Abstract
mRNA vaccines against SARS-CoV-2 have revolutionized vaccine development, but their immunological mechanisms are not fully understood. Here, we investigate injection site responses of mRNA vaccines by generating a comprehensive single-cell transcriptome profile upon lipid nanoparticle (LNP) or LNP-mRNA challenge in female BALB/c mice. We show that LNP-induced stromal pro-inflammatory responses and mRNA-elicited type I interferon responses dominate the initial injection site responses. By tracking the fate of delivered mRNA, we discover that injection site fibroblasts are highly enriched with the delivered mRNA and that they express IFN-β specifically in response to the mRNA component, not to the LNP component of mRNA vaccines. Moreover, the mRNA-LNP, but not LNP alone, induces migratory dendritic cells highly expressing IFN-stimulated genes (mDC_ISGs) at the injection site and draining lymph nodes. When co-injected with LNP-subunit vaccine, IFN-β induces mDC_ISGs at the injection site, and importantly, it substantially enhances antigen-specific cellular immune responses. Furthermore, blocking IFN-β signaling at the injection site significantly decreases mRNA vaccine-induced cellular immune responses. Collectively, these data highlight the importance of injection site fibroblasts and IFN-β signaling during early immune responses against the mRNA vaccine and provide detailed information on the initial chain of immune reactions elicited by mRNA vaccine injection.
Subject terms: RNA vaccines, Innate immunity, Mouse
The initial immune response following mRNA vaccine injection is not entirely clear. Here, the authors comprehensively profile injection site responses using single-cell transcriptomics in a mouse model showing activation of major axes of innate immune responses upon and the role of IFN-β to promote cellular immunity.
Introduction
The mRNA vaccine has emerged as an attractive vaccine platform for cancer immunotherapy and infectious disease prevention because of its ability to induce robust cellular and humoral immune responses1–3. Indeed, US Food and Drug Administration–approved mRNA vaccines against spike proteins of SARS-CoV-2 induce robust production of spike-specific neutralizing antibodies and potent T-cell responses, typically weeks after vaccine injections4–9. Strong innate immune responses preceding these protective adaptive immune responses feature a wave of type I interferon (IFN) responses that culminate within a day after mRNA vaccine injection, as demonstrated in draining lymph nodes (dLNs) in mice10 and human peripheral blood11. Although these studies have revealed some important features of mRNA vaccine–triggered immunity, early immune responses at the injection site have not been systematically investigated.
The mRNA vaccines consist of two key components: mRNA encoding the antigenic protein of interest and ionizable lipid nanoparticles (LNPs) that can efficiently deliver the mRNA into the cytoplasm of target cells12,13. Thanks to its built-in adjuvanticity, the mRNA vaccine platform does not require additional adjuvants14. Recent studies have shown that LNPs not only enable efficient cellular delivery of mRNA molecules but also provide the strong adjuvanticity required for the development of adaptive immune responses15,16. The ionizable lipid component of the LNP is crucial for the adjuvanticity, and interleukin (IL)−6 production triggered by the LNP is required for efficient induction of follicular helper T-cell and germinal center B-cell reactions16.
Meanwhile, a strong immune response provoked by exogenous mRNA17,18 has been regarded as a major obstacle for this therapeutic platform, as it hinders cellular translation of targeted antigens19,20. The advent of base modification12 strategies has efficiently circumvented this issue, and with the other major technological advances in reducing immune activations20, the mRNA components are now even referred to as immuno-silent14,21 because of the absence of type I IFN induction in vitro. However, recent studies with SARS-CoV-2 mRNA vaccines (BNT162b2, Pfizer) have demonstrated strong induction of type I IFN responses in vivo after mRNA vaccine injections10,11. In addition, the cell-mediated immune responses induced by mRNA vaccine (BNT162b2, Pfizer) are highly dependent on Ifnar and Mda5 genes in vaccinated mouse models10. These results suggest a potential contribution of mRNA components to the proper immunogenicity required for the robust induction of cell-mediated immune responses. Thus, further investigations into the precise immunological role of each component in mRNA vaccine are needed.
Here, through construction of a single-cell transcriptome atlas of the mRNA vaccine injection site, we have deeply profiled and analyzed the initial immune responses to the mRNA vaccines, offering systematic analysis of injection site responses that these vaccines induce. We identified major target cells of mRNA vaccine components at the injection site and their associated transcriptional changes. By comparing transcriptional responses elicited by empty LNP or LNP + mRNA injection, we systematically evaluated the differential adjuvant effects of two major components of mRNA vaccines: mRNA and LNP. The extensive transcriptomic survey conducted here, encompassing 91,601 single-cell transcriptome profiles of the injection site (n = 83,094) and dLNs (n = 8507), provides blueprints of the initial innate immune response to mRNA vaccines at the injection site, identifies the contributions of each vaccine component to adjuvanticity, and provides mechanistic frameworks for future development of more efficient vaccines based on this platform.
Results
Single-cell atlas of mRNA vaccine injection-site responses
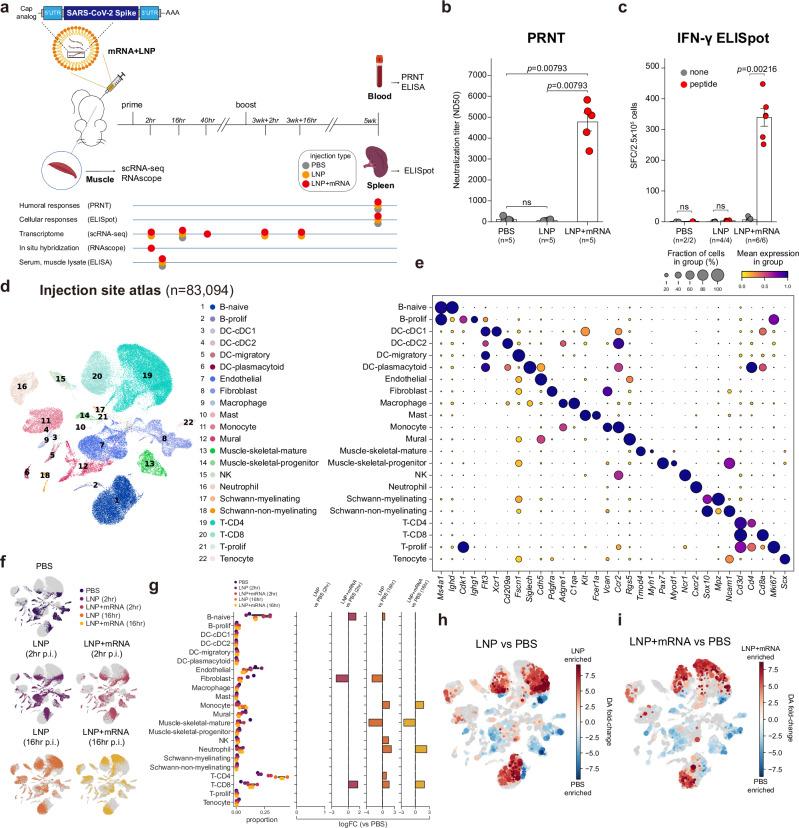
To construct the single-cell transcriptome atlas of mRNA vaccine injection-site responses, we immunized mice with mRNA vaccine, a nucleoside-modified mRNA formulated in LNPs that encodes the SARS-CoV-2 spike glycoprotein stabilized in its prefusion conformation (Fig. 1a and Supplementary Fig. 1a, b). The vaccines were administered twice via intramuscular injection, with an interval of 3 weeks between injections (prime and boost shot). From 2 to 40 h after the injections, we resected the anterior thigh muscles where the mRNA vaccines were administered. Resected muscle tissues were both mechanically and chemically digested to yield single-cell suspensions that were then used to construct the single-cell RNA sequencing library. We used saline (phosphate-buffered saline; PBS) and empty LNP (formulated without the mRNA component of the vaccine) injections as controls, and the anterior thigh muscles of PBS-injected and empty LNP–injected mice also were resected for single-cell RNA sequencing. To validate the efficacy of the mRNA vaccine mouse model, we collected blood and spleen samples from the immunized mice 2 weeks after the boost shot (5 weeks after the prime dose). The plaque reduction neutralization (PRNT) assay and IFN-γ enzyme-linked immunospot (ELISpot) assay were used to validate the antigen-specific humoral immune responses and cellular responses, respectively22. We confirmed the robust induction of neutralizing antibodies (Fig. 1b) and cellular responses against the spike proteins (Fig. 1c) in the vaccinated mice.
Fig. 1. Single-cell atlas of mRNA vaccine injection-site responses.
a Experimental and sample preparation schemes used in this study. b Serum titers of neutralizing antibodies measured with the PRNT assay, showing median neutralizing doses (ND50) values compared between the groups with biological replicates. Data are presented as mean values ±SEM. c Results from IFN-γ ELISpot assay. Spleen cells (2.5 × 105 cells/well) were challenged with negative control (none), or a peptide pool of SARS-CoV-2 spike protein (peptide), and the number of IFN-γ spot-forming cells (SFCs) was counted and displayed. Average counts of SFCs from technical duplicates are plotted, and the displayed data points indicate biological replicates. Data are presented as mean values ±SEM. d Uniform manifold approximation and projection (UMAP) representation of injection site scRNA-seq data annotated with cell types. e A dot plot representation of cell types and their representative marker genes identified in the injection-site scRNA-seq data. Dot sizes are proportional to the fraction of cells expressing marker genes, and dot colors represent min-max normalized average expression levels of marker genes in each cell type. f Injection site scRNA-seq data are shown according to the treatment conditions and p.i. time points. g Proportions of cell types in each treatment time and p.i. time point. The left-most panel shows raw portions of each cell type, and the panels on the right side indicate results from differential cell composition analysis. Only the log-fold change values passing the false discovery rate cut-off (<0.05) are shown in the bar graphs. h, i Results from differential abundance testing are overlaid on cellular landscape. Red colors indicate local cell neighborhoods enriched in (h) LNP treatment samples (2 h, 16 h), and (i) LNP + mRNA treatment samples (2 h, 16 h), against PBS-treated samples. Dot sizes are proportional to the sizes of the cellular neighborhoods. All statistical evaluations were conducted with two-tailed Mann–Whitney U tests (ns: nonsignificant). Schematic images were created with BioRender.com.
