Abstract
The importance of coupling the process of endocytosis to factors that regulate actin dynamics has been clearly demonstrated in yeast, and many proteins involved in these mechanisms have been identified. Sla1p is a well-characterized yeast protein that binds both to activators of actin dynamics, Las17p and Pan1p, and to cargo proteins, such as the pheromone receptor Ste2p. Previously, we reported that the Lsb5 protein plays a role in endocytosis in yeast and that it localizes to the plasma membrane. Lsb5p has a similar structure to the GGA [Golgi-localized, γ-ear-containing, Arf (ADP-ribosylation factor)-binding] family of proteins with an N-terminal VHS [Vps27p (vacuolar protein sorting protein 27), Hrs, Stam] domain and a GAT (GGA and Tom1) domain. It does not, however, contain either a γ-adaptin ear or a clathrin-binding motif. In the present study, we have further defined its interaction site with both Sla1p and with Las17p, two regulators of actin dynamics. The site of interaction with Sla1p involves the Sla1 HD1 (homology domain 1), which also was shown previously to interact with the pheromone receptor Ste2p. We also demonstrate hitherto unknown interactions between Lsb5p and the active form of the yeast Arf3 protein, and with ubiquitin. Finally, we demonstrate a requirement for Arf3p expression in order to localize Lsb5p to the correct cortical site in cells. Taken together, our data provide further evidence for the role of Lsb5p in membrane-trafficking events at the plasma membrane and also demonstrate for the first time an interaction of Arf3 with the endocytic machinery in yeast.
Keywords: ADP-ribosylation factor 3 (Arf3), actin dynamics, endocytosis, GGA and Tom1 domain (GAT domain), Golgi-localized, γ-ear-containing, ADP-ribosylation-factor-binding protein (GGA protein), Saccharomyces cerevisiae
Abbreviations: Arf, ADP-ribosylation factor; Arp2/3, actin-related protein complex 2/3; GFP, green fluorescent protein; GGA protein, Golgi-localized, γ-ear-containing, Arf-binding protein; GAT, GGA and Tom1; GST, glutathione S-transferase; HD, homology domain; IPTG, isopropyl β-D-thiogalactoside; RFP, red fluorescent protein; VHS, Vps27p (vacuolar protein sorting protein 27), Hrs, Stam; WASP, Wiskott–Aldrich syndrome protein; WH, WASP homology
INTRODUCTION
There is a growing body of evidence that demonstrates a requirement for a dynamic actin cytoskeleton to facilitate the process of endocytosis. Much of the initial evidence for a role for actin in the process of endocytosis derives from studies in yeast which demonstrate that strains mutated in actin-binding proteins cause concomitant defects both in cortical actin organization and in endocytosis (reviewed in [1]). Despite some controversy, the growing consensus supports the idea of evolutionary conservation of the endocytic machinery, and many studies using mammalian cells are now indicating the importance of cortical actin in facilitating endocytosis [2,3]. Currently, the precise mechanisms in the endocytic process for which actin is required are not completely defined. However, recent studies indicate that there are likely to be multiple stages, involving distinct activators of actin dynamics, for which there is a requirement for actin reorganization [4].
Previous work in our laboratory, and that from other researchers, has characterized the cortical protein Sla1p in the regulation of actin dynamics and in coupling this role to the functioning of the endocytic machinery [5–9]. While Sla1p does not yet have a direct mammalian homologue, it does have a similar domain composition, and a comparable spectrum of interactions with the protein CIN85 (Cbl-interacting protein of 85 kDa)/CD2AP (CD2-associated protein)/CMS (Cas ligand with multiple Src homology 3 domains) found to be involved in endocytosis in mammalian cells [10–13]. A combination of in vitro and in vivo studies in yeast reveals that Sla1p localizes with the endocytic machinery, but it functions to regulate actin dynamics through its interactions with Arp2/3 (actin-related protein complex 2/3) activator proteins [8]. Two-hybrid approaches led us to identify Lsb5p as an Sla1p interactor [14]. Lsb5p has also been listed in a report of proteins interacting with the yeast WASP (Wiskott–Aldrich syndrome protein) homologue Las17p [15]. Sequence analysis reveals that Lsb5p contains a number of recognizable motifs reminiscent of membrane-trafficking proteins (Figure 1A). At its N-terminus is a region that has homology with VHS [Vps27p (vacuolar protein sorting protein 27), Hrs, Stam] domains. It also contains a putative Arf (ADP-ribosylating factor)-interacting GAT [GGA (Golgi-localized, γ-ear-containing, Arf-binding) protein and Tom1] domain and a motif termed NPF (Asn-Pro-Phe), which, in other proteins, has been demonstrated to interact with EH (Eps homology) domains [16]. VHS and GAT domains are found as adjacent domains in a recently recognized family of proteins termed the GGA proteins (reviewed in [17,18]). However, this family of proteins also contains a γ-adaptin ear domain and a clathrin-binding motif, neither of which is identifiable in Lsb5p. Furthermore, the GGA proteins do not contain NPF motifs. The GGA proteins have been shown to function largely within the endoplasmic reticulum and Golgi apparatus [19,20].
