→Ala) derivative (KA) [55–57] or the two smaller ΔBD and ΔE2F derivatives. The adenovirus-encoded mRNAs exhibit greater apparent mobility in formaldehyde-containing denaturing gels compared with endogenous mRNAs. Protein expression was also confirmed for each of the expression vectors with anti-HA epitope immunoblotting of infected extracts, as shown in Figure 1(C).
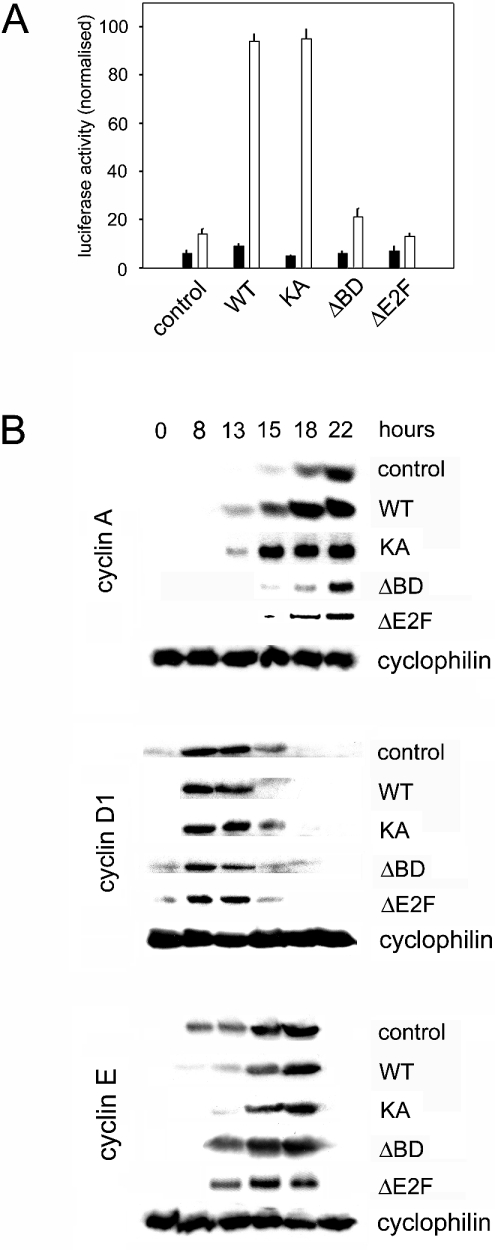
Next, we tested whether these mutations had functional consequences for Brd2-dependent transcriptional activation. We have demonstrated previously in NIH/3T3 cells that full-length recombinant Brd2 (WT) transactivates luciferase promoter constructs of cyclin A in concert with oncogenic ras or certain ras effectors [57]. In the present study, we extended the analysis with the same functional assay, transfecting each CMV promoter-driven Brd2 plasmid construct into NIH/3T3 cells and assaying its ability to transactivate a cyclin A promoter luciferase reporter construct. Consistent with our previous report and as shown in Figure 2(A), ras co-expression synergized with WT Brd2 to stimulate transcriptional activation of the cyclin A promoter (open bars). KA Brd2 produced strong activation with co-expressed ras, equal to WT Brd2. Without ras, WT Brd2 did not activate transcription (Figure 2A, closed bars), also in agreement with previous results [57], and the same was true for KA Brd2. Interestingly, mutation of the active-site lysine residue of the WT kinase to alanine in the KA derivative did not alter its ability to synergize with ras and transactivate the cyclin A luciferase reporter, suggesting that intrinsic kinase activity is not important for Brd2 transactivation function of cyclin A under these conditions. None of the other domain mutants transactivated the cyclin A promoter as well as did WT or KA Brd2. These results were consistent with our published observation that transgenic mice that constitutively express in the B cell lineage either WT Brd2 (Tg) or KA Brd2 (Tgmut) both develop an apparently identical B cell lymphoma and leukaemia at the same rate [58], suggesting that intrinsic Brd2 kinase activity is not essential for lymphomagenesis, but rather that some other property of Brd2 is more important for oncogenic transformation [58].
Figure 2. Brd2-dependent transcriptional activation.
(A) NIH/3T3 cells were transfected with a luciferase reporter for the cyclin A promoter and WT or mutants of Brd2 either alone (closed bars) or with Ki-ras (open bars). Luciferase units were normalized for protein (n=2). (B) Quiescent, serum-starved Rat1 cells were infected with either empty-vector adenovirus (control) or adenovirus constructs that expressed WT Brd2 protein (WT), or mutants (KA, ΔBD or ΔE2F). Cell-cycle entry was initiated with addition of serum. Cell extracts recovered at progressive times after the addition of serum (hours) were assayed by immunoblot for cyclin A, cyclin D1 or cyclin E protein expression. Cyclophilin B was assayed as a loading control. Only one of the loading controls is shown for each of the cyclin immunoblots; loading was confirmed to be equal in all experiments (results not shown).
Brd2-dependent cell-cycle acceleration
We have shown previously that, in addition to co-expression of activated ras, serum also provides a mitogenic signal that works through Brd2 to stimulate E2F-dependent transcription [57], suggesting that Brd2 might participate in multiprotein complexes as a general effector of mitogenic signals, as has been suggested for Mediator transcription [21,22], coupling these signals to the cell-cycle machinery. On the basis of the results of Figure 2(A) and our previously published observations, we hypothesized that transcriptional up-regulation of the endogenous cyclin A gene might occur in a Brd2-dependent manner, which might contribute to Brd2-dependent cell-cycle progression. We therefore tested the ability of Brd2 constructs to accelerate the Rat1 cell cycle after serum starvation and cell-cycle re-entry upon serum stimulation of the quiescent cells. As shown in Figure 2(B), immunoblotting of endogenous cyclin A protein revealed that the timing of its synthesis was accelerated in response to overexpressed WT and KA Brd2, in agreement with the transcriptional activation of the cyclin A luciferase reporter data in Figure 2(A). ΔBD and ΔE2F Brd2 gave the same result as the negative control, suggesting that these derivatives had no transcriptional activity on the endogenous gene. Early in the cell cycle, cyclin A protein accumulation was detected in cells infected with WT and KA Brd2-expressing adenoviruses, but not with ΔBD Brd2, ΔE2F Brd2 or control adenoviruses. However, cyclin A accumulation was detected later in all cases, consistent with normal S-phase progression; by 22 h after serum stimulation, the levels were similar across all the cases. Cyclin A expression normally increases throughout the S-phase and reaches a peak during late S/G2; cyclin A is known to play an important and essential role in both the G1→S transition and S-phase progression. We did not continue to measure the accumulation of cyclin A protein beyond 22 h since the cell-cycle experiments were conducted in the presence of the mitotic inhibitor nocodazole, which trapped the cells in M-phase approximately 24 h after serum stimulation. We included this trap to check whether some of the cells completed the cell cycle and re-entered the G1 phase, which would have affected the cell counts for G1 and G2/M compartments. Overall, these results suggested that Brd2 might function at least in part on a pathway that targets cyclin A as a transcriptional endpoint and that S-phase events might be affected by Brd2 expression.