To delineate cellular responses elicited by mRNA vaccine injection, we deeply profiled the single-cell transcriptome of the vaccine injection site, consisting of 83,094 single-cell profiles. The single-cell transcriptome data encompass 22 different cell types in muscle tissues, including T and B cells, dendritic cells, neutrophils, monocytes, endothelial cells, and fibroblasts (Fig. 1d, e). We found that both mRNA vaccine and empty LNP injection provoked substantial shifts in the overall landscape of single-cell transcriptome data (Fig. 1f). Differential cell composition analysis23 revealed that the injection of either empty LNP or mRNA vaccine led to prominent increases in CD8 T cell, neutrophil and monocyte populations, 16 h after injection (Fig. 1g). Differential cell neighborhood analysis24 indicated diverging transcriptomic alterations in most of the cell types at the injection site (Fig. 1h, i). Overall, these results reaffirm the strong immunogenic effects of the LNPs16,25, which leads to multicellular responses at the injection site.
Major axes of transcriptional responses elicited by mRNA vaccine injection
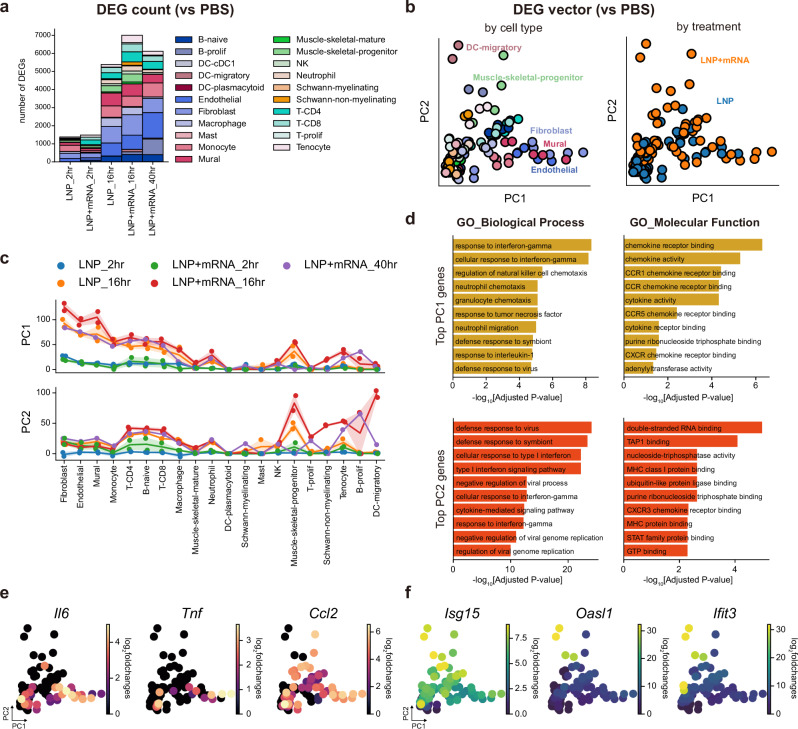
To profile cell type–specific responses elicited by the injections, we conducted differential gene expression analysis on all the different cell types in muscle tissue by comparing their gene expression patterns with that of the saline-injected sample. By counting the number of differentially expressed genes (DEGs) for each treatment condition and time point, we discovered that transcriptional responses at the injection site culminated at around 16 h after mRNA vaccine injection (Fig. 2a). To identify the common axes of transcriptional responses across different cell types, we conducted principal component analysis (PCA) on the DEG vectors, which consisted of log-fold changes in individual genes calculated on each cell type in each condition (compared with the PBS sample), primarily based on the 16 h sample reactions where the responses peaked (Fig. 2b). Interestingly, this analysis showed the diversification of early innate responses between different cell types in muscle tissue. The PC1 axis represented the responses in stromal cells, namely fibroblasts, endothelial cells, and mural cells, whereas the PC2 axis mainly featured responses in migratory dendritic cells (mDCs) (Fig. 2b, c). Intriguingly, we found that the PC2 responses in mDCs were highly specific to LNP-mRNA-injected samples, in contrast to the PC1 axis responses commonly seen in both the empty-LNP and mRNA-LNP–injected muscle tissues (Fig. 2c and Supplementary Fig. 2a–c). Additionally, we found that the earliest phases of immune responses were similar between the prime and the boost shots, in terms of both the major cell type compositions and the major axes of transcriptional responses (Supplementary Fig. 3a–d).
Fig. 2. Major axes of the transcriptional responses elicited by mRNA vaccine injection.
a Bar plots showing the number of cell type–wise DEGs in each treatment condition and time point (vs PBS injected sample). b Scatter plot representation of DEG vectors. Each dot represents a DEG vector of a cell type in each sample (vs PBS injected). The x and y axes represent each DEG’s projection on the PC1 and PC2 axes of the maximal responses (16 h p.i. response after mRNA vaccine injection). Dot colors denote cell type identities (left) and treatment conditions (right). c Line plots showing PC1 (top) and PC2 (bottom) projections of DEG vectors. Colors of lines and dots represent treatment conditions and p.i. times of DEG vectors. Line plot indicates mean values, and the error bands indicate 95% confidence intervals. d, Bar plots representing the results from pathway enrichment analysis, which was conducted on the top 100 genes contributing to PC1 (top panel) or PC2 axes (bottom panel). Biological Process (left) and Molecular Function (right) gene sets from the Gene Ontology (2023) were used for functional enrichment analysis. Adjusted P values (two-tailed, Benjamini-Hochberg adjustment) from the enrichment analysis were log-transformed, and their negative values are represented on the x axis of each bar plot. e, f DEG vectors plotted according to their PC1 projections (x-axis) and PC2 projections (y-axis). Dot colors represent log2 fold changes of (e) pro-inflammatory cytokine genes or (f) IFN-stimulated genes in the DEG vectors.
Pathway enrichment analysis revealed that the top PC1 loading genes were functionally associated with immune cell chemotaxis, whereas antiviral and type I IFN response genes were highly associated with the PC2 axis (Fig. 2d and Supplementary Fig. 4a, b). In detail, we discovered that transcriptional induction of inflammatory cytokine genes, such as Il6, Tnf, and Ccl2, was explained mainly by the PC1 axis (Fig. 2e, Supplementary Fig. 4c and Supplementary data 1). In contrast, the type I IFN-responsive genes, including Isg15, Oasl1, and Ifit326–28, were highly associated with the PC2 axis (Fig. 2f, Supplementary Fig. 4c and Supplementary data 1). To summarize, the global transcriptional changes evoked by the mRNA-LNP vaccine could be abstracted into two major axes: PC1, representing strong inflammatory responses in stromal cell populations at the injection site, mostly driven by the LNP component of the mRNA vaccine; and PC2, featuring type I IFN responses in mDCs, highly specific to mRNA component of the mRNA vaccine.
mRNA vaccine transcripts are enriched in injection-site fibroblasts
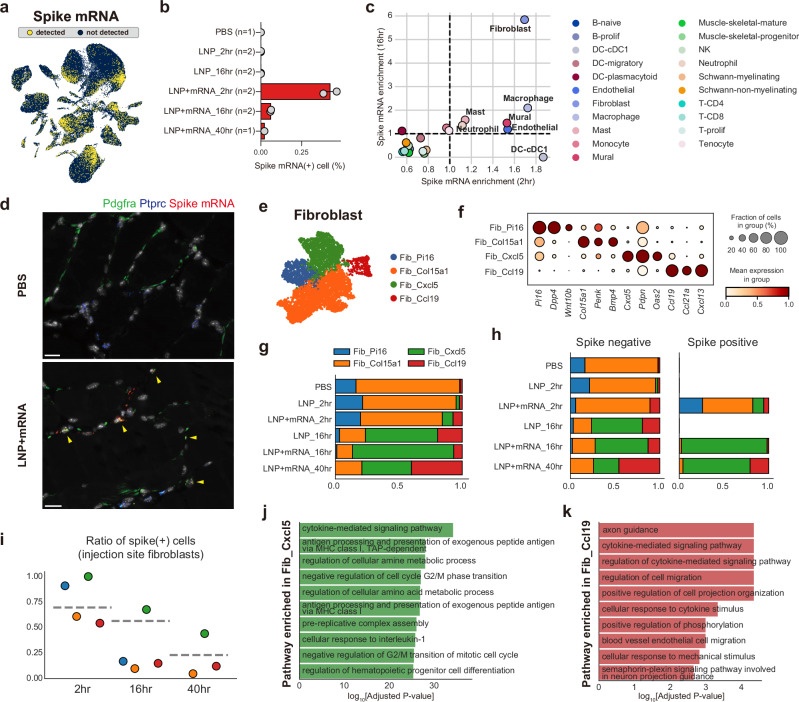
To identify the cellular dynamics of mRNA vaccine transcripts at the injection site, we investigated the spike mRNA content of each cell by mapping the single-cell transcriptome sequences to the custom-built spike open reading frame reference (Fig. 3a). Spike mRNAs were not detected in either the saline- or empty-LNP–injected samples, whereas 2%–46% of cells in the mRNA-LNP–injected samples were spike-positive (Fig. 3b). Detection rates decreased with post-injection (p.i.) time, possibly due to the degradation of the spike mRNA molecules. To profile cellular tropisms for the mRNA vaccines at the injection site, we compared the average spike mRNA counts in each cell type. The spike mRNAs were highly enriched in stromal cells including endothelial cells, pericytes, and fibroblasts, and also in myeloid cells at 2 h p.i. samples, whereas lymphoid cells and other structural cells contained relatively low amounts of the target mRNA transcripts (Fig. 3c). Of note, we found that fibroblasts were highly enriched with the spike mRNAs after 16 h, unlike any other cell types. The enrichment of spike mRNA in fibroblasts was further confirmed using RNA in situ hybridization (ISH) images (Fig. 3d).