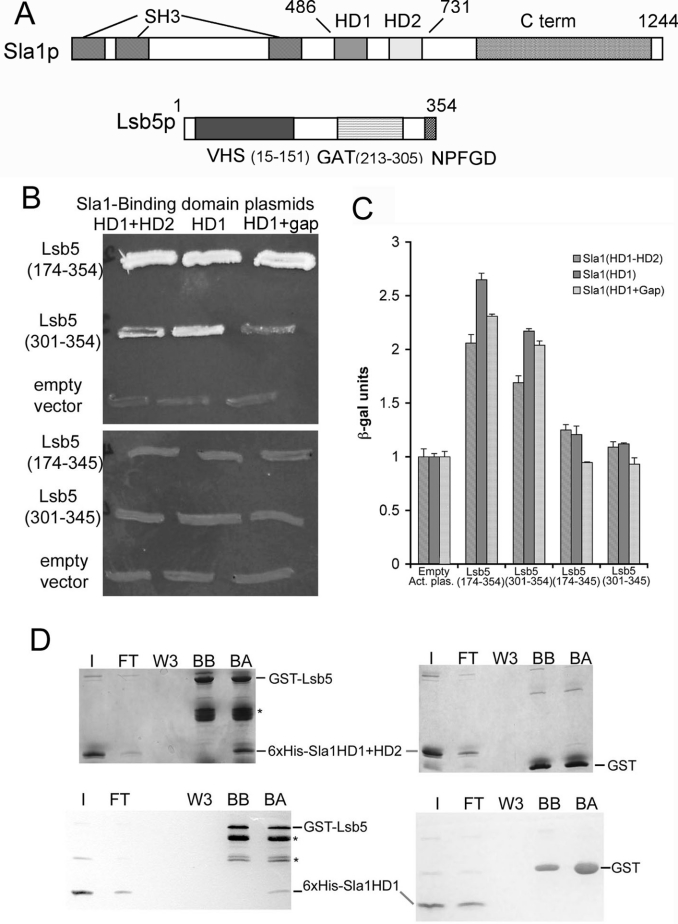
Figure 1. Interactions between Sla1p and Lsb5p.
(A) Schematic diagram depicting domain structures of Sla1p and Lsb5p. Sla1p residues 486–731 were used in the original yeast two-hybrid screen to isolate interacting Lsb5p. C-term, C-terminal; SH3, Src homology 3. (B) Yeast two-hybrid analysis showing growth on plates lacking histidine, adenine, uracil and leucine of strains carrying different combinations of bait and prey plasmids. The upper panel depicts interactions between Sla1p domains and the original Lsb5 plasmid [Lsb5 (174–354)], which includes the GAT domain. The plasmid Lsb5 (301–354) lacks the GAT domain. The lower panel reveals the effect of deleting the C-terminal nine amino acids, including the NPF motif, when co-transformed with the same Sla1 plasmids. (C) The same combinations of plasmids were analysed for interactions using a β-galactosidase assay. Data shown are from triplicate samples and are normalized to the empty activation plasmid reading for each case. Results are means±S.E.M. (D) Biochemical analysis of the Sla1p–Lsb5p interaction. His6-tagged Sla1 (HD1+HD2) (upper panels) or His6-tagged Sla1 (HD1 alone) (lower panels) was incubated with GST–Lsb5p on beads or with GST–beads alone. Binding of both fragments to Lsb5p, but not to GST, was detected. I, input; FT, flow through; W3, third wash; BB, beads before incubation; BA, beads after incubation with His6-tagged protein.