Intrinsic kinase activity
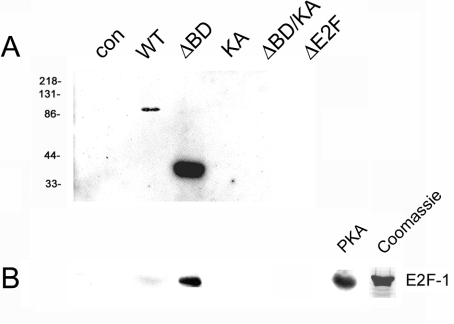
We demonstrated previously that mutation of the putative catalytic Lys-574 to alanine (K574A) eliminates intrinsic kinase activity in bacterially expressed, recombinant protein that had been modified with HeLa nuclear extract and ATP [55]. To determine whether the bromodomains could regulate kinase activity, we used the ingel kinase assay to test each of the present derivatives for in vitro kinase activity after anti-HA immunoprecipitation from transfected 293T cells. Surprisingly, we observed that the ΔBD mutant had a high level of autophosphorylation activity after immunoprecipitation from transfected 293T cells, compared with the same amount of WT protein, as shown in Figure 3(A). Deletion of the C-terminal E2F-association and kinase domain (ΔE2F) produced a mutant with no detectable kinase activity, as expected, similar to the KA mutant. We hypothesized that deletion of the bromodomains in the ΔBD mutant had removed an autoinhibitory domain from the kinase and produced a constitutively active mutant. To confirm that the putative catalytic Lys-574 was responsible for this activity, we generated a bromodomain deletion/K574A double mutant (ΔBD/KA) and tested its ability to autophosphorylate under the same conditions. As predicted, this mutant had no detectable autophosphorylation activity, as shown in Figure 3(A).
Figure 3. Phosphorylation of E2F-1 by Brd2.
(A) Autophosphorylation in vitro by in gel kinase assay of the HA-tagged Brd2 derivatives WT, ΔBD, KA, ΔBD/KA or ΔE2F. Derivatives were recovered by immunoprecipitation with anti-HA monoclonal antibody from infected cells, resolved in an SDS-containing polyacrylamide gel, denatured with guanidine, renatured with buffer and incubated with [γ-32P]ATP for autophosphorylation [55]. (B) Recombinant E2F-1 was phosphorylated in vitro with the same derivatives, then resolved by SDS/PAGE and visualized with autoradiography. As a positive control, E2F-1 was phosphorylated under the same conditions with the catalytic subunit of PKA. Coomassie Blue-stained E2F-1 protein is shown as the loading control.
Our previous observation that Brd2 exists in a multiprotein complex that contains E2F proteins [57] suggested that these proteins might be substrates for Brd2 kinase activity. Accordingly, we expressed His6-tagged recombinant human E2F-1 protein in bacteria and purified it for use as a kinase substrate in an in vitro phosphorylation assay. We used an anti-HA antibody to immunoprecipitate recombinant WT, KA, ΔBD and ΔE2F HA–Brd2 proteins from transfected 293T cells to determine if they could phosphorylate E2F-1. We observed that WT protein exhibited very weak phosphorylation of E2F-1 and ΔBD exhibited significant E2F-1 phosphorylation (Figure 3B), consistent with the strong autophosphorylation activity of immunoprecipitated ΔBD protein (Figure 3A). As a positive control, we showed that the catalytic subunit of PKA robustly phosphorylated E2F-1 protein in vitro (Figure 3B). We confirmed that the K574A mutation also eliminated the E2F-1 phosphorylation activity in the ΔBD/KA mutant isolated from transfected 293T cells. Whereas ΔBD Brd2 catalysed the incorporation of 49.4 nmol of PO43−·min−1·mg−1 (±0.5; n=3) into E2F-1, ΔBD/KA Brd2 incorporated only 0.14 nmol of PO43−·min−1·mg−1 (±0.1; n=3). Background phosphorylation in the absence of Brd2 was <0.05 nmol of PO43−·min−1. These observations confirm that Brd2, similar to TAFII250 [29], possesses intrinsic kinase activity.
ΔBD and ΔE2F functional mutants
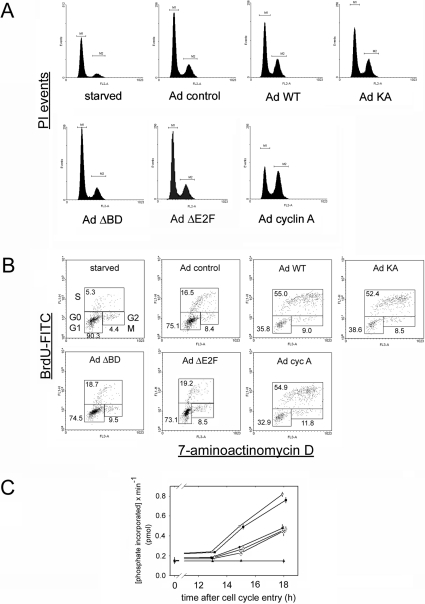
The observed Brd2-dependent transcriptional activation of the cyclin A locus suggested that expression of these derivatives should create forward pressure on the cell cycle. We therefore performed an analysis of cell-cycle kinetics with each of the derivatives. Rat1 cells infected with adenovirus that overexpresses WT or KA Brd2 showed greater cell-cycle progression compared with cells infected with the same MOI (multiplicity of infection) of empty-vector control adenovirus as shown in Figure 4(A). As a positive control, cells infected with the same MOI of adenovirus that overexpresses cyclin A showed cell-cycle progression beyond that observed with Brd2 alone. Consistent with transcriptional activation data, expression of the ΔBD and ΔE2F derivatives did not accelerate the cell cycle beyond the control. Therefore deletion of the bromodomains ablates the cell-cycle acceleration phenotype even as it converts the protein into a constitutively active kinase. To obtain more detailed information about the cell-cycle kinetics under the influence of each of the adenovirus constructs, we performed an analysis of BrdU incorporation during cell-cycle progression. Figure 4(B) shows that, at 18 h after serum stimulation to initiate cell-cycle entry, cells infected with adenoviruses that encode WT and KA constructs exhibited greater DNA synthesis compared with cells infected with control, ΔBD or ΔE2F adenovirus constructs. As a negative control, starved cells exhibited only marginal BrdU labelling and, as a positive control, cells infected with the cyclin A-expressing adenovirus showed significant BrdU incorporation. The complete cell-cycle kinetics are summarized for a representative experiment in Table 1.