Fig. 3. Enrichment of mRNA vaccine transcripts in injection-site fibroblasts.
a Spike mRNA-detected cells depicted on the UMAP plot of the injection-site scRNA-seq atlas. Yellow colored dots indicate spike mRNA-detected cells (spike mRNA count >1). b Fraction of spike mRNA-detected cells in the injection-site scRNA-seq data in each treatment and time point condition. Data are presented as mean values ±SEM. c Identification of spike mRNA–enriched cell types. Average spike mRNA counts (log-transformed) of the cell types were normalized across the cell types in each sample. The normalized values of spike mRNA (spike mRNA enrichment) in each time point are represented on the x-axis (enrichment in 2 h p.i.) and y-axis (enrichment in 16 h p.i.) of the scatter plot. Dot colors represent cell type identities. d Representative RNA-ISH image acquired from saline-injected (top) or mRNA-vaccine (bottom) 16 h p.i. muscle tissues. The images were selected from biological replicates of mRNA-vaccine injected samples (n = 2) and control samples (n = 2). Green, blue, and red indicate detection signals for Pdgfra, Ptprc, and Spike mRNA transcripts, respectively. Yellow arrowheads indicate fibroblasts with the spike mRNAs. Scale bars indicate 20 µm. e, f Fibroblast populations in the injection-site scRNA-seq atlas are represented in (e) a UMAP plot, and (f) their representative markers are depicted with a dot plot. g Fractions of fibroblast subpopulations according to treatment conditions and p.i. time points. h Fractions of fibroblast populations in spike mRNA–detected or non-detected fibroblasts from each treatment condition. For PBS- or LNP-treated samples, all fibroblasts were negative for mRNA spike, so not plotted on the spike-positive side. i Ratio of spike-positive cells in each fibroblast population. The y-axis of the plot indicates fractions of spike mRNA–detected cells in each fibroblast population, and the x-axis of the plot indicates p.i. time points after mRNA vaccine treatment. j, k Results from pathway enrichment analysis (GO_Biological Process), conducted on the DEGs of (j) Fib_Cxcl5 and (k) Fib_Ccl19. DEGs (vs rest) were defined as genes with adjusted P < 0.05 and log2FC > 1. P values were calculated from two-tailed Wilcoxon rank-sum test and were adjusted with Benjamini-Hochberg method.
To investigate the enrichment of target mRNA in fibroblasts in more detail, we isolated fibroblasts and re-clustered them. Injection-site fibroblasts were clustered into four different populations corresponding to previously reported fibroblast subtypes from the fibroblast atlas study29, which identified Pi16+ and Col15a1+ fibroblasts as global fibroblast populations, Cxcl5+ fibroblasts as muscle damage–related fibroblasts, and Ccl19+ fibroblasts as fibroblastic reticular cells (Fig. 3e, f and Supplementary data 2). Proportions of the global fibroblast populations (Fib_Pi16 and Fib_Col15a1) substantially decreased after vaccine injection, whereas Fib_Cxcl5 and Fib_Ccl19, which were barely observable in the saline-injected muscle, dominated the fibroblast population at the injection site at least from 16 h p.i. (Fig. 3g). In a comparison of cluster compositions in spike- and spike+ fibroblasts, we found that Fib_Cxcl5 fibroblasts were highly enriched with spike+ fibroblasts (Fig. 3h, i). The absence of Fib_Cxcl5 at the baseline (PBS) and the low fraction of Fib_Cxcl5 in the 2 h spike+ fibroblasts strongly suggest that the spike mRNA induces characteristics of these damage-related fibroblasts29,30, rather than the specific uptake of mRNA by Fib_Cxcl5.
To identify characteristics of these vaccine-induced fibroblast populations (Fib_Cxcl5 and Fib_Ccl19), we conducted pathway enrichment analysis on significantly upregulated genes in each cluster. The marker genes of Fib_Cxcl5 fibroblasts were highly correlated with cytokine-mediated signaling pathways, MHC class I antigen presentation pathways, and cellular responses to IL-1 (Fig. 3j and Supplementary data 2), whereas Fib_Ccl19 fibroblasts highly expressed cytokine-related and cell migration regulation–related genes (Fig. 3k). These results suggest that these fibroblasts are closely involved in cytokine-mediated signaling networks elicited by mRNA vaccine injection.
To systematically evaluate alterations in cytokine-mediated signaling networks elicited by mRNA vaccine injection, we conducted differential cell-to-cell interaction analysis31 on our single-cell transcriptome data. The interaction analysis suggested strong induction of outgoing signals in fibroblasts, especially at >40 h after injection (Supplementary Fig. 5a). The analysis also revealed specific induction CCL signaling pathways in vaccine-injected fibroblasts (Supplementary Fig. 5b). By investigating expression levels of individual CCL genes, we found that many of them, including Ccl5, Ccl2, Ccl7, Ccl4, and Ccl19, were upregulated after either LNP or LNP+mRNA injection (Supplementary Fig. 5c). These results indicate that the LNP components in the mRNA vaccine could increase expression of chemokine genes in fibroblasts. We also noted that these genes were differentially expressed in fibroblast sub-clusters (Supplementary Fig. 5d). Fib_Cxcl5 highly expressed Ccl2, Ccl5, Ccl7, and Ccl8, whereas Fib_Ccl19 specifically expressed Ccl19, Ccl21a, and Ccl9. The CCL chemokines expressed by Fib_Cxcl5 largely consisted of myeloid-recruiting chemokines, whereas the Fib_Ccl19 chemokines were those mainly involved in lymphocyte recruitment (Supplementary Fig. 5e).
The robust induction of Fib_Cxcl5 and Fib_Ccl19 in the empty LNP–injected muscle tissue (Fig. 3g) strongly suggests that the induction of these two inflammatory fibroblasts at the injection site depends on the LNP component of the vaccine. Trajectory analysis32,33 on the LNP-injected fibroblasts revealed a trajectory branch stretching from Fib_Pi16, a presumed fibroblast progenitor29, to Fib_Ccl19 (Supplementary Fig. 6a–c). Alignment of fibroblasts on the trajectory pseudotime demonstrated sequences of Fib_Pi16, Fib_Col15a1, Fib_Cxcl5, and Fib_Ccl19 clusters, which correlated well with p.i. time points (Supplementary Fig. 6d). Fibroblast progenitor markers, such as Cd34 and Ly6a29,34,35, decreased along the trajectory axis, whereas known markers of peripheral Ccl19+ fibroblasts36,37, including Ccl19, Cxcl10, and Cd74, gradually increased across the axis (Supplementary Fig. 6d). Pathway enrichment analysis revealed that TNF-ɑ– and IL-1–responsive genes were enriched in genes that positively correlated with the pseudotime (Supplementary Fig. 6e), in accordance with results from a previous study37. Overall, the detailed analysis of injection-site fibroblasts revealed target mRNA enrichment in these cells and the emergence of fibroblast populations highly expressing pro-inflammatory cytokines in response to the LNP component of the mRNA vaccine.
mRNA-specific induction of IFN-β in injection-site fibroblasts
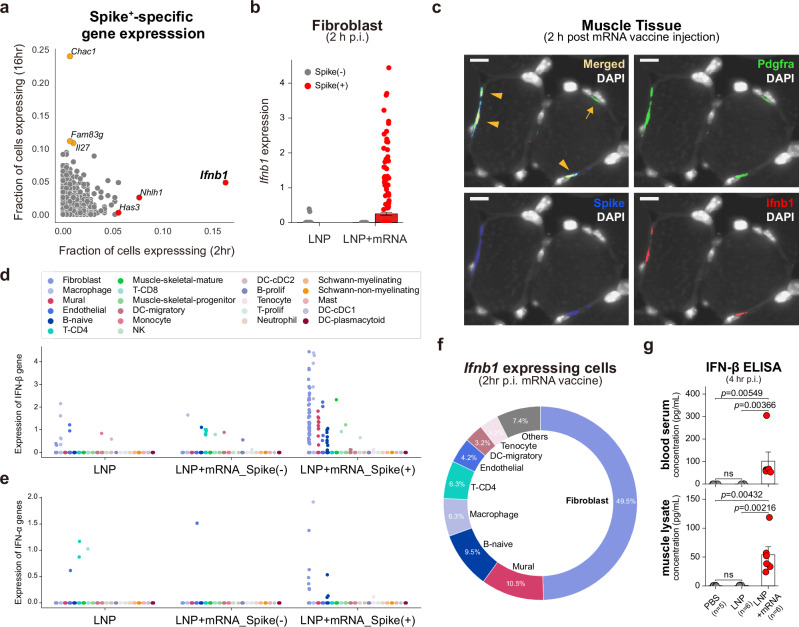
To identify cellular programs in fibroblasts that specifically respond to the mRNA component of the vaccine, we surveyed for genes that can be found only in the target mRNA-detected cells. First, we fetched a list of genes that are expressed in less than 1% of spike- fibroblasts. We then profiled their expression in spike+ fibroblasts from 2 h p.i. samples and 16 h p.i. samples, respectively. Interestingly, we found that the IFN beta gene, Ifnb1, was exclusively expressed in spike+ fibroblasts in the 2 h p.i. samples (Fig. 4a). The absence of Ifnb1 transcripts in empty LNP–injected fibroblasts highly suggests that this type I IFN gene responded specifically to the mRNA molecules within the vaccine (Fig. 4b). To validate this spike-specific induction of Ifnb1 in fibroblasts, we conducted RNA-ISH on the vaccine-injected muscle tissue. ISH images showed that Pdgfra+Spike+ double-positive cells were largely Ifnb1+, whereas Pdgfra+Spike- cells were negative for Ifnb1 transcripts (Fig. 4c and Supplementary Fig. 7a–c), which strongly supports mRNA-specific induction of Ifnb1 transcripts in fibroblasts. We further looked for Ifnb1-expressing cells in cell types other than fibroblasts using the scRNA-seq data from mRNA vaccine 2 h p.i. samples (Fig. 4d) and investigated expression of IFN-α genes, the other major type I IFN, in the same scRNA-seq dataset (Fig. 4e). IFN-α transcripts were barely expressed in 2 h p.i. scRNA-seq data, and ~50% of the Ifnb1-expressing cells were fibroblasts (Fig. 4f), which is remarkable considering the relatively low fractions of fibroblasts in the total population (<5% of total cells in 2 h p.i. scRNA-seq data; Fig. 1g).