We have described previously a role for Lsb5p in endocytic events [14], and proposed that yeast has at least two definable endocytic pathways and that Lsb5p is an important adaptor-like protein in one of these. In the present study, we have investigated the interactions of Lsb5p with Sla1p and Las17p in more detail. We have also identified Arf3p as a new binding partner for Lsb5p, providing the first evidence of a role for Arf3p in the endocytic process in yeast. Our data contribute to the hypothesis that Lsb5p is a novel adaptor-like protein which functions in membrane-trafficking processes through interactions with Arf3p and regulators of the actin cytoskeleton.
MATERIALS AND METHODS
Materials
Unless stated otherwise, chemicals were obtained from BDH/Merck or Sigma. Media were from Melford Laboratories (yeast extract, peptone and agar) or Sigma (minimal synthetic medium and amino acids). Molecular-biology reagents were purchased from Qiagen (Miniprep and Maxiprep kits and pQE31 plasmid), Taq polymerase was from Bioline and restriction enzymes were from New England Biolabs.
Yeast growth, transformation and strains
Yeast strains and plasmids used in this study are listed in Tables 1 and 2 respectively. Unless stated otherwise, cells were grown with rotary shaking at 30 °C in liquid YPD medium (1% yeast extract, 2% Bacto-peptone and 2% glucose, supplemented with 40 μg/ml adenine), except for strains carrying plasmids which were grown on appropriate selective media.
Table 1. Yeast strains used in the present study.
| Strain | Genotype | Ref/origin |
|---|---|---|
| PJ69-2A | MATatrp1-901, leu2-3,112, ura3-52, his3Δ200, gal4Δ, gal80Δ, GAL2::ADE2, GAL1::HIS3 | Clontech |
| PJ69-4a | MATatrp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ | [34] |
| PJ69-4α | MATα trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ | [34] |
| KAY446 | MATahisΔ1, leu2Δ,urs3Δ, met15Δ | Invitrogen |
| KAY583 | MATahis3Δ, leu2Δ, ura3Δ, met15Δ, ycl034wΔ::KanMx | Invitrogen |
| KAY659 | MATahis3Δ, leu2Δ, ura3Δ, met15Δ, arf3Δ::KanMx | Invitrogen |
| KAY684 | Mat α Sac6RFP::KanMx, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0 | [23] |
| KAY687 | Mat α Snf7RFP::KanMx, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0 | [23] |
| KAY686 | Mat α Anp1RFP::KanMx, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0 | [23] |
Table 2. Plasmids used in the present study.
UTR, untranslated region.
| Name | Description | Ref/origin |
|---|---|---|
| pGBDU-C1 and C3 | Two-hybrid plasmid, Ura-marked | [34] |
| pGAD-C1 | Two-hybrid plasmid, Leu-marked | [34] |
| pKA142 | pGEX4T1 | Amersham |
| pKA260 | Nucleotides 1460–2192 of Sla1 corresponding to region HD1+HD2 were amplified by PCR. The product was then cloned into pGBDU-C3. | [14] |
| pKA263 | Nucleotides 522–1065 of Lsb5, including the GAT domain to end of gene in pGAD-C1 | [14] |
| pKA267 | pGEX2T+full-length Lsb5 amplified by PCR with flanking BamHI and BglII restriction sites | [14] |
| pKA273 | pQE31 His-tagging vector (Qiagen) with nucleotide region 1460-2192 of Sla1, which was subcloned using restriction sites BamHI and PstI from pKA260 | This study |
| pKA312 | PGBDU-C1+LSB5(1–1062) | This study |
| pKA316 | PEGFP-C2+LSB5 | This study |
| pKA325 | PGAD-C1+LAS17(292–536) isolated from screen | |
| pKA342 | pKA260 with stop codon introduced at nucleotide 1733 of Sla1 using oligonucleotides oKA329 and 330 | This study |
| pKA343 | pKA260 with stop codon at nucleotide 1927 using oligonucleotides oKA331 and 332 | This study |