Figure 4. Effects of Brd2 expression on cell cycle.
(A) Cell-cycle profiles. Serum-starved, quiescent Rat1 cells were infected with empty vector adenovirus (Ad control), adenovirus that expresses WT Brd2 (Ad WT), kinase mutant Brd2 (Ad KA), bromodomain-deleted Brd2 (Ad ΔBD), E2F-complex association domain-deleted Brd2 (Ad ΔE2F), or murine cyclin A (Ad cyclin A). Cells were stimulated with 3.5% serum to initiate cell-cycle entry, then harvested by trypsinization 18 h after serum stimulation, fixed with 70% ethanol, stained with propidium iodide and subjected to FACS for measurement of DNA content. Gated events (20000) were recorded on the FL2 channel of the cytometer with the detector in linear mode. (B) Complete cell-cycle kinetics. BrdU-stained cells in G0/G1, S and G2/M phases of the cell cycle were defined by electronic gating. Distributions are shown for the 18 h time point. BrdU incorporation was detected with FITC-conjugated antibody (BrdU-FITC) and was compared with DNA content detected with an intercalating agent (7-aminoactinomycin D). Percentages of gated events are shown. (Table 1 summarizes kinetics for 8, 13 and 18 h time points.) (C) Cyclin A-associated cdk activity. Cell extracts were assayed for cyclin A-associated cyclin-dependent kinase activity as determined by the histone H1 kinase activity in cyclin A co-immunoprecipitated immunocomplexes at indicated times after serum stimulation. Standard deviations for n=3 are shown. ●, control; ∇, WT; ■, KA; ◇, ΔBD; ×, ΔE2F; ▲, preimmune serum immunoprecipitation.
Table 1. Cell-cycle kinetics.
Results represent the percentage of gated cells.
| Phase | |||
|---|---|---|---|
| G0/G1 | S | G2/M | |
| Starved | 90.3 | 5.3 | 4.4 |
| 8 h after the addition of serum | |||
| Ad control | 88.8 | 5.7 | 5.5 |
| Ad Brd2 (WT) | 88.1 | 5.4 | 6.5 |
| Ad Brd2 (KA) | 89.8 | 4.8 | 5.4 |
| Ad Brd2 (ΔBD) | 89.6 | 5.2 | 5.2 |
| Ad Brd2 (ΔE2F) | 88.5 | 5.5 | 5.1 |
| Ad cyclin A | 85.1 | 9.0 | 5.9 |
| 13 h after the addition of serum | |||
| Ad control | 78.8 | 11.7 | 9.5 |
| Ad Brd2 (WT) | 78.4 | 11.4 | 10.5 |
| Ad Brd2 (KA) | 77.1 | 12.5 | 10.4 |
| Ad Brd2 (ΔBD) | 79.6 | 10.2 | 10.2 |
| Ad Brd2 (ΔE2F) | 78.2 | 11.0 | 10.1 |
| Ad cyclin A | 56.0 | 21.1 | 22.9 |
| 18 h after the addition of serum | |||
| Ad control | 75.1 | 16.5 | 8.4 |
| Ad Brd2 (WT) | 35.8 | 55.0 | 9.0 |
| Ad Brd2 (KA) | 38.6 | 52.4 | 8.5 |
| Ad Brd2 (ΔBD) | 74.5 | 18.7 | 9.5 |
| Ad Brd2 (ΔE2F) | 73.1 | 19.2 | 8.5 |
| Ad cyclin A | 32.9 | 54.9 | 11.8 |
The activity of cyclin/cdk complexes drives cell-cycle progression and attenuation of cdk2 activity arrests cells [77]. We therefore expected to find a functional link between Brd2-dependent increase in cyclin A protein levels and increased cell-cycle progression, in the form of increased cyclin A-associated cdk2 activity [78,79]. Extracts of cells treated as above were immunoprecipitated with cyclin A antibody under low-stringency (co-immunoprecipitation) conditions and cyclin A-associated histone H1 kinase activity was determined with a cdk assay kit. As shown in Figure 4(C), greater cyclin A/cdk activity was mobilized in WT and KA Brd2-infected cells compared with cells infected with ΔBD, ΔE2F Brd2 or control adenoviruses, consistent with expectation. Since increased cyclin A-associated cdk activity drives E2F-regulated S-phase progression [79], we concluded that Brd2 overexpression under these conditions had stimulated the machinery that controls the cell cycle.