Fig. 4. mRNA-specific induction of IFN-β in injection-site fibroblasts.
a Expression profiles of spike- non-expressed genes in spike+ fibroblasts. Fibroblasts without spike mRNA transcripts from the injection-site scRNA-seq data were merged, and genes with an expressing cell ratio <0.01 were designated as the non-expressed genes in spike- fibroblasts. The expression profiles of the spike- non-expressed genes were evaluated in spike+ fibroblasts from either 2 h p.i. or 16 h p.i. samples of mRNA vaccine injection, and their expressing cell ratios are denoted on the x-axis (2 h p.i.) and the y-axis (16 h p.i.) of the scatter plot. b Ifnb1 expression (log-transformed) in each of the injection-site fibroblasts from 2 h p.i. samples of LNP or LNP+mRNA injection. Red indicates spike+ fibroblasts (n = 289 from LNP+mRNA injection), and gray dots represent spike- fibroblasts (n = 1601 from LNP injection, n = 43 from LNP + mRNA injection). Data are presented as mean values ±SEM. c Representative RNA-ISH image acquired from mRNA-vaccine 2 h p.i. muscle tissue. Images were selected from biological replicates of mRNA-vaccine injected samples (n = 4). Green, blue, and red indicate detection signals for Pdgfra, Spike, and Ifnb1 transcripts, respectively. Orange arrowheads indicate co-localization of the fluorescent signals, which implies spike+ fibroblasts expressing Ifnb1 transcripts, and the orange arrow indicates a spike- fibroblast not expressing Ifnb1 transcript. Scale bars indicate 10 µm. d, e Expression of (d) IFN-β transcripts (Ifnb1 gene) and (e) IFN-α transcripts (sum of the 14 IFN-α transcripts) in the injection-site scRNA-seq data from 2 h p.i. samples of LNP and LNP+mRNA. The 2 h p.i. injection site cells were split according to the treatment condition, detection of spike mRNA, and cell type identities. The y-axis represents log-transformed counts of the transcripts. f The fraction of cell types in the cells with Ifnb1 transcripts. g Measured levels of IFN-β in (top) blood and (bottom) muscle samples using ELISA. All the samples were collected 4 h after injections. Data are presented as mean values ±SEM. Statistical evaluations were conducted with two-tailed Mann–Whitney U tests (ns: nonsignificant).
To identify cellular programs associated with the induction of Ifnb1, we conducted gene regulatory network analysis38,39 on the 2 h p.i. fibroblast population. Comparison of regulon activities across treatment conditions revealed enrichment of NF-κB regulon activities in LNP-treated fibroblasts and enrichment of STAT1/2 and IRF regulon activities in the mRNA vaccine–treated fibroblasts (Supplementary Fig. 8a). IRF3 and IRF7 are known positive regulators of type I IFN transcription40,41, so we evaluated the regulon activities of these transcription factors in detail. We found that the inferred activities of Irf7 were highly specific to the spike+ cells among the fibroblasts in mRNA vaccine–injected muscle, whereas the activities of Irf3 were minimal (Supplementary Fig. 8b, c). These results suggest Irf7-dependent induction of Ifnb1 in early injection-site fibroblasts. We also discovered that the activity of IFN-stimulated gene factor 3 (ISGF3), which consists of STAT1/2 and IRF941, was spike-specific (Supplementary Fig. 8d–f), whereas NF-κB regulon activities were robust in both LNP- and LNP+mRNA-injected fibroblasts (Supplementary Fig. 8g–k). To validate our findings from the transcriptome data, we measured protein levels of IFN-β in vaccinated mice using enzyme-linked immunosorbent assay (ELISA) 4 h after the injections. We found no significant increase in blood and muscle levels of IFN-β in mice injected with empty LNP, in contrast to the mice injected with the mRNA vaccine, which showed substantial rises in blood and muscle IFN-β levels (Fig. 4g). Overall, these data suggest that injection-site fibroblasts produce IFN-β specifically in response to the mRNA component of the mRNA vaccine.
mRNA-specific induction of type I IFN responses in mDCs
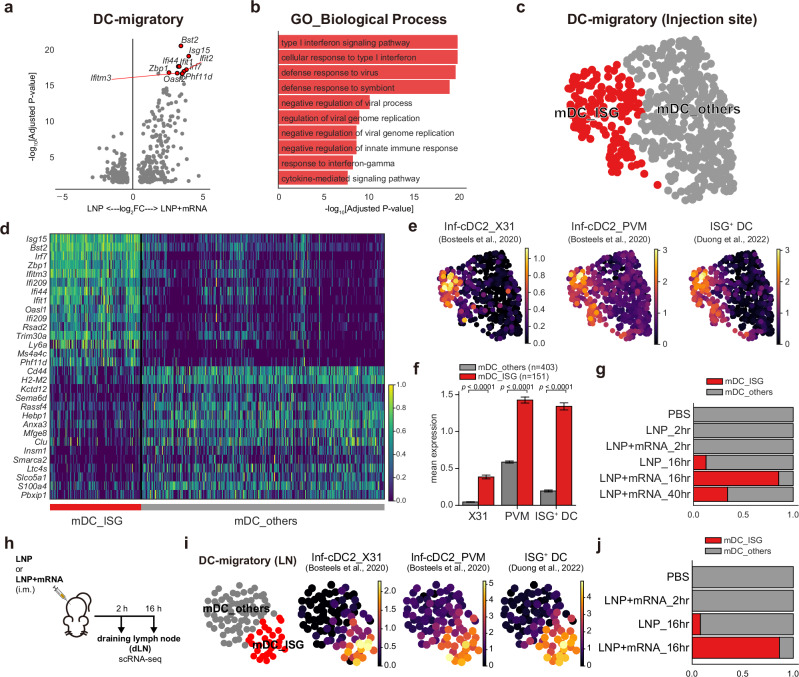
Our analysis of mRNA vaccine injection-site responses revealed two major axes of transcriptional responses: PC1, representing inflammatory responses in stromal cells that are robust even in empty LNP–injected samples, and PC2, representing type I IFN responses attributable to the mRNA component (Fig. 2b–d). Given that discrepancies between the empty LNP and mRNA vaccines on the PC2 axis were highest in the mDCs (Fig. 2c), we isolated scRNA-seq data of these cells from 16 h p.i. muscle tissue and directly compared their gene expression patterns (Fig. 5a and Supplementary Data 3). The results revealed upregulation of type I IFN response–related genes in mRNA vaccine samples (Fig. 5b). Accordingly, sub-clustering of mDCs revealed a distinct cell cluster that was highly enriched in IFN-stimulated genes, which we designated as mDC_ISGs (Fig. 5c, d). The cells in the mDC_ISG cluster highly expressed transcriptional signatures of the IFN-responsive dendritic cells, which are efficient in priming T-cell responses under virus infection42 and tumor43 conditions (Fig. 5e, f). The differences in the fraction of mDC_ISGs in mRNA vaccine–injected muscle tissue (~85.4%, 16 h p.i.) and empty LNP–injected muscle tissue (~12.6%, 16 h p.i.) strongly suggested mRNA-specific induction of these IFN-responsive mDCs at the injection site (Fig. 5g).
Fig. 5. mRNA-specific induction of type I IFN responses in mDCs.
a Volcano plot showing DEGs between the mDCs from 16 h p.i. injection-site scRNA-seq data of LNP and LNP+mRNA injection. The x-axis represents log2FC of genes (positive value indicates higher expression in LNP + mRNA), and the y-axis represents −log10 values of the adjusted P values (two-tailed, Benjamini-Hochberg adjustment). b Top 10 terms from the pathway enrichment analysis (GO_Biological Process) on the upregulated DEGs of LNP+mRNA (adjusted P < 0.05 and log2FC > 1). c UMAP representation of mDC clusters. d Marker genes of the mDC clusters are displayed on a heatmap, with rows indicating representative genes and columns corresponding to cells in each mDC cluster. e, f Average expression of the transcriptional signatures of IFN-responsive dendritic cells from previous studies42,43, displayed (e) on the UMAP plot of mDC populations or (f) as a bar plot. Data are presented as mean values ±SEM. Statistical evaluations were performed with the Mann–Whitney U test (two-tailed, ****P < 1 × 10−4). g Fraction of mDC clusters in each injection-site scRNA-seq sample. h Experimental scheme: Mice were given intramuscular injections of LNP or LNP + mRNA and the draining lymph nodes were collected 2 or 16 h after the shot. i Average expression of the IFN-responsive dendritic cell signatures in lymph node mDC populations, as in (e). j Fraction of mDC clusters in each dLN scRNA-seq data. Schematic images were created with BioRender.com.
To further track the fate of these mDC_ISGs, we additionally conducted scRNA-seq analysis on the dLNs, i.e., the ipsilateral inguinal and iliac lymph nodes of the vaccine-injected muscles (Supplementary Fig. 9a–d). We were able to identify the mDC_ISG cluster among the mDC population in dLNs (Fig. 5h). We confirmed that the mDC_ISGs in the lymph nodes highly expressed features of IFN-responsive dendritic cells, like mDC_ISGs from the injection site (Fig. 5i). Notably, the proportion of mDC_ISGs was substantially higher in the mRNA vaccine–injected samples compared with the empty LNP–injected sample (16 h p.i.) (Fig. 5j). This result strongly indicates that the mRNA-specific induction of mDC_ISGs robustly propagates to the dLNs, where adaptive cell-mediated immunity originates44, possibly through cell migration or draining of interstitial fluids containing either the unabsorbed vaccine components or secreted cytokines from the injection site. To summarize, we have identified mRNA-specific induction of IFN-responsive mDCs, both at the injection sites and at the dLNs.