| pKA346 | pGAD-C1+Lsb5(301–354). Lsb5 from nucleotide 901 was cloned after PCR amplification from genomic DNA using oligonucleotides oKA346 and 347 | This study |
| pKA365 | pGAD-Las17(292–422). Stop codon in pKA325 introduced using oKA323 and 324. | This study |
| pKA366 | pGAD-Las17(292–384). Stop codon in pKA325 introduced using oKA325 and 326. | This study |
| pKA367 | pQE31 His-tagging vector with nucleotides 1460–2192 of Sla1, subcloned using restriction sites BamHI and PstI from pKA342. | This study |
| pKA369 | pGEX6P2+Arf3 lacking the N-terminal 14 amino acids | S. Munro |
| pKA370 | pGEX6P2+Arf3 lacking the N-terminal 14 amino acids and GDP-locked (T31N) | S. Munro |
| pKA371 | pGEX6P2+Arf3 lacking the N-terminal 14 amino acids and GTP-locked (Q71L) | S. Munro |
| pKA372 | pRS316+GFPArf3 | S. Munro |
| pKA384 | pNB701. pRS316 plasmid modified for GFP tagging under CUP1 control. | N. Bryant |
| pKA409 | pKA263 with stop codon introduced at nucleotide 1036 of Lsb5 using oligonucleotides oKA443 and 444 | This study |
| pKA410 | pKA346 with stop codon introduced at nucleotide 1036 of Lsb5 using oligonucleotides oKA443 and 444 | This study |
| pKA416 | pKA384+Lsb5. Lsb5 PCR amplified using oligonucleotides oKA441 and 442 with 3′-CUP1 and 5′-UTR flanking from a plasmid containing full-length Lsb5. | This study |
Transformations were performed using lithium acetate as described in [21]. The yeast strain pJ69-2A was transformed with both activation and bait plasmids at the same time, and cells were grown on plates lacking uracil and leucine. Yeast strains pJ69-4a and pJ69-4α were transformed with either the bait or the activation plasmid, and were grown accordingly on plates lacking uracil or leucine. The two transformed strains were then crossed on YPD plates first and later grown on plates lacking uracil and leucine. Interaction between bait and activation domains was observed on plates lacking histidine, uracil, leucine and adenine or by β-galactosidase assay (performed as described in [21]).
Cloning and in vitro mutagenesis
GFP (green fluorescent protein)–LSB5 was cloned into plasmid pKA384 by PCR using oligonucleotides oKA441 and oKA442 which placed a CUP1 promoter and UTR (untranslated region) sequence at the 3′ and 5′ end of the gene respectively. The PCR product was co-transformed with linearized plasmid pKA384 into strain KAY446 and grown on plates lacking uracil. The plasmid was rescued as follows: 1 ml of overnight culture was spun down at 1500 g for 5 min, and the pellet was washed first in 1 ml of TE (Tris/EDTA), then resuspended in 500 μl of buffer S (100 mM potassium phosphate, pH 7.2, 10 mM EDTA, 50 mM 2-mercaptoethanol and 50 μg/ml zymolase), incubated for 30 min at 37 °C before adding 100 μl of lysis buffer (25 mM Tris/HCl, pH 7.5, 25 mM EDTA and 2.5% SDS), followed by brief vortex-mixing and incubation at 65 °C for 30 min. A 166 μl volume of 3 M potassium acetate was added, and DNA was ethanol-precipitated before resuspending in 40 μl of water. DNA (1–2 μl) was then used for electrotransformation into bacterial KC8 cells. Plasmids were then extracted from KC8 cells using a Qiagen Miniprep kit, according to the manufacturer's instructions.
Mutagenesis of plasmids pKA260, pKA263, pKA325 and pKA346 to generate Sla1(HD1 alone) (where HD is homology domain) and Sla1(HD1+gap region) (from pKA260), Lsb5Δnpf (from pKA263 and pKA346); and Las17 truncations (from pKA325), was carried out according to the manufacturer's protocol for the QuikChange Site-Directed Mutagenesis kit (Stratagene) using oligonucleotides as listed in Table 3.
Table 3. Oligonucleotides used in the present study.