Promoter association and HAT activity
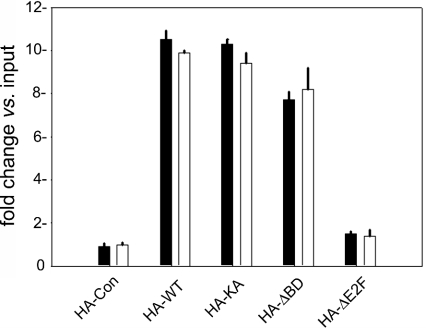
We next wished to determine whether deletion of any of the functional domains of Brd2 had an impact on the ability of the protein to associate physically with the cyclin A promoter. We have published evidence that the C-terminal portion of Brd2 mediates association of the protein with a multiprotein complex that contains E2F proteins [57]; this complex could also provide an anchor point to the promoter DNA. A similar structure–function relationship has been reported for the double bromodomain protein Rsc4 [42]. The N-terminal bromodomains of Brd2 also suggest a point of contact with promoter chromatin and have been reported to associate selectively with acetylated histone H4 [63]. To test whether these points of contact are redundant and whether domain-deleted mutants of Brd2 can still function, we infected Rat1 cells again with Brd2 and its derivatives as above and performed ChIP with a biotinylated anti-HA monoclonal antibody at two time points after serum stimulation, 8 h (early in G1 phase) and 18 h (in mid-S-phase). We used real-time PCR to amplify cyclin A promoter DNA with primers that we had validated for the rat cyclin A promoter. As shown in Figure 5, we observed antibody-specific enrichment of the recombinant WT, KA and ΔBD proteins in association with cyclin A promoter chromatin at 8 h (closed bars) and at 18 h (open bars), but not the ΔE2F protein, at either time. It was interesting to note that the ΔBD mutant protein was present at the cyclin A promoter, despite its apparent lack of transcriptional activity under the above conditions.
Figure 5. ChIP of Brd2 derivatives in association with the cyclin A promoter.
Chromatin was immunoprecipitated with biotinylated anti-HA primary monoclonal antibody and streptavidin coupled with agarose. Real-time PCR amplification determined the enrichment of rat cyclin A promoter DNA, expressed as fold change over input chromatin (n=2). ChIP was performed at two time points, 8 h (closed bars) and 18 h (open bars), after serum stimulation. Non-specific IgG controls in each case defined a baseline below HA-Con enrichment (results not shown).
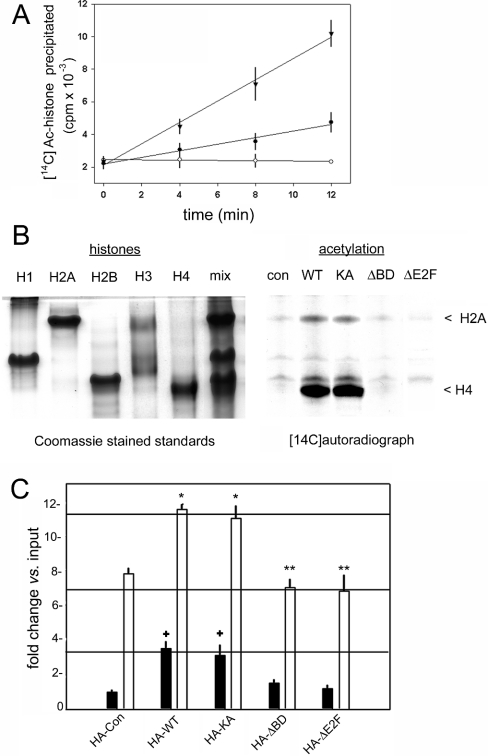
These results, and the established link between E2F and chromatin remodelling in the control of cyclin A transcription [1–5], prompted us to test whether, in addition to E2F proteins [57], the Brd2 multiprotein complex also contains histone modification activities. After its nuclear translocation, Brd2 associates with an approx. 330 kDa multiprotein complex [56,57]. We hypothesized that this complex includes histone modification activities involved in the regulation of transcription. To determine whether HAT activity associates with Brd2, nuclear extracts were co-immunoprecipitated with rabbit polyclonal antibody against Brd2 and the incorporation of [14C]acetyl-CoA into mixed histones was assayed. Brd2 immune co-immunoprecipitation complexes contained HAT activity, whereas preimmune co-immunoprecipitation complexes did not, as shown in Figure 6(A). Recombinant yeast Gcn5 served as a positive control for histone acetylation [36]. Recombinant Brd2 alone does not have HAT activity (results not shown), suggesting that the immunocomplex contains a Brd2-associated HAT. We then co-immunoprecipitated HA-epitope-tagged WT Brd2 and the mutants and assayed the immunocomplexes to determine their ability to acetylate a mixture of purified histones in vitro. WT and KA Brd2 associated with a complex that contains a histone H4-specific HAT, with weak activity against histone H2A; no detectable HAT activity was associated with ΔBD Brd2, ΔE2F Brd2 or the empty vector adenovirus control (Figure 6B).
Figure 6. Brd2-associated HAT activity.
(A) Endogenous HAT activity in cell extracts. Control immunoprecipitation was performed with preimmune serum (○), Brd2 immunoprecipitation was performed with rat cross-reactive anti-Brd2 rabbit polyclonal antibody [55] (●) and recombinant yeast Gcn5 was a positive control (▼). Incorporation of [14C]acetate into a histone mixture comprising equal amounts of each of the nucleosomal core histones and histone H1 was determined as trichloroacetic acid-precipitable 14C-radioactivity. S.E.M. for n=2 is shown. (B) Rat1 cells were infected with adenovirus constructs and cell extracts were assayed at 18 h after serum stimulation. HAT activity associated with recombinant proteins was co-immunoprecipitated with an anti-HA antibody. (B) Left panel: histone standards, separated by Triton X-100/urea/acetic acid PAGE [96], either individually or in an equimolar mixture (mix) and stained with Coomassie Blue. Right panel: autoradiogram of incorporation of [14C]acetate from [14C]acetyl-CoA into histone mix. (C) ChIP of acetylated histone H4 in association with the cyclin A promoter. Chromatin was immunoprecipitated with pan-anti-acetyl histone H4 antibody and Protein A/G–agarose. Real-time PCR amplification determined the enrichment of cyclin A promoter DNA, expressed as fold change over input chromatin (n=2). ChIP was performed at two time points, 8 h (closed bars) and 18 h (open bars), after serum stimulation. *, ** and ‘+’ indicate statistically indistinguishable means as determined by Student's t test, P<0.05.