IFN-β at the injection site guides cellular immune responses against mRNA vaccine
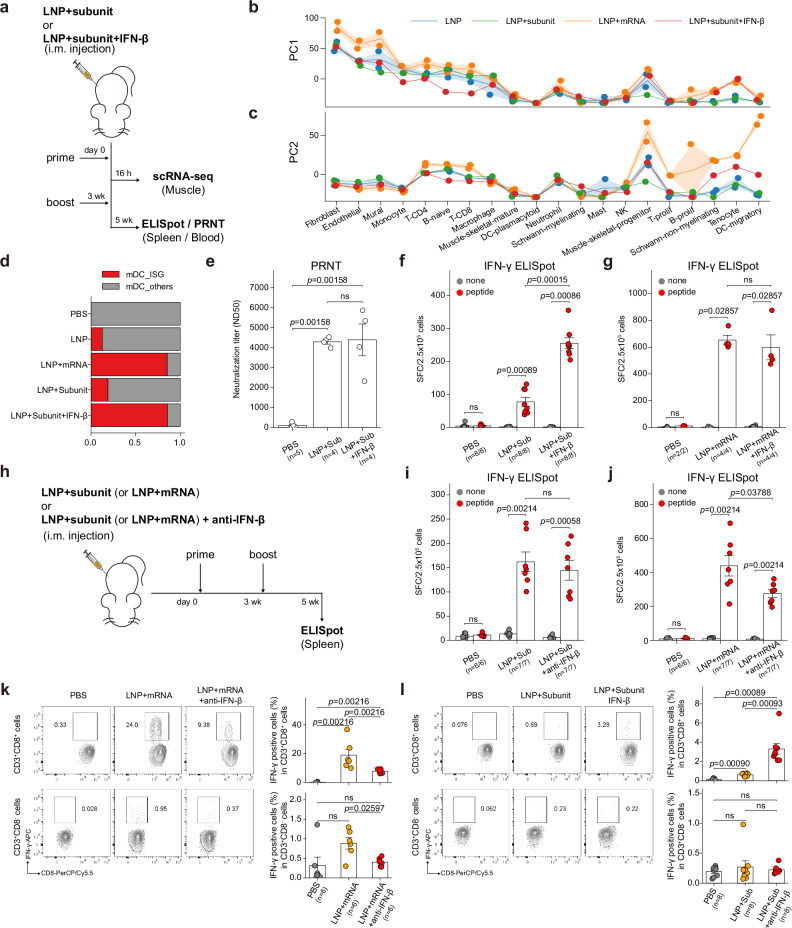
In this study, we have identified two mRNA-specific responses at the earliest phase of mRNA vaccine injection. In the first phase, injection-site fibroblasts expressed IFN-β specifically in response to the mRNA component of the vaccine within ~2 h after the injection. In the second phase, mRNA-specific induction of mDC_ISGs, which feature strong type I IFN responses, was enriched at the injection site and at the dLNs at around ~16 h after injection. Therefore, we postulated that the IFN-β at the injection site, which is induced by the mRNA component, specifically induces type I IFN responses in the mDC population. To evaluate the sole effect of injection-site IFN-β, we used a protein subunit of spike proteins, instead of mRNA transcripts, as a measure to deliver the antigen information. We compared the transcriptional responses of LNP+subunit and LNP+subunit+IFN-β at the injection site (16 h p.i.) by generating an additional scRNA-seq dataset (Fig. 6a). To evaluate the multicellular responses in these newly generated scRNA-seq data, we used the same analysis framework (cell-wise DEG vectors projected on the principal component space).
Fig. 6. Injection-site IFN-β guides cellular immune responses against mRNA vaccine.
a Experimental and sample preparation scheme: Mice were immunized with intramuscular injection of LNP+subunit or LNP+subunit+IFN-β. Two injections were given (prime and boost shot) with 3 weeks between shots. Injection-site muscles were resected 16 h after the prime shot, and spleen tissue for the ELISpot assay was sampled 2 weeks after the boost shot. b, c PC projections of the cell type–wise DEG vectors (vs PBS injection) of 16 h p.i. samples. (b) PC1 projections and (c) PC2 projections of the DEG vectors are plotted as line plots. Line plot indicates mean values, and the error bands indicate 95% confidence intervals. d Fraction of injection-site mDC clusters in each treatment condition. e PRNT assay conducted on mice intramuscularly injected with PBS, LNP+subunit, or LNP+subunit+IFN-β. Results from IFN-γ ELISpot assay. f, g Effects of IFN-β i.m. co-injection were evaluated in (f) LNP+subunit and (g) LNP+mRNA (mRNA vaccine) vaccination strategies. h Experimental scheme: Mice were immunized with intramuscular injection of LNP+subunit or LNP+mRNA, with or without anti-IFN-β blocking antibody. Shots were given twice and the spleen samples for ELISpot assay were collected 2 weeks after the boost shot. i, j Results from the IFN-γ ELISpot assay. Effects of IFN-β blocking antibody i.m. co-injection were evaluated in (i) LNP+subunit and (j) LNP+mRNA vaccination strategies. k, l Antigen-specific T cell responses at 35 days after vaccination as determined by flow cytometry analysis. Effects of (k) IFN-β blockade and (l) IFN-β addition are evaluated in (top panels) CD8+ and (bottom panels) CD8- T cells. All the data in bar plots are presented as mean values ±SEM. All statistical evaluations were conducted with two-tailed Mann–Whitney U tests (ns: nonsignificant). Schematic images were created with BioRender.com.
First, we discovered that the PC1 responses were robustly induced in all LNP-injected samples (LNP, LNP+mRNA, LNP+subunit, and LNP+subunit+IFN-β) (Fig. 6b). This result reassures that the PC1 responses, which feature strong stromal inflammatory responses, are largely LNP-driven. Interestingly, we discovered that PC2 responses, which feature type I IFN responses in the mDC population, were partially rescued in the IFN-β–added samples (red line; Fig. 6c), whereas PC2 responses in mDCs from LNP- or LNP+subunit-injected samples remained negligible (blue and green line; Fig. 6c). We also profiled the fraction of mDC_ISGs in each condition and found that it was fully rescued in IFN-β–co-injected samples (Fig. 6d). These results indicate that IFN-β at the injection site induces strong type I IFN responses in mDC populations, which can be identified as the induction of mDC_ISGs at the single-cell population level.
Conventional dendritic cells with type I IFN responses are highly efficient in priming CD8 T-cell responses in anti-tumor43 and anti-viral immunity42. Likewise, in the context of mRNA vaccine immunity, impaired antigen-specific CD8 T-cell responses in Ifnar−/− and Mda5−/− mice have been reported10. Thus, we hypothesized that the IFN-β at the injection site promotes robust induction of antigen-specific cellular immune responses. We compared the effects of LNP+subunit and LNP+subunit+IFN-β immunizations for their induction of antigen-specific cellular immune responses at 2 weeks after the boost shot (Fig. 6a). First, we found that the production of neutralizing antibodies was not affected by the IFN-β co-injection (Fig. 6e). Notably, we discovered that co-injection of IFN-β substantially enhanced cellular immune responses in LNP+subunit vaccination strategy (Fig. 6f). Moreover, the intramuscular co-injection of IFN-β elevated antigen-specific cellular immune responses also in the alum-adjuvanted vaccination strategy (Supplementary Fig. 10a), and even in the adjuvant-free strategy (Supplementary Fig. 10b). These results suggest LNP-independent mechanisms of IFN-β in the elevation of cellular immune responses. Intriguingly, the effect of IFN-β co-administration was negligible in LNP+mRNA vaccination (Fig. 6g), possibly due to the saturation of type I IFN responses by the endogenous IFN-β produced at the injection site in response to the mRNA components of the mRNA vaccine (Fig. 5b–g).
Finally, we evaluated the effect of blocking of IFN-β signaling in these vaccination strategies (Fig. 6h). In LNP+subunit vaccination strategy, the vaccine-induced cellular immune responses were not affected by intramuscular co-injection of IFN-β blocking antibodies (Fig. 6i). Notably, the cellular immune responses against spike proteins were significantly decreased by the intramuscular co-injection of IFN-β blocking antibodies in LNP+mRNA vaccination strategy (Fig. 6j), in contrast to the intraperitoneal pretreatment of IFN-β blocking antibodies which did not incur any difference (Supplementary Fig. 10c). We further validated the effect of IFN-β addition and blockade by measuring antigen-specific induction of IFN-γ responses in T cell subsets after vaccination (Supplementary Fig. 11). The results from the flow cytometry analysis reaffirmed that the blockade of IFN-β at the injection site significantly reduces antigen-specific CD8 T cell responses in mRNA vaccination (Fig. 6k), and the addition of IFN-β in LNP+subunit vaccination substantially enhances antigen-specific CD8 T cell responses (Fig. 6l). Collectively, these results indicate that injection-site IFN-β can promote antigen-specific cellular immune responses, and it is also responsible for the cellular immune responses induced by mRNA vaccines.
Discussion
The immunological mechanisms of mRNA vaccines have been systematically addressed in several recent studies10,11,14. The earliest immune responses at the vaccine injection site had not previously been systematically assessed, however, and the source of type I IFNs eliciting a tide of global type I IFN responses in blood11 or lymph nodes10 has remained elusive. Here, by deeply profiling injection-site cellular responses at single-cell resolution, we systematically analyzed the initial transcriptional responses elicited by mRNA vaccine injection. To identify major axes in transcriptional responses across cell types, we conducted PCA on the DEG vectors representing transcriptional changes in each cell type. From the PCA, we found that the LNP-driven PC1 response, featuring inflammatory responses in the stromal cells, and the mRNA-driven PC2 response, featuring type I IFN responses in mDCs, dominate the earliest immune responses at the mRNA vaccine injection site. To the best of our knowledge, this work is the first systematic dissection of injection-site responses elicited by mRNA vaccines.
Target organ and cell types for mRNA delivery can differ according to the chemical properties of the nanoparticles used, representing a major research interest in developing LNP-based drugs25,45–47. In the context of intramuscular injection of mRNA vaccines, antigen production is anticipated to be maximized at the injection site and at the dLNs48. The fate of mRNAs delivered to the dLNs was investigated in a recent study10, which showed that the exogenous spike mRNAs were enriched in monocytic myeloid cells. In the current work, we showed that spike mRNAs are highly enriched in fibroblasts at the injection site. In addition, inflammatory fibroblasts (Fib_Cxcl5 and Fib_Ccl19) emerge at the injection site in response to the LNP component of the vaccine and express diverse chemokine genes that can recruit immune cells to the injection site. Inflammatory fibroblasts and their role in various inflammatory diseases have been reported in several recent studies29,36,37,49, including the role of Ccl19+ fibroblasts36,37,50,51, but their role and even their existence at the mRNA vaccine injection site have not been reported previously. Collectively, both the high-throughput survey on the fate of delivered mRNA and the comparative cell-to-cell signaling analysis in this study underscore the role of injection-site fibroblasts in mRNA vaccine immunity.