| Name | Sequence (5′→3′) |
|---|---|
| oKA323 | ACAGGCCACTGGAAGAAGAGTCTAGACACCACCACCTCCTCCAAG |
| oKA324 | CTTGGAGGAGGTGGTGGTGTCTAGACTCTTCTTCCAGTGGCCTGT |
| oKA325 | CTTTGGATATCAGACAAATATCTAGATGTCATCTCCACCCCCTCC |
| oKA326 | GGAGGGGGTGGAGATGACATCTAGATATTTGTCTGATATCCAAAG |
| oKA329 | CAAGAGACTCTGAAAGAGAAATCTAGAAAGGAGATTGAAGGAGCAGG |
| oKA330 | CCTGCTCCTTCAATCTCCTTTCTAGATTTCTCTTTCAGAGTCTCTTG |
| oKA331 | CAAATACGACGTCCGTACCACTCTAGACAGCCGAAAGCAGCAATAAC |
| oKA332 | GTTATTGCTGCTTTCGGCTGTCTAGAGTGGTACGGACGTCGTATTTG |
| oKA346 | CGTCGTGAATTCAGCAGCGATGAAAGTGATC |
| oKA347 | CGTCGTGGATCCTTCGAGCAAGGCCAAGTT |
| oKA441 | GATATTAAGAAAAACAAACTGTACAATCAATCAATCAATCATCACATAAAGCTGCTGGCATGGGGTTTCTTTCGGATCA |
| oKA442 | ATTATAACGTATTAAATAATATGTGAAAAAAGAGGGAGAGTTAGATAGGATCAAATTTTGTTATGATCACCG |
| oKA443 | CAGCCGTCCACGATCTTCTAGACTTTCGGTGATCATAAC |
| oKA444 | GTTATGATCACCGAAAGTCTAGAAGATCGTGGACGGCTG |
Protein purification
His6-tagged Sla1(HD1+HD2) was expressed and purified from 100 ml of BL21 bacterial culture on Ni-NTA (Ni2+-nitrilotriacetate) resin column as described in the QIAexpressionist™ handbook (Qiagen; http://www1.qiagen.com/literature/handbooks/PDF/Protein/Expression/QXP_QIAexpressionist/1024473_QXPHB_0603.pdf). GST (glutathione S-transferase) alone and GST–Lsb5 fusion protein were induced from 200 ml of BL21 bacterial culture for 2 h at 37 °C by the addition of 0.5 mM of IPTG (isopropyl β-D-thiogalactoside) and then purified by washing the cell pellet in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.76 mM KH2PO4) twice, resuspended in PBS containing protease inhibitors (0.5 mg/ml leupeptin, aprotinin, chymostatin, pepstatin A and 1 mM PMSF) and 5 μg/ml lysozyme, sonicated, incubated with 1 μM MnCl2, 10 μM MgCl2 and 10 mg/ml DNaseI followed by a 45000 rev./min spin in a TLA100 ultracentrifuge. The supernatant was incubated with 0.5 ml of glutathione–Sepharose 4B beads (Amersham Biosciences) for 1 h at 4 °C with rotation. Beads were pelleted at 900 g for 3 min and washed three times in PBS with 1% (v/v) Triton X-100. GST–Arf3 fusion proteins were induced from 1 litre of BL21 bacterial culture for 4 h at 25 °C by the addition of 0.1–0.2 mM of IPTG. Cells were pelleted at 4000 g for 20 min and resuspended in buffer A1 [1% Triton X-100, 5 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM PMSF and 1 μl/ml protease inhibitor mixture (stock 0.5 mg/ml leupeptin, aprotinin, pepstatin A and chymostatin), in 1×PBS], homogenized 10–15 times, sonicated five to ten times for 15 s. Lysates were spun at 16500 g for 10 min at 4 °C. Supernatant was then incubated with 0.75 ml of glutathione–Sepharose 4B beads for 30 min at 4 °C on rotation. Beads were washed with 10 ml of buffer A1, followed by a wash with 5 ml of buffer A2 [5 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM PMSF and 1 μl/ml protease inhibitor mixture (stock 0.5 mg/ml leupeptin, aprotinin, pepstatin A and chymostatin), in 1×PBS], and resuspended in 5 ml of buffer A3 [20 mM Tris/HCl, pH 8, 110 mM KCl, 5 mM 2-mercaptoethanol, 1 mM PMSF and 1 μl/ml protease inhibitor mixture (stock 0.5 mg/ml leupeptin, aprotinin, pepstatin A and chymostatin), in water].
Protein interactions
For the Sla1p–Lsb5p interaction, 40 μl of GST or GST–Lsb5p fusion protein on glutathione–Sepharose 4B beads was incubated with 80 μl of eluted His6-tagged Sla1(HD1+HD2) protein in binding buffer (50 mM Hepes, pH 7.5, 100 mM KCl and 1 mM EDTA, pH 8, to a total volume of 400 μl), for 2.5 h at 4 °C with rotation. The beads were spun down at 250 g for 3 min and washed three times with 10 bed vol. of binding buffer with 15 min rotation at 4 °C for each wash. Bound proteins were eluted in sample buffer and separated on 10% [for Sla1(HD1+HD2)] and 15% [for Sla1(HD1 alone)] polyacrylamide/SDS gels before transfer on to PVDF membranes for analysis.