Finally, we hypothesized from the results of Figures 5 and 6(B) that the cyclin A promoter probably undergoes a Brd2-dependent increase in histone acetylation that could account in part for its transcriptional activation (Figure 2). We isolated chromatin at 8 and 18 h after serum stimulation as described for Figure 5, and instead of performing immunoprecipitation with anti-HA antibody, we used anti-pan-acetyl histone H4 antibody and amplified cyclin A promoter DNA using real-time PCR. With infected control cells (HA-Con), we first confirmed an increase in cyclin A promoter acetylation (represented as fold change versus input chromatin in Figure 6C) of histone H4 at 18 h, when many of the cells have moved into S-phase (Figure 6C, open bars) and the promoter is transcriptionally active. Acetylation at 18 h was compared with acetylation at 8 h, when most of the cells are in G1 phase (closed bars) and the promoter is transcriptionally silent. Then we compared each of the Brd2 derivatives with these controls. As expected, expression of both WT and KA constructs increased the acetylation of histone H4 at 18 h, but expression of neither ΔBD nor ΔE2F proteins increased the acetylation of histone H4 above control levels. Curiously, G1 phase acetylation of histone H4 was increased above basal levels with the expression of WT and KA constructs, suggesting that early nucleosome modification might prime the locus for later transcription.
DISCUSSION
We have been developing a model wherein Brd2 provides a ‘scaffolding’ or bridging function [11] for cyclin A transactivation during S-phase, anchoring or recruiting HAT activity to the promoter through stable, multiprotein nuclear complexes. These Brd2 complexes contain E2F transcription factors, which are known to transactivate the cyclin A promoter. E2F-dependent cyclin A transactivation in S-phase is essential for cell-cycle progression in response to mitogenic signalling. Yet, histone acetylation of the cyclin A promoter, probably mediated in part through double bromodomain proteins, such as the basal transcription factor TAFII250 [29,65] and its intrinsic [37] or recruited HAT activity, is also essential for cyclin A transcription [1,3] and for cell-cycle progression [10,80]. To explore this dual mechanism of regulation of the cyclin A locus, we undertook a domain analysis of Brd2. The analysis was grounded in the observation that constitutive expression of Brd2 up-regulates cyclin A transcription and destabilizes the cell cycle, leading to malignancy in a mouse model [58]. We hypothesized that both the N-terminal bromodomains of Brd2, motifs that are known to associate with acetyl-lysine substituents of nucleosomal histones, and its C-terminal domain, which mediates interaction with multiprotein complexes that contain E2F proteins, are required for transcription activation function. Deletion of the N-terminal bromodomains of Brd2 in the ΔBD mutant abolishes its transcriptional activation of the cyclin A promoter, yet ChIP (Figure 5) shows the derivative is nevertheless still present at the promoter. This result suggests that, surprisingly, Brd2 can bind to the cyclin A promoter through its associated E2F proteins, rather than through bromodomain-mediated binding to acetylated nucleosomes. The hypothesis was confirmed with reciprocal deletion of the E2F complex-binding domain; the ΔE2F mutant is neither present at the promoter nor does it have transactivation function, leading to the conclusion that both domains are required for function. Taken together with our previous results, the present results suggest that Brd2 plays a role in both E2F-regulated transcription and histone modification of the cyclin A promoter.
A question has emerged as to whether the reported kinase activity of Brd2 is intrinsic [55], as in the case of the similar bromodomain-containing factor TAFII250 [29,81], or associated [82]. One group reported inability to detect recombinant Brd2-dependent phosphorylation of substrates in transfected cell extracts [83]. Another group did detect Brd2-dependent phosphorylation, but was unable to eliminate it as expected using site-directed mutagenesis to inactivate the kinase active site [82]. This question is resolved herein with the construction of the ΔBD mutant of Brd2, which expresses constitutively active intrinsic kinase function and loses activity on mutation of the putative catalytic lysine residue in the double mutant. E2F-1, which is present in Brd2 multiprotein complexes [57], is an in vitro substrate for Brd2 kinase activity. However, the functional significance of the ability of ΔBD Brd2 to phosphorylate E2F-1 is not yet clear, since transcriptional activation of the locus does not depend on this ability, but rather on Brd2 recruitment of HAT activity and Brd2 participation in E2F-containing complexes.
The observation that a histone H4-specific HAT protein is present in the Brd2 complex, rather than a mixture of HATs with overlapping specificities, rules out recruitment of Gcn5, for example, but would be consistent with recruitment of a MYST family member [84], named for MOZ, Ybf2/Sas3 and Tip60, to participate in cyclin A transcriptional control. These results also suggest a mechanistic explanation for the apparently indistinguishable B cell lymphoma and leukaemia that arises in Tg and Tgmut mice and the similar in vitro proliferative phenotype of transgenic B cells [58]; both WT and KA derivatives explored in the present study behaved similarly with respect to cell-cycle progression, mobilization of cyclin A and association with the histone H4-specific HAT. The kinase activity of an oncogene is usually associated with its neoplastic transforming ability; however, the result in mice is not surprising upon consideration of the Brd2 multi-protein complex, which is recruited through the C-terminal tail of Brd2 [57]. The catalytic Lys-574 is not located in the C-terminal domain that is required for the association with the E2F-containing complex, and its point mutation to an alanine residue is insufficient to abolish association with the complex. It follows that the ΔBD or ΔE2F Brd2 mutants probably do not exhibit these phenotypes and are probably not leukaemogenic if constitutively expressed in the B cells of transgenic mice.
The observation that Brd2 does not possess intrinsic HAT activity is consistent with the divergence between the BET class of bromodomain-containing proteins, to which Brd2 belongs, and the Gcn5 superfamily members that do have HAT activity [85]. Identification of the Brd2-associated HAT and the timing of its association with Brd2 will be informative. Given that transcriptional repression of cyclin A is maintained by HDAC-associated RB–E2F complexes through the G1 phase [1–3,47–50], but alleviated as RB dissociates from the complex, we speculate that HDAC activity is exchanged for a putative Brd2-recruited HAT. We are at present characterizing the other components of the complex using an MS approach and have found in 293T cells that the Brd2 multiprotein complex contains Swi/Snf family members (G. V. Denis and A. Sinha, unpublished work). Therefore it seems probable that recruited chromatin-remodelling machines also contribute to Brd2-dependent transcriptional activation. The presence of Brd2 at the cyclin A promoter during both G1 phase, when the locus is transcriptionally repressed, and S-phase, when it is activated, supports a switching model wherein Brd2 ‘scaffolding’ of HDAC activity in G1 is switched to HAT activity in S-phase. Swi/Snf complexes can perform different functions, depending on the promoter context, and sometimes work in collaboration with HAT/HDAC exchange [86–90]. As the histone code hypothesis suggests, recruitment of a HAT to acetylate histone H4 might promote the subsequent recruitment of bromodomain-containing HATs such as Gcn5 to modify further the cyclin A promoter nucleosomes and recruit Swi/Snf [15,39].