Our investigation of the spike mRNA content in fibroblasts revealed that Ifnb1, the gene encoding IFN-β, is specifically expressed in spike mRNA+ fibroblasts, especially in 2 h p.i. tissue. Ifnb1 transcripts were barely expressed in fibroblasts of empty-LNP–injected muscle tissue, which strongly suggests that Ifnb1 expression in spike+ fibroblasts is specifically induced by the mRNA component of the mRNA vaccine. Since the PC2 responses, featuring type I IFN responses in mDCs, were highly specific to LNP+mRNA–injected samples, we postulated that IFN-β at the injection site induces strong type I IFN responses in the mDC population (identified as mDC_ISGs at cluster level). Accordingly, we discovered that supplementing the mRNA-free LNP injection with IFN-β rescued induction of the mDC_ISG population at the injection site. Moreover, the addition of IFN-β to the LNP+subunit injections significantly enhanced antigen-specific cellular immune responses, in agreement with a recent report describing significantly impaired antigen-specific CD8 T-cell responses in Ifnar -/- and Mda5 -/- mice10,52. We also discovered that the cellular immune responses induced by mRNA vaccines are dependent on the injection site IFN-β. The mechanisms of how IFN-β at the injection site fosters robust development of antigen-specific cellular responses are not clearly demonstrated here, a limitation of this study. Nonetheless, findings from recent studies42,43 showing that type I IFN signaling in dendritic cells can promote antigen-specific CD8 T-cell responses, are consistent with the potential role of mDC_ISGs in the enhancement of cellular immune responses.
LNPs and mRNA are the two major components of the mRNA vaccine. The immunogenic role of LNPs has drawn more attention because of their strong adjuvanticity15,16,48,53. A recent study16 regarding the adjuvanticity of LNPs showed that LNPs alone can act as a highly potent protein vaccine adjuvant through the production of IL-6, which plays a pivotal role in LNP-induced germinal center reactions. Accordingly, the PC1 reactions identified in our study, featuring strong stromal induction of inflammatory cytokines, including IL-6, TNF-ɑ, and MCP-1, were robustly induced in both LNP+mRNA and empty LNP injections. In contrast to the effects of LNPs, the immune responses elicited by mRNA were generally considered as an obstacle in mRNA therapeutics because it would hamper target deliveries54,55. In this study, we discovered that the transcriptional induction of IFN-β at the injection site was highly specific to the mRNA component of the vaccine, and it was further validated by measuring serum levels of IFN-β, which was detectable only in LNP+mRNA–injected mice. Considering both the significant enhancement of cellular responses in IFN-β–co-injected mice from LNP+subunit strategy and the substantial decrease in cellular responses in IFN-β-blocked mice from LNP+mRNA vaccination strategy, these results highlight the role of the mRNA component of mRNA vaccines as a potent adjuvant that can boost cellular immune responses against the vaccine antigen.
In summary, our detailed analysis of the mRNA vaccine injection site not only identified major axes of initial immune responses but also identified major targets of mRNA vaccine delivery. We found that fibroblasts at the injection site are highly enriched with delivered mRNA and dramatically increase chemokine expression in response to the vaccine injection. Furthermore, our systematic comparison of the major components of the mRNA vaccine – LNP and mRNA – both reaffirmed the strong adjuvanticity of the LNP component and highlighted the overlooked contribution of the mRNA component to the proper development of cellular immune responses, through the induction of IFN-β at the injection site. Overall, these results provide novel insights into the mechanisms of vaccine immunity that could prove useful for developing more efficient mRNA vaccines.
Methods
Mice and immunization
Female BALB/c mice aged 4–6 weeks were purchased from Samtako Bio Korea. Mice were housed and bred in the specific pathogen–free mouse facility, where temperature, humidity, and light cycle were controlled. The animal protocol used in this study was reviewed and approved by the Institutional Animal Care and Use Committee of the Korea Disease Control and Prevention Agency (KCDC-IACUC-22-004). Animal experiments were conducted in an animal biosafety level 2 facility. Vaccines were administered into the muscles of the hindlimb. Mice were immunized with one of the following conditions: PBS, empty LNP (equivalent to lipid content of 5 μg mRNA-LNP), 5 μg mRNA-LNP, 10 μg subunit vaccine+empty LNP (equivalent to lipid content of 5 μg mRNA-LNP), 5 μg subunit vaccine, or 5 μg subunit vaccine+Alum. For IFN-β co-injection strategies, 0.5 μg of recombinant IFN-β (12401-1, PBL assay science) was co-administered with the injections. For IFN-β neutralization, BALB/c mice were co-injected intramuscularly with 0.3 μg of anti-IFN-β antibody (Clone HD β-4A7; Leinco Technologies) along with 5 μg mRNA or 10 μg subunit vaccines. Two shots were given 3 weeks apart. Blood samples and spleens were collected at 5 weeks after the first vaccination for analysis of humoral and cellular responses, respectively.
Preparation of empty LNP, mRNA-LNP, and recombinant subunit vaccines
Spike (S) antigen encoded by mRNAs used in this study was from SARS-CoV-2 isolate 2019-nCoV/USA-WA1/2020 (GenBank: MN985325). A human codon–optimized S sequence (IDT codon-optimization tool)56 with two proline substitutions (K986P/V987P)57–59 was synthesized and cloned into an mRNA production plasmid. A linearized DNA template containing the open reading frame flanked by 5′ and 3′ untranslated regions3 (UTRs) (5′ UTR: GGGAAAUAAGAGAGAAAAGAAGAGUAAGAAGAAAUAUAAGAGCCACC; and 3′ UTR: UGAUAAUAGGCUGGAGCCUCGGUGGCCAUGCUUCUUGCCCCUUGGGCCUCCCCCCAGCCCCUCCUCCCCUUCCUGCACCC GUACCCCCGUGGUCUUUGAAUAAAGUCUGA) and 120-nucleotide poly(A) tails were produced by PCR amplification. mRNAs were synthesized in vitro using T7 RNA polymerase–mediated transcription with complete replacement of uridine by N1-methyl-pseudouridine-5′-triphosphate (m1Ψ, N-1081, TriLink). Capping of the in vitro–transcribed mRNAs was performed a co-transcriptionally using the trinucleotide cap1 analog (m7(3’OMeG)(5’)ppp(5’)(2’OMeA)pG, N-7413, TriLink). After in vitro transcription, the DNA template was removed by DNase, and the final mRNA product was purified using RNase-free SPRI magnetic beads (A63987, Beckman). RNA integrity was analyzed by automated electrophoresis (Supplementary Fig. 1a, b, Agilent TapeStation). RNA concentration was spectrophotometrically quantified, and the sample was stored at −80 °C prior to LNP formulation. LNPs were prepared using NanoAssemblr Benchtop Instrument (Precision Nanosystems Inc). Ionizable lipids (ALC0315, 2036272-55-4, SINOPEG), cholesterol (C3045-100G, Sigma-Aldrich), distearoylphosphatidylcholine (DSPC, 850365 P, Avanti), and PEG-lipid (ALC-0159, 1849616-42-7, SINOPEG) were dissolved in ethanol, and mRNA was diluted in 10 mM citrate (pH 3). The molar ratio of the lipid components was ALC-0315:DSPC:cholesterol:ALC-0159 = 46.3:9.4:42.7:1.6. The final ionizable lipid:RNA weight ratio was 10:1, and the final volume ratio was 1:3. The LNPs were formulated by microfluidic mixing of the prepared solutions at a 12 ml/min flow rate. The resulting LNPs were diluted in a 40-fold volume of 1× PBS and concentrated via ultrafiltration (Amicon® Ultra-15 Centrifugal Filter Unit, UFC9010). mRNA-loaded LNPs were further characterized for physical properties. The hydrodynamic size was measured by DLS using a Malvern Nano-ZS zetasizer (Malvern Instruments Ltd) to confirm mean particle diameter and the polydispersity index. RNA concentration and encapsulation efficiency were confirmed using the RiboGreen Assay (ThermoFisher Scientific, Waltham, MA, USA). The recombinant SARS-CoV-2 spike protein was composed with a consensus sequence of 537 isolates belonging to the delta variant in Korea, containing six proline mutations for stability. The protein was produced by a mammalian expression system using the CHO-S cell line and purified using a HisTrapTM FF Crude column.
Cell and virus harvest
Vero E6 cells were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin. Cells were cultured at 37 °C under 5% CO2 in an incubator. The first human-isolated SARS-CoV-2 strain in Korea (BetaCoV/Korea/KCDC03/2020, NCCP 43326) was passaged and titrated as PFU in Vero E6 cells. All experiments involving live virus were conducted in a biosafety level 3 facility following the recommended safety precautions and measures.
Plaque reduction neutralization assay (PRNT)
PRNT was performed to analyze neutralizing antibodies to the mRNA vaccine. PRNT titers were determined using the highest serum dilution that inhibited >50% of the number of plaques in the absence of test serum. Serum samples from 5 weeks after the first vaccination were heat-inactivated for 30 min at 56 °C prior to use. The prepared serum samples from immunized mice were serially diluted from 1:10 to 1:5120 in a serum-free medium. Virus–serum mixtures were prepared by 50 PFUs (for mouse serum) of SARS-CoV-2 with the diluted serum samples, and the mixtures were incubated at 37 °C for 1 h. Vero E6 cells in 12-well plates (2 × 105 cells/well) were inoculated with the virus–serum mixtures, and the plates were incubated at 37 °C under 5% CO2 for 1 h. After virus absorption, a 1.2% agar overlay medium was added, and the plates were incubated at 37 °C under 5% CO2 for 2 days for cell fixation. Plaques were counted after crystal violet mixture staining.
IFN-γ enzyme-linked immunospot assay (ELISpot)
ELISpot was performed using splenocytes isolated from mice at 5 weeks after the first vaccination (2 weeks after the second vaccination). IFN-γ–secreting cells were detected using the mouse IFN-gamma ELISpot kit (XEL485, R&D System). The assays were performed according to the manufacturer’s protocol. Splenocytes (2.5 × 105 cells/well) were spread in the 96-well polyvinylidene fluoride–backed microplate coated with a monoclonal antibody specific for mouse IFN gamma (890894, R&D System). Splenocytes were stimulated with 100 ng/well SARS-CoV-2 Spike Glycoprotein-crude (RP30020, GeneScript). Tests were conducted with a negative control containing only medium and a positive control (Cell Stimulation Cocktail; 00-4970-93, eBioscience). Cells were incubated in a 37 °C incubator for 18–24 h. The plate from which cells were removed was washed with a wash buffer (895308, R&D Systems), and a biotinylated monoclonal antibody specific for mouse IFN-gamma (890895, R&D Systems) was added to each well and incubated for 2 h at room temperature on a rocking platform. After washing, plates were incubated with streptavidin conjugated to alkaline phosphatase (895358, R&D Systems) for 2 h. Incubated plates were developed using BCIP/NBT Substrate (895867, R&D Systems) for 1 h at room temperature. Spots were counted using an ImmunoSpot reader.