For the Arf3–Lsb5 interaction, Lsb5p was cleaved from the GST tag by incubation with 4 μl of thrombin in 1 ml of PBS for 2 h and 40 min with rotation. A 100 μl volume of the cleaved Lsb5p protein (0.2 mM) was incubated with 50 μl of glutathione–Sepharose 4B beads, containing 10 μl of GST–Arf3p or GST alone beads, in binding buffer to a total volume of 200 μl, for 3 h at 4 °C with rotation. Beads were washed three times in binding buffer and bound protein was eluted in sample buffer and separated on an SDS/10% polyacrylamide gel before transfer on to PVDF membrane for analysis.
For the ubiquitin–Lsb5 interaction, 50 μl of 10 mg/ml ubiquitin–agarose resin (AG Scientific) or mouse IgG–agarose (Santa Cruz Biotechnology) was incubated with 10 μl of 0.2 mM Lsb5p, in binding buffer to a total volume of 200 μl, for 2 h at 4 °C with rotation. Washes, elution and analysis were performed as described above.
For ubiquitin–Lsb5p–Arf3p interaction, ubiquitin was obtained as a His6-tagged fusion protein and as a GST fusion protein (BioMol International). For the first study, GST–Arf3 was prepared as described above. It was then incubated with Lsb5 (cleaved from GST–Lsb5) which was pre-incubated with 2 μl of 1 mg/ml His6–ubiquitin. Beads were then washed and separated on an SDS/14% polyacrylamide gel as described above. For the second study, GST–ubiquitin was incubated with Arf3 (cleaved from GST–Arf3) and Lsb5 (cleaved from GST–Lsb5). Beads were then washed and separated on an SDS/10% polyacrylamide gel as described above.
Antibody preparation
Polyclonal rabbit antisera raised against Sla1p and GST–Lsb5p were partially purified by Protein A–Sepharose (Amersham Biosciences) and eluted with 100 mM glycine, pH 2.5, neutralized with 10 mM Tris/HCl, pH 7.5. The anti-GST–Lsb5 antibody was also cleaned with GST-bound glutathione–Sepharose 4B beads to remove antibody raised against GST. The antibodies were used in a 1:500 and 1:2000 respectively.
Mammalian cell culture and transfection
Plasmid pKA316 was prepared using a Qiagen Maxiprep kit. It was transfected into subconfluent COS7 cells using Lipofectamine™ (Invitrogen) according to the manufacturer's instructions. Cells were fixed on coverslips using formaldehyde before being visualized as described below. For co-localization with tubulin, after fixation, cells on coverslips were incubated with monoclonal mouse anti-tubulin antibodies (Sigma) at a dilution of 1:1000 and then detected using TRITC (tetramethylrhodamine β-isothiocyanate)–anti-mouse secondary antibodies (Vector) at a dilution of 1:100. Cells were visualized as described below.
Microscopy techniques
Cells expressing GFP–Arf3 (pKA372) or CUP–GFP–LSB5 (pKA416) were grown overnight in synthetic medium lacking uracil. They were re-inoculated to low density and grown for a further 6 h to mid-exponential phase of growth (D600 of 0.2). Strains transformed with pKA416 were induced with 500 μM of CuSO4 before visualization. Cells were viewed with an Olympus BX-60 fluorescence microscope with a 100 W mercury lamp and an Olympus 100X Plan-NeoFluar® oil-immersion objective. Images were captured using a Roper Scientific Micromax 1401E cooled CCD (charge-coupled device) camera using IPLab Software (Scanalytics, Fairfax, VA, U.S.A.) on an Apple Macintosh G4 computer.