In preliminary experiments, we observed that doubling the number of delivered adenovirus particles that express WT Brd2 could cause cell-cycle retardation, rather than acceleration, relative to adenovirus control (results not shown). In addition, serum stimulation at concentrations above 3.5% (v/v) tended to decrease differences between Brd2 overexpression in experiment and control, with earlier entry into S-phase in both cases, suggesting that the ‘strength’ of the mitogenic signalling component of serum stimulation must be proportional to the signal amplification that Brd2 overexpression provides, in order for cell-cycle acceleration to be a useful endpoint. This principle has been invoked to explain aspects of scaffold-facilitated MAP kinase signalling [91] that involve the MAP kinase scaffold protein Ste5 [92].
The role of BET family bromodomain proteins in other phases of the cell cycle is not yet fully understood. Gene disruption studies in mice have established that Brd4 [70,71] associates with acetylated chromatin in mitotic chromosomes [73]. Intriguingly, association of Brd2 with the LANA (latent nuclear antigen) protein of Kaposi's sarcoma herpesvirus/human herpesvirus-8 [82], which tethers the viral genome to mitotic chromosomes [93], may have transcriptional activation or chromatin-remodelling functions [94] and, intriguingly, transactivates cyclin A [95]. The function of Brd2 during these late stages of the cell cycle requires additional investigation. Related, and important, unresolved questions are whether BET proteins, including Brd2, are essential participants in basic transcription and whether they perform specialized functions in individual tissues or whether they are functionally redundant with other bromodomain-containing transcription factors or co-activators, such as TAFII250 or CBP. The close structural similarity between Brd2 and TAFII250 implies that Brd2 may substitute for TAFII250 function in certain contexts. Although TAFII250 is essential for viability, it is not known if Brd2 is also essential. The cell-cycle phenotype of a brd2(−/−) null mouse, which we plan to generate, will address this question and may illustrate tissue-specific and cell-cycle-specific components of the Brd2 multiprotein complex.
Our results explore the mediating role that Brd2 plays in the integration of mitogenic signal transduction with cell-cycle progression, through a dual mechanism of direct transcription activation in association with E2F proteins and recruitment of chromatin-remodelling machines, to participate in the transactivation of key genes responsible for mitogenesis, such as cyclin A.
Acknowledgments
We thank J.M. Blanchard (Institut de Génétique Moléculaire, Montpellier, France) for the cyclin A reporter, F. J. Giordano (Yale University, New Haven, CT, U.S.A.) for adenovirus expression plasmids, H. Cohen (Boston University School of Medicine, Boston, MA, U.S.A.) for 293T cells, C. Vaziri (Boston University School of Medicine) for help with adenovirus technology (including green fluorescent protein-expressing adenovirus), R. Sternglanz (Stony Brook University, New York, NY, U.S.A.) for an expression vector for recombinant yeast Gcn5, D. McDevit (Boston University School of Medicine) for help with ChIP and J. Tumang (Boston University School of Medicine) for help with real-time PCR. We thank members of the Cancer Research Center, B. Nikolajczyk and T. Rothstein of the Immunobiology Unit, both at Boston University School of Medicine, and G. Schnitzler of Tufts University School of Medicine for helpful criticism. This work was supported by United States Public Health Service grants CA84193 (D.V.F.), CA75107 and CA102889 (G.V.D.) from the National Cancer Institute.
References
- 1.Stiegler P., De Luca A., Bagella L., Giordano A. The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter. Cancer Res. 1998;58:5049–5052. [PubMed] [Google Scholar]
- 2.Knudsen K. E., Fribourg A. F., Strobeck M. W., Blanchard J.-M., Knudsen E. S. Cyclin A is a functional target of retinoblastoma tumor suppressor protein-mediated cell cycle arrest. J. Biol. Chem. 1999;274:27632–27641. doi: 10.1074/jbc.274.39.27632. [DOI] [PubMed] [Google Scholar]
- 3.Siddqui H., Solomon D. A., Gunawardena R. W., Wang J. Y., Knudsen E. S. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol. Cell. Biol. 2003;23:7719–7731. doi: 10.1128/MCB.23.21.7719-7731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulze A., Zerfass K., Spitkovsky D., Middendorp S., Berges J., Helin K., Jansen-Durr P., Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spitkovsky D., Schulze A., Boye B., Jansen-Durr P. Down-regulation of cyclin A gene expression upon genotoxic stress correlates with reduced binding of free E2F to the promoter. Cell Growth Diff. 1997;8:699–710. [PubMed] [Google Scholar]
- 6.Angus S. P., Fribourg A. F., Markey M. P., Williams S. L., Horn H. F., DeGregori J., Kowalik T. F., Fukasawa K., Knudsen E. S. Active RB elicits late G1/S inhibition. Exp. Cell Res. 2002;276:201–213. doi: 10.1006/excr.2002.5510. [DOI] [PubMed] [Google Scholar]
- 7.Stiegler P., Giordano A. The family of retinoblastoma proteins. Crit. Rev. Eukaryotic Gene Exp. 2001;11:59–76. [PubMed] [Google Scholar]
- 8.Nevins J. R. The Rb/E2F pathway and cancer. Human Mol. Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 9.Sherr C. J., McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 10.Cress W. D., Seto E. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 11.Denis G. V. Bromodomain motifs and ‘scaffolding’? Front. Biosci. 2001;6:1065–1068. doi: 10.2741/a668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn S. The role of TAFs in RNA polymerase II transcription. Cell (Cambridge, Mass.) 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 13.Hampsey M. A SAGA of histone acetylation and gene expression. Trends Genet. 1997;13:427–429. doi: 10.1016/s0168-9525(97)01292-4. [DOI] [PubMed] [Google Scholar]
- 14.Grant P. A., Sterner D. E., Duggan L. J., Workman J. L., Berger S. L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 15.Hassan A. H., Prochasson P., Neely K. E., Galasinski S. C., Chandy M., Carrozza M. J., Workman J. L. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell (Cambridge, Mass.) 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 16.Peterson C. L., Workman J. L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 17.Muchardt C., Yaniv M. When the SWI/SNF complex remodels the cell cycle. Oncogene. 2001;20:3067–3075. doi: 10.1038/sj.onc.1204331. [DOI] [PubMed] [Google Scholar]