Serum and lysate enzyme-linked immunosorbent assay (ELISA)
Mice were inoculated with PBS, Empty LNP, and LNP+spike mRNA, and 4 h after the injections, mouse blood serum and muscle samples were obtained. The muscle sample at the injection site was homogenized with 4 times the amount of muscle volume in PBS and homogenized with speed 8500 rpm, 5 cycles of 20 s, 15 s break between cycles according to the Precellys animal tissue protocol (Precellys lysing kit, CK28R 2 ml). Serum and lysate ELISA were performed on homogenized muscle samples according to the protocol in Mouse IFN-β ELISA Kit, High Sensitivity (42410-1, PBL). Standards and samples on the plate were shaken 650 rpm for 1 h. After washing, antibody solution was added, and the plate was incubated at 650 rpm for 30 min. HRP solution was added after washing and the plate was incubated at 650 rpm for 10 min. We added TMB substrate solution after washing step and incubated plate in a dark place for 10 min. Stop solution was added to stop the reaction, and the plate was read at 450 nm.
Flow cytometry analysis
Spleens were collected 35 d after prime (14 d after boost) and cells were isolated by mechanical disruption using a gentleMAX machine. After filtering with strainers, red blood cells were removed using an ACK lysis buffer. Cells were suspended in RPMI containing 10% FBS and 1% P/S and seeded at 1 × 106 cells per 100 µL in a 96-well U-shaped plate. Cells were stimulated with 100 ng/well SARS-CoV-2 Spike Glycoprotein-crude (RP30020, GeneScript) in complete RPMI 1640 medium. GolgiPlug (51-2301KZ, BD) was added to each well and incubated in a 37 °C incubator for 18 to 24 h. After collecting the stimulated cells in a tube, cells were washed with PBS. For Live/Dead staining, cells were stained with Aqua Fluorescent Reactive Dye (1:300 dilution; L34965, Invitrogen) and incubated for 30 min at 4 °C. After washing steps, cells were stained with following surface marker antibodies: anti-CD3 (1:200 dilution; APC/Cyanine7; 100222, BioLegend), anti-CD8 (1:200 dilution; PerCP/Cyanine5.5; 100734, BioLegend), for 30 min at 4 °C, and FC Receptor block antibody anti-CD16/32 (1:500 dilution; 156604, BioLegend) was added to Flow cytometry staining buffer (00-4222-26, eBioscience). For intracellular cytokine staining, cells were washed with the Cytofix/Cytoperm solution (51-2090KZ, BD) added staining buffer according to Cytofix/CytopermTM Plus (555028, BD) protocol, and the cells were incubated for 40 min at 4 °C and washed twice with 1X Perm/Wash buffer (51-2091KZ, BD). We added anti-IFN-γ (1:100 dilution, APC, 554413, BD) antibodies to the cells and incubated for 1 h at 4 °C. Finally, cells were washed with 1X Perm/Wash buffer and resuspended with the Flow cytometry staining buffer. Data were acquired on a CytoFLEX LX flow cytometer (Beckman Coulter) and analyzed using FlowJo analysis software v10.
RNA in situ hybridization assay (RNA-ISH)
Resected muscle tissues were formalin-fixed for 24 h, paraffin-embedded with tissue processor (TP1020, Leica), and sliced at 5 µm onto SuperFrost Plus adhesion slides (J1800AMNZ, ThermoFisher) with a rotary microtome (RM2235, Leica). The formalin-fixed paraffin-embedded slices were deparaffinized and pretreated according to the manufacturer’s instructions (RNAscope H2O2 and Protease Reagents; 322381, ACD). ISH was performed with the RNAscope kit (RNAscope Multiplex Fluorescent Detection Reagents v2, 323110, ACD; RNAscope 4-Plex Ancillary Kit for Multiplex Fluorescent Kit v2, 323120, ACD), using the following RNAscope probes: Mm-Pdgfra-C3 (480661-C3, ACD), Mm-Ifnb1-C4 (406531-C4, ACD), Mm-Ptprc (318651, ACD) and V-SARS-CoV-2-opt-O1-C2 (1196611-C2: custom probe, ACD). For fluorescent staining, the following dyes were used: OPAL 520 (1487001KT, Akoya), OPAL 570 (1488001KT, Akoya), and OPAL 690 (1497001KT, Akoya). Fluorescent detection was performed using a digital fluorescence slide scanner (Pannoramic SCAN II, 3D HisTech).
Single-cell RNA sequencing (scRNA-seq)
Muscles resected from the vaccine-injected site were mechanically dissected with scissors and rinsed with 1× PBS. For chemical digestion, we incubated samples with Type II collagenase (LS004177, Worthington) at 37 °C for 40 min with gentle shaking. To increase cell yield and viability, we additionally incubated cells with Type II collagenase (LS004177, Worthington) and Dispase II (04942078001, Roche) at 37 °C for 20 min with gentle shaking. Finally, we passed the digestions 10 to 12 times through an 18-gauge needle on a 5-ml syringe. To remove extracellular matrices, we filtered the digestions through a 40-µm strainer. The pass-throughs were centrifuged at 525 × g for 5 min at room temperature. For removal of red blood cells (RBCs), cell pellets were treated with RBC lysis buffer (eBioscience™ 1X RBC Lysis Buffer; 00-4333-57, Thermo) for 3 min. After RBC lysis, cells were washed and resuspended with 10 ml of complete Roswell Park Memorial Institute (RPMI) medium. To remove debris entirely, cells were filtered with Flowmi Cell Strainers with a porosity of 40 µm (136800040, Merck).
Draining iliac lymph nodes and draining inguinal lymph nodes near the injection sites were collected and digested with Collagenase D (11088866001, Roche) at 37 °C for 20 min. Subsequently, samples were smashed on a 40-μm strainer to create a single-cell suspension. Cells were resuspended and treated with RBC lysis buffer (eBioscience™ 1X RBC Lysis Buffer; 00-4333-57, ThermoFisher) for 3 min and washed with RPMI medium before being resuspended in 10 ml complete RPMI and counted with hemocytometers.
To prepare single-cell suspensions for scRNA-seq, cell pellets were resuspended up to 106 cells per milliliter of 0.04% bovine serum albumin (130-091-376, Miltenyi Biotec). scRNA-seq libraries were constructed with Chromium Next GEM Single Cell 3’ Library Kit v3.1 (1000268, 10x Genomics). Single-cell gel beads in emulsions were generated using a Chromium Controller (100171, 10x Genomics), with a targeted cell recovery of 10,000 cells. After reverse transcription, cDNAs were pre-amplified with 11 reaction cycles. Fragmentation, adaptor ligation, and SPRIselect processes were conducted according to the manufacturer’s guidelines. Fragment sizes of the constructed libraries were measured with Bioanalyzer (2100, Agilent Technologies). Sequencing of the constructed libraries was performed with Novaseq 6000 (Illumina), with a per-sample library size of approximately 100 Gb.
Count matrices were generated by the CellRanger (v.6.0.2) pipeline, using mm10 reference, and downstream analysis was conducted based on the Scanpy60 pipeline (v.1.9.1). Low-quality cells meeting any one of the following criteria were excluded in the downstream analysis: (1) UMI gene count <1000, (2) number of genes detected <500, (3) number of genes detected >7000, and (4) fraction of mitochondrial genes >10%. Predicted doublets identified by Scrublet61 (v.0.2.3) also were removed. Raw counts were normalized up to 10,000 counts per cell and log-transformed (log1p). A total of 2645 highly variable genes were selected with the highly_variable_genes function in the Scanpy package. Subsequently, the expression matrix of the highly variable genes was standardized using the scale function of the preprocessing module in the Scanpy package. PCA (number of PCs = 50) was conducted based on the scaled expression values of highly variable genes. Neighborhood graphs of the cells were constructed with the BBKNN62, and we used the UMAP function in the tool module of the Scanpy package to acquire UMAP embeddings. For spike mRNA detection, raw sequence files were aligned to a custom-built spike mRNA reference using CellRanger (v.6.0.2), and the spike mRNA counts of each cell were imported to the processed scRNA-seq data for further analysis.
Differential abundance testing and differentially expressed gene analysis (DEG)
Differential cell composition testing was conducted with scCODA23, a Bayesian model for differential cell composition analysis in single-cell transcriptome data. The cut-off value for the false discovery rate (FDR) was set to 0.05, and only the log-fold changes below the FDR cut-off value were used. Differential cell abundance testing at a local community level was conducted using Milo24, with alpha value set to 0.1. DEG analysis was performed using the rank_genes_groups function in Scanpy (v.1.9.1) package. Log-normalized counts of the cells were compared using the Wilcoxon rank-sum method, and P values were corrected with the Benjamini-Hochberg method. Genes with adjusted P < 0.05 and log2FC > 1 were considered as DEGs.
Principal component assay (PCA) on DEG vectors
Using the saline-injected sample as a reference, we conducted DEG analysis on each cell type in each treatment sample. The saline-injected sample contained 20 cell types (excluding DC-cCDC1 and DC-cDC2, which had cell counts less than 3), and our initial analysis contained nine treatment samples (two samples each from 2 h and 16 h p.i. samples of LNP or LNP+mRNA injection and one sample from 40 h p.i. LNP+mRNA). Accordingly, we conducted 178 (20 × 9 - 2; one treatment sample had cell counts <3 for two cell types) independent DEG analyses. DEG vectors were constructed in two steps. First, we fetched log-fold change (logFC) vectors and adjusted P value vectors of genes from the DEG results. Second, for the genes with adjusted P > 0.05, values in the logFC vectors were replaced with 0 to ensure robustness of the logFC values.
To focus on identification of shared responses among different cell types, we first analyzed the DEG vectors from the 16 h p.i. LNP+mRNA (mRNA vaccine) sample, where the overall transcriptional responses peaked (Fig. 2a). The zero imputed DEG vectors of 16 h p.i. mRNA vaccine injections were concatenated into a single matrix, with dimensions of 40 × 36601 (number of total genes). PCA was conducted on the merged 16 h DEG matrix using scikit-learn (v.1.2.2). Using the weight vector matrix from the PCA (Supplementary data 1), we projected the whole DEG vector matrix, which had 178 × 36601 dimensions, onto the PC1 and PC2 axes, which yielded a 178 × 2 dimension projection matrix (Fig. 2c, d). Computationally, we used the transform function in the scikit-learn PCA object, which is fitted with the 16 h DEG matrix, for PC projections of the whole DEG vectors.