RESULTS
Defining the interacting sites in Sla1p and Lsb5p
In our previous study, we reported that Lsb5p interacts with the central homology domains, HD1 and HD2, of Sla1p, encompassed by residues 486–731 (Figure 1A) [14]. These central domains are the most highly conserved regions of Sla1p when sequences from several fungal species are compared. It has previously been reported that HD1 interacts with the α-factor pheromone receptor Ste2p, making Sla1p an endocytic adaptor protein, since it interacts both with cargo proteins and with the endocytic machinery itself [22]. Further evidence for the role of Sla1p in Ste2p uptake was from the demonstration that deletion of SLA1 is inhibitory to internalization of Ste2p [8,22]. The sequence that the Sla1p HD1 motif was shown to interact with is the NPFXD motif found in a number of integral membrane proteins that transit the plasma membrane. Interestingly, although not postulated to be an integral membrane protein, a similar sequence is found near the C-terminus of Lsb5p (residues 346–350). In the present study, we investigated which of the homology domains was involved in Sla1p binding to Lsb5p. We also determined whether the NPF sequence in Lsb5p is required for the interaction. Two-hybrid bait plasmids were constructed expressing the full Sla1p HD1-gap-HD2 sequence, HD1-gap or HD1 alone. These were co-expressed with the C-terminal half of Lsb5p (residues 174–354) to determine whether there was an interaction using two-hybrid growth assays on plates. As shown in Figure 1(B), all of the Sla1p fusions, including the HD1 sequence alone, were able to interact with Lsb5p. The portion of Lsb5p used in the assay included the GAT domain. To determine whether GAT played a role in the interaction, a construct was generated in which the activation domain sequence used in the two-hybrid assay was ligated to the sequence of Lsb5 downstream of GAT (residues 301–354). As shown in Figure 1(B), the HD1 domain of Sla1p was able to interact with both Lsb5p-(174–354) and Lsb5p-(301–354), but the interaction appeared to be weaker. A possible role for the NPF motif of Lsb5p in the interaction with Sla1 was investigated by generating a truncation mutant of Lsb5p. The Lsb5p-interacting plasmids were mutagenized to generate both Lsb5-(174–345) and Lsb5-(301–345) lacking the NPF motif and the extreme four C-terminal amino acids. As shown (Figure 1B), there was no interaction with Sla1p when the NPF motif was deleted. Thus the extreme C-terminus, including the NPF motif, of Lsb5p is necessary for the interaction with HD1 of Sla1p, while the GAT domain of Lsb5p appears to enhance the interaction. These data were verified by detecting interactions using a β-galactosidase assay (Figure 1C).
To show that the interaction between Lsb5p and Sla1p is direct and not mediated by intermediary proteins, recombinant Sla1p fragments of HD1+HD2 and HD1 alone were purified and incubated with GST–Lsb5p beads. In agreement with the yeast two-hybrid data, the Sla1p HD1 region alone is necessary and sufficient to support the interaction with Lsb5p (Figure 1D).
Lsb5p interactions with Las17p
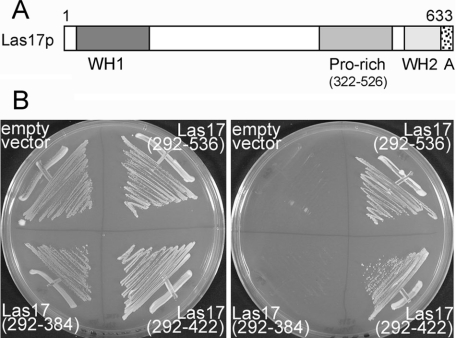
Lsb5p was first described as a Las17p-binding protein, and an interaction was shown between full-length Las17p and residues 40–213 of Lsb5p [15]. We conducted a two-hybrid screen using strain pJ69-2A with full-length Lsb5p on the bait plasmid. Plasmids expressing interacting fusions were sequenced, and over 40% (15 of 36) were shown to be Las17p. Clones containing Lsb5p- and Las17p-interacting partners dominated the first 25 colonies that grew when selected directly on plates lacking histidine and adenine (as well as uracil and leucine) to select for activation (20 of 25), indicating that the interaction between Las17p and Lsb5p is relatively strong and very reproducible. The smallest fragment obtained was Las17p-(292–536), which encompasses the polyproline region of Las17p, but does not include the WH (WASP homology) 1 domain, the actin-binding WH2 domain or the acidic region at the extreme C-terminus (Figure 2A). As well as interacting with full-length Lsb5p, we also obtained a positive yeast two-hybrid interaction, albeit slightly weaker, when the Las17 clone was co-expressed with Lsb5 residues 1–156, which encompasses just the VHS domain of Lsb5p (results not shown). In vitro mutagenesis was then used to introduce stop codons within the Las17p sequence. Two truncations were made, each introducing a stop codon at gaps in the tracts of polyproline that dominate this region of Las17p. Using this approach, we were able to observe two-hybrid interactions between Lsb5p and a region of Las17p between residues 292 and 422, but not with a construct encompassing Las17p-(292–384) (Figure 2B), suggesting that the sequence between 384 and 422 of Las17p is responsible for the interaction with Lsb5p.
Figure 2. Interaction of Lsb5p and Las17p.