- 18.Katsani K. R., Mahmoudi T., Verrijzer C. P. Selective gene regulation by SWI/SNF-related chromatin remodeling factors. Curr. Top. Microbiol. Immunol. 2003;274:113–141. doi: 10.1007/978-3-642-55747-7_5. [DOI] [PubMed] [Google Scholar]
- 19.Wang W. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr. Top. Microbiol. Immunol. 2003;274:143–169. doi: 10.1007/978-3-642-55747-7_6. [DOI] [PubMed] [Google Scholar]
- 20.Myers L. C., Gustafsson C. M., Bushnell D. A., Lui M., Erdjument-Bromage H., Tempst P., Kornberg R. D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y. W., Veschambre P., Erdjument-Bromage H., Tempst P., Conaway J. W., Conaway R. C., Kornberg R. D. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuras L., Borggrefe T., Kornberg R. D. Association of the Mediator complex with enhancers of active genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haynes S. R., Dollard C., Winston F., Beck S., Trowsdale J., Dawid I. B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeanmougin F., Wurtz J.-M., Le Douarin B., Chambon P., Losson R. The bromodomain revisited. Trends Biochem. Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 25.Winston F., Allis C. D. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 26.Horn P. J., Peterson C. L. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front. Biosci. 2001;6:D1019–D1023. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- 27.Dorr A., Kiermer V., Pedal A., Rackwitz H. R., Henklein P., Schubert U., Zhou M. M., Verdin E., Ott M. Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J. 2002;21:2715–2723. doi: 10.1093/emboj/21.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mujtaba S., He Y., Zeng L., Yan S., Plotnikova O., Sachchidanand Sanchez R., Zeleznik-Le N. J., Ronai Z., Zhou M. M. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson R. H., Ladurner A. G., King D. S., Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 30.Owen D. J., Ornaghi P., Yang J. C., Lowe N., Evans P. R., Ballario P., Neuhaus D., Filetici P., Travers A. A. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matangkasombut O., Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 32.Hudson B. P., Martinez-Yamout M. A., Dyson H. J., Wright P. E. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- 33.Dhalluin C., Carlson J. E., Zeng L., He C., Aggarwal A. K., Zhou M.-M. Structure and ligand of a histone acetyltransferase bromodomain. Nature (London) 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 34.Ogryzko V. V., Schiltz O. L., Russanova V., Howard B. H., Nakatani Y. The transcriptional co-activators p300 and CBP are histone acetyltransferases. Cell (Cambridge, Mass.) 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 35.Bannister A. J., Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 36.Kuo M.-H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D. Transcription-linked acetylation by Gcn5p of histone H3 and H4 at specific lysines. Nature (London) 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 37.Mizzen C. A., Yang X. J., Kokubo T., Brownell J. E., Bannister A. J., Owen-Hughes T., Workman J., Wang L., Berger S. L., Kouzarides T., et al. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell (Cambridge, Mass.) 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 38.Gansheroff L. J., Dollard C., Tan P., Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Syntichaki P., Topalidou I., Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature (London) 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- 40.Tamkun J. W., Deuring R., Scott M. P., Kissenger M., Pattatucci A. M., Kaufman T. C., Kennison J. A. Brahma – a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SWI2/SNF2. Cell (Cambridge, Mass.) 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 41.Nicolas R. H., Goodwin G. H. Molecular cloning of polybromo, a nuclear protein containing multiple domains including five bromodomains, a truncated HMG-box, and two repeats of a novel domain. Gene. 1996;175:233–240. doi: 10.1016/0378-1119(96)82845-9. [DOI] [PubMed] [Google Scholar]
- 42.Kasten M., Szerlong H., Erdjument-Bromage H., Tempst P., Werner M., Cairns B. R. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 44.Agalioti T., Chen G., Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell (Cambridge, Mass.) 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 45.Trouche D., Le Chalony C., Muchardt C., Yaniv M., Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 47.Brehm A., Miska E. A., McCance D. J., Reid J. L., Bannister A. J., Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 48.Magnaghi-Jaulin L., Groisman R., Naguibneva I., Robin P., Lorain S., Le Villain J. P., Troalen F., Trouche D., Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 49.Ferreira R., Magnaghi-Jaulin L., Robin P., Harel-Bellan A., Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H. S., Gavin M., Dahiya A., Postigo A. A., Ma D., Luo R. X., Harbour J. W., Dean D. C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell (Cambridge, Mass.) 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 51.Burns L. G., Peterson C. L. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol. Cell. Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell (Cambridge, Mass.) 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 53.Beck S., Hanson I., Kelly A., Pappin D. J. C., Trowsdale J. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Seq. 1992;2:203–210. doi: 10.3109/10425179209020804. [DOI] [PubMed] [Google Scholar]
- 54.Rachie N. A., Seger R., Valentine M. A., Ostrowski J., Bomsztyk K. Identification of an inducible 85-kDa nuclear protein kinase. J. Biol. Chem. 1993;268:22143–22149. [PubMed] [Google Scholar]
- 55.Denis G. V., Green M. R. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10:261–271. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- 56.Guo N., Faller D. V., Denis G. V. Activation-induced nuclear translocation of RING3. J. Cell Sci. 2000;113:3085–3091. doi: 10.1242/jcs.113.17.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Denis G. V., Vaziri C., Guo N., Faller D. V. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Diff. 2000;11:417–424. [PMC free article] [PubMed] [Google Scholar]