Gene set enrichment analysis and signature scoring
Gene set enrichment analysis was performed using the enrichr function in the GSEApy63 package (v.1.0.4). For upregulated or downregulated DEGs, we used genes with adjusted P < 0.05 in each direction. To calculate gene set signature scores, we calculated average expression (log-transformed normalized counts) of genes included in each gene set. Signatures of IFN-responsive dendritic cells were acquired from two previous studies42,43.
Cell–cell communication network analysis
We used CellChat31 (v.1.6.1) for cell-to-cell communication network analysis. From the processed scRNA-seq data, scRNA-seq data generated from mRNA vaccine–injected or saline-injected muscle tissues were extracted and converted to Seurat64 RDS files. The extracted data were split according to the p.i. time and converted to CellChat objects using cell type annotation as identities. For interaction analysis, we used the Secreted Signaling module from the CellChatDB mouse database. We filtered communications (minimum number of cells ≥10), and the contributions (outgoing signaling strength) of each cell type to the inferred communication networks were compared across the p.i. time points.
Trajectory inference analysis
We used Monocle333,65 for trajectory inference analysis. Fibroblasts from LNP-injected samples were isolated from the processed scRNA-seq data and converted into a Seurat RDS file. Trajectory graphs were constructed with the learn_graph function in Monocle3. Pseudotimes were calculated with order_cells function in Monocle3, using Fib_Pi16, a presumed global fibroblast population29, as a root node. Among the trajectory branches, a branch spanning to the Fib_Ccl19 cluster was selected by the choose_graph_segments function in Monocle3.
Gene regulatory network analysis
We used the Python implementation of the SCENIC (pySCENIC)38,39 pipeline (v.0.12.0) for gene regulatory network analysis on the injection-site fibroblasts from 2 h p.i. muscle tissues. Briefly, co-expression modules in the scRNA-seq data were identified with the Grnboost2-Arboreto pipeline38,66. Subsequently, cis-regulatory motif analysis was conducted using cisTarget67 approaches and databases (https://resources.aertslab.org/cistarget/), from which we used 10 kb upstream ~ 10 kb downstream motif annotation data (mm10_v10) to identify regulons. Regulon activities in each cell were scored with AUCell methods.
Statistical analysis
Statistical analysis for scRNA-seq data was performed as described above. The rest of the statistical analysis were conducted using the statannot (v.0.2.3) package in Python, which uses the stats module in scipy (v.1.7.1) package. P values were determined with Mann–Whitney U tests (two-tailed). No statistical method was used to predetermine sample size, and no data were excluded from the analysis. Data generation, collection, and analysis were not randomized. Data distribution was not formally tested, and we used non-parametric tests for statistical analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank BioMedical Research Center Core Facility at KAIST GSMSE for technical support in single-cell library construction and fluorescent imaging. The authors acknowledge the Korea Research Environment Open Network (KREONET) service and the usage of the Global Science Experimental Data Hub Center (GSDC) provided by Korea Institute of Science and Technology Information (KISTI). This work was supported by the Korea Disease Control and Prevention Agency (Grants 4800-6634-329; the intramural research number: 2022-NI-001-00) (J.Y.), MD-PhD/Medical Scientist Training Program from the Korea Health Industry Development Institute (KHIDI) (S.K.), HR21C0198030102002 (J.-E.P.) from KHIDI, National Research Foundation Grant NRF-2021M3A9I402444712 (J.-E.P.), NRF-2021R1C1C101009412 (J.-E.P.), and POSCO Science Fellowship (J.-E.P.).
Author contributions
Conceptualization: D.K., H.L., Y.-J.K., V.N.K., J.-E.P., and J.Y. Methodology and Investigation: S.K., J.H.J., M.K., Y.L., Y.-H.H., M.P., C.H.L., T.L., J.-A.L., J.-E.P., and J.Y. Data analysis and visualization: S.K. Administration and Supervision: D.K., H.L., Y.-J.K., V.N.K., J.-E.P., and J.Y. Writing-draft: S.K. Writing-editing: S.K., J.H.J., M.K., Y.-M.K., J.-E.P., and J.Y.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Raw single-cell RNA sequencing data generated in this study have been deposited in the Gene Expression Omnibus database under accession code GSE239574. Processed single-cell RNA sequencing data are available through the figshare repository (10.6084/m9.figshare.24547210.v2). Source data are provided with this paper.
Code availability
The codes used for data analysis are available via Zenodo (https://zenodo.org/records/11203839).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Seongryong Kim, Ji Hyang Jeon.
Contributor Information
Jong-Eun Park, Email: jp24@kaist.ac.kr.
Jinah Yeo, Email: jinahyeo@korea.kr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51411-9.
References
- 1.Pardi, N. et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med.215, 1571–1588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardi, N. et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature543, 248–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richner, J. M. et al. Modified mRNA vaccines protect against Zika virus infection. Cell169, 176 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Corbett, K. S. et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin, U. et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature586, 594–599 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med.383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med.384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahin, U. et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature595, 572–577 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Laczkó, D. et al. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in Mice. Immunity53, 724–732.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, C. et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat. Immunol.23, 543–555 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arunachalam, P. S. et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature596, 410–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karikó, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Cullis, P. R. & Hope, M. J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther.25, 1467–1475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobiyama, K. & Ishii, K. J. Making innate sense of mRNA vaccine adjuvanticity. Nat. Immunol.23, 474–476 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Swaminathan, G. et al. A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccine34, 110–119 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Alameh, M.-G. et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity54, 2877–2892.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni, H. et al. Extracellular mRNA induces dendritic cell activation by stimulating tumor necrosis factor-alpha secretion and signaling through a nucleotide receptor. J. Biol. Chem.277, 12689–12696 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Karikó, K., Ni, H., Capodici, J., Lamphier, M. & Weissman, D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem.279, 12542–12550 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Karikó, K., Muramatsu, H., Ludwig, J. & Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res.39, e142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov.17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbeke, R. et al. Co-delivery of nucleoside-modified mRNA and TLR agonists for cancer immunotherapy: restoring the immunogenicity of immunosilent mRNA. J. Control. Release266, 287–300 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Nuñez, N. G. et al. High-dimensional analysis of 16 SARS-CoV-2 vaccine combinations reveals lymphocyte signatures correlating with immunogenicity. Nat. Immunol.24, 941–954 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Büttner, M., Ostner, J., Müller, C. L., Theis, F. J. & Schubert, B. scCODA is a Bayesian model for compositional single-cell data analysis. Nat. Commun.12, 6876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol.40, 245–253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater.6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, M. S., Kim, B., Oh, G. T. & Kim, Y.-J. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat. Immunol.14, 346–355 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Perng, Y.-C. & Lenschow, D. J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol.16, 423–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fensterl, V. & Sen, G. C. Interferon-induced Ifit proteins: their role in viral pathogenesis. J. Virol.89, 2462–2468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buechler, M. B. et al. Cross-tissue organization of the fibroblast lineage. Nature593, 575–579 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Yaghi, O. K. et al. A discrete ‘early-responder’ stromal-cell subtype orchestrates immunocyte recruitment to injured tissue. Nat. Immunol. 10.1038/s41590-023-01669-w (2023). [DOI] [PMC free article] [PubMed]
- 31.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol.32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joe, A. W. B. et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol.12, 153–163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soliman, H. et al. Pathogenic potential of Hic1-expressing cardiac stromal progenitors. Cell Stem Cell26, 459–461 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Kinchen, J. et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell175, 372–386.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korsunsky, I. et al. Cross-tissue, single-cell stromal atlas identifies shared pathological fibroblast phenotypes in four chronic inflammatory diseases. Med3, 481–518.e14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc.15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Honda, K. et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature434, 772–777 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol.32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bosteels, C. et al. Inflammatory Type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity52, 1039–1056.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duong, E. et al. Type I interferon activates MHC class I-dressed CD11b+ conventional dendritic cells to promote protective anti-tumor CD8+ T cell immunity. Immunity55, 308–323.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bousso, P. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nat. Rev. Immunol.8, 675–684 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol.15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krienke, C. et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science371, 145–153 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Liu, S. et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater.20, 701–710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verbeke, R., Hogan, M. J., Loré, K. & Pardi, N. Innate immune mechanisms of mRNA vaccines. Immunity55, 1993–2005 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol.20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cupovic, J. et al. Adenovirus vector vaccination reprograms pulmonary fibroblastic niches to support protective inflating memory CD8+ T cells. Nat. Immunol.22, 1042–1051 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma, F. et al. Single cell and spatial sequencing define processes by which keratinocytes and fibroblasts amplify inflammatory responses in psoriasis. Nat. Commun.14, 3455 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlee, M. & Hartmann, G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol.16, 566–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han, X. et al. Adjuvant lipidoid-substituted lipid nanoparticles augment the immunogenicity of SARS-CoV-2 mRNA vaccines. Nat. Nanotechnol. 10.1038/s41565-023-01404-4 (2023). [DOI] [PubMed]
- 54.Karikó, K. & Weissman, D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel.10, 523–532 (2007). [PubMed] [Google Scholar]
- 55.Hajj, K. A. & Whitehead, K. A. Tools for translation: non-viral materials for therapeutic mRNA delivery.Nat. Rev. Mater.2, 1–17 (2017). [Google Scholar]
- 56.Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pallesen, J. et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl Acad. Sci. USA114, E7348–E7357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirchdoerfer, R. N. et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep.8, 15701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst.8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polański, K. et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics36, 964–965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang, Z., Liu, X. & Peltz, G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics39, btac757 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol.36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moerman, T. et al. GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics35, 2159–2161 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Imrichová, H., Hulselmans, G., Atak, Z. K., Potier, D. & Aerts, S. i-cisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res.43, W57–W64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Raw single-cell RNA sequencing data generated in this study have been deposited in the Gene Expression Omnibus database under accession code GSE239574. Processed single-cell RNA sequencing data are available through the figshare repository (10.6084/m9.figshare.24547210.v2). Source data are provided with this paper.
The codes used for data analysis are available via Zenodo (https://zenodo.org/records/11203839).