- 58.Greenwald R., Tumang J. R., Sinha A., Currier N., Cardiff R. D., Rothstein T. L., Faller D. V., Denis G. V. Eμ-BRD2 transgenic mice develop B cell lymphoma and leukemia. Blood. 2004;103:1475–1484. doi: 10.1182/blood-2003-06-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leone G., DeGregori J., Sears R., Jakoi L., Nevins J. R. Myc and Ras collaborate in inducing accumulation of active cyclin E/cdk2 and E2F. Nature (London) 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 60.Sylvester A. M., Chen D., Krasinski K., Andres V. Role of c-fos and E2F in the induction of cyclin A transcription and vascular smooth muscle cell proliferation. J. Clin. Invest. 1998;101:940–948. doi: 10.1172/JCI1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winston J. T., Coats S. R., Wang Y.-Z., Pledger W. J. Regulation of the cell cycle machinery by oncogenic ras. Oncogene. 1996;12:127–134. [PubMed] [Google Scholar]
- 62.Florence B., Faller D. V. You bet-cha: a novel family of transcriptional regulators. Front. Biosci. 2001;6:D1008–D1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 63.Kanno T., Kanno Y., Siegel R. M., Jang M. K., Lenardo M. J., Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 64.Florence B., McGinnis W. A genetic screen of the Drosophila X chromosome for mutations that modify Deformed function. Genetics. 1998;150:1497–1511. doi: 10.1093/genetics/150.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wassarman D. A., Sauer F. TAFII250: a transcription toolbox. J. Cell Sci. 2001;114:2895–2902. doi: 10.1242/jcs.114.16.2895. [DOI] [PubMed] [Google Scholar]
- 66.Lygerou Z., Conesa C., Lesage P., Swanson R. N., Ruet A., Carlson M., Sentenac A., Seraphin B. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 1994;22:5332–5340. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chua P., Roeder G. S. Bdf1, a yeast chromosomal protein required for sporulation. Mol. Cell. Biol. 1995;15:3685–3696. doi: 10.1128/mcb.15.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matangkasombut O., Buratowski R. M., Swilling N. W., Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 69.Pamblanco M., Poveda A., Sendra R., Rodriguez-Navarro A., Perez-Ortin J. E., Tordera V. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 2001;496:31–35. doi: 10.1016/s0014-5793(01)02397-3. [DOI] [PubMed] [Google Scholar]
- 70.Dey A., Ellenberg J., Farina A., Coleman A. E., Maruyama T., Sciortino S., Lippincott-Schwartz J., Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol. Cell. Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Houzelstein D., Bullock S. L., Lynch D. E., Grigorieva E. F., Wilson V. A., Beddington R. S. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maruyama T., Farina A., Dey A., Cheong J., Bermudez V. P., Tamura T., Sciortino S., Shuman J., Hurwitz J., Ozato K. A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol. 2002;22:6509–6520. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U.S.A. 2000;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Becker T. C., Noel R. J., Coats W. S., Gomez-Foix A. M., Alam T., Gerard R. D., Newgard C. B. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994;43:161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 75.McGrory J., Bautista D., Graham F. L. A simple technique for the rescue of early region I mutations into infectious human adenovirus type 5. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 76.Mizzen C. A., Brownell J. E., Cook R. G., Allis C. D. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol. 1999;304:675–697. doi: 10.1016/s0076-6879(99)04041-0. [DOI] [PubMed] [Google Scholar]
- 77.Ohtsubo M., Theodoras A. M., Schumacher J., Roberts J. M., Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S transition. Mol. Cell. Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pagano M., Pepperkok R., Verde F., Ansorge W., Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pagano M., Draetta G., Jansen-Durr P. Association of cdk2 kinase with the transcription factor E2F during S phase. Science. 1992;255:1144–1147. doi: 10.1126/science.1312258. [DOI] [PubMed] [Google Scholar]
- 80.Dunphy E. L., Johnson T., Auerbach S. S., Wang E. H. Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol. Cell. Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dikstein R., Ruppert S., Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell (Cambridge, Mass.) 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 82.Platt G. M., Simpson G. R., Mittnacht S., Schulz T. F. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rhee K., Brunori M., Besset V., Trousdale R., Wolgemuth D. J. Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J. Cell Sci. 1998;111:3541–3550. doi: 10.1242/jcs.111.23.3541. [DOI] [PubMed] [Google Scholar]
- 84.Utley R. T., Cote J. The MYST family of histone acetyltransferases. Curr. Top. Microbiol. Immunol. 2003;274:203–236. doi: 10.1007/978-3-642-55747-7_8. [DOI] [PubMed] [Google Scholar]
- 85.Dutnall R. N., Tafrov S. T., Sternglanz R., Ramakrishnan V. Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell (Cambridge, Mass.) 1998;94:427–438. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
- 86.Heinzel T., Lavinsky R. M., Mullen T.-M., Soderstrom M., Laherty C. D., Torchia J., Yang W.-M., Brard G., Ngo S. D., Davie J. R., et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 87.Davie J. R. Covalent modifications of histones: expression from chromatin templates. Curr. Opin. Genet. Dev. 1998;8:173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 88.Saleh M., Rambaldi I., Yang X. J., Featherstone M. S. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 2000;20:8623–8633. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McKinsey T. A., Zhang C. L., Olson E. N. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 90.Kraus W. L., Wong J. Nuclear receptor-dependent transcription with chromatin. Is it all about enzymes? Eur. J. Biochem. 2002;269:2275–2283. doi: 10.1046/j.1432-1033.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- 91.Levchenko A., Bruck J., Sternberg P. W. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burack W. R., Shaw A. S. Signal transduction: hanging on a scaffold. Curr. Opin. Cell Biol. 2000;12:211–216. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- 93.Ballestas M. E., Chatis P. A., Kaye K. M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 94.Mattsson K., Kiss C., Platt G. M., Simpson G. R., Kashuba E., Klein G., Schulz T. F., Szekely L. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J. Gen. Virol. 2002;83:179–188. doi: 10.1099/0022-1317-83-1-179. [DOI] [PubMed] [Google Scholar]
- 95.Duro D., Schulze A., Vogt B., Bartek J., Mittnacht S., Jansen-Durr P. Activation of cyclin A gene expression by the cyclin encoded by human herpesvirus-8. J. Gen. Virol. 1999;80:549–555. doi: 10.1099/0022-1317-80-3-549. [DOI] [PubMed] [Google Scholar]
- 96.Waterborg J. H. Acid-urea-triton polyacrylamide gels for histones. In: Walker J. M., editor. The Protein Protocols Handbook. Totowa, NJ: Humana; 1996. pp. 91–100. [Google Scholar]