(A) A schematic diagram depicting the domains of Las17p. A, acidic region. (B) Yeast two-hybrid analysis on plates lacking histidine, adenine, uracil and leucine of strains carrying an Lsb5 bait plasmid and different combinations of Las17 prey plasmids. Interaction was found between Lsb5p and a region of the proline-rich (Pro-rich) region in Las17p.
Localization of GFP–Lsb5p
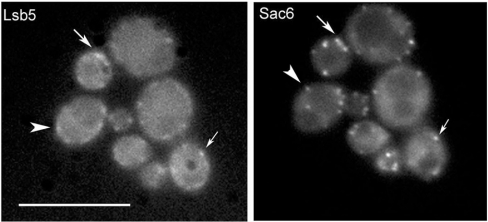
In order to determine whether the interaction between Lsb5p and either Las17p or Sla1p is functionally important for localization in vivo, a construct was generated in which the sequence of Lsb5p was fused to that of GFP in a plasmid containing a copper-inducible promoter. This was transformed into wild-type cells (KAY446), and expression of GFP–Lsb5p was induced for 6 h. As shown in Figure 3, in wild-type cells, the GFP–Lsb5p staining is largely punctate and at the cell cortex. As well as spots, there also appear to be elongated cortical patches which appear as smooth lines of cortical fluorescence. This pattern of localization is very similar to the staining that we obtained previously when we integrated the sequence for a 13×myc tag on the 3′ end of LSB5 and localized the protein using indirect immunofluorescence [14]. In order to determine whether Lsb5p shows co-localization with known organelle markers, a strain expressing GFP–Lsb5p was crossed with strains expressing Sac6p–RFP (red fluorescent protein) (cortical actin marker), Snf7p–RFP (endosomal marker) and Anp1p (Golgi apparatus marker) [23]. These strains were then visualized under appropriate filters to determine whether staining patterns overlapped. Merging of images indicated that there was no overlap in localization of Lsb5p with Snf7p or Anp1p. However, as shown in Figure 3, some localization with Sac6p could be observed (indicated by arrows), although there are clearly spots that also contain either Lsb5p or Sac6p alone (arrowheads). To determine whether either Sla1p or Las17p is required for localization of Lsb5p, pKA416 was transformed into cells lacking Sla1 and Las17. As reported previously [14], the absence of Sla1 causes a reduction, but not complete abrogation, of Lsb5 localization to the cell cortex. Similarly, cells lacking Las17 expression were clearly aberrant and carried some morphological and actin organization defects. However, GFP–Lsb5p was observed at the cell cortex, mostly in punctate structures resembling those in wild-type cells (results not shown). Therefore Las17p is not required for Lsb5p localization.
Figure 3. Co-localization of Lsb5p and Sac6p.
Yeast strain KAY446 expressing a GFP–Lsb5 plasmid was crossed with KAY684 carrying integrated Sac6–RFP, as described in the Materials and methods section. Arrows indicate regions of overlap of the staining patterns, indicating some co-localization. Other spots of Lsb5p or Sac6p fluorescence are clearly not overlapping (arrowheads), indicating that they exist in distinct complexes at the cell cortex. Scale bar, 10 μm.
Lsb5p interacts with active Arf3p and ubiquitin
The central region of Lsb5 contains a domain with significant, but low-level, homology with the previously described GAT domain. In the GGA family of proteins, the GAT domain has been shown to interact with Arf proteins [20,24]. In particular, the yeast Gga1p and Gga2p proteins interact with both Arf1p and Arf2p [20]. Yeast Arf3p, however, does not interact with the GGA proteins, and it appears to localize to the plasma membrane [25]. These earlier studies by Huang et al. [25] also suggested that Arf3p does not play a role in endocytosis. However, these studies investigated the deletion in a wild-type background. As we have shown in our previous study, deletion of lsb5 alone does not confer any endocytic defect on cells, so it may be that the role of Arf3p in endocytosis is also masked by an overlapping or redundant pathway [14]. Because Lsb5p contains a putative GAT domain that could bind Arf proteins, and because Arf3p, like Lsb5p, is localized to the plasma membrane, we made recombinant proteins to determine whether Lsb5p is able to interact with active or inactive Arf3p. As shown in Figure 4(A), Lsb5 interacts strongly with the active form of Arf3 and only weakly with the inactive form, demonstrating a direct interaction of Arf3 with a protein involved in endocytosis in yeast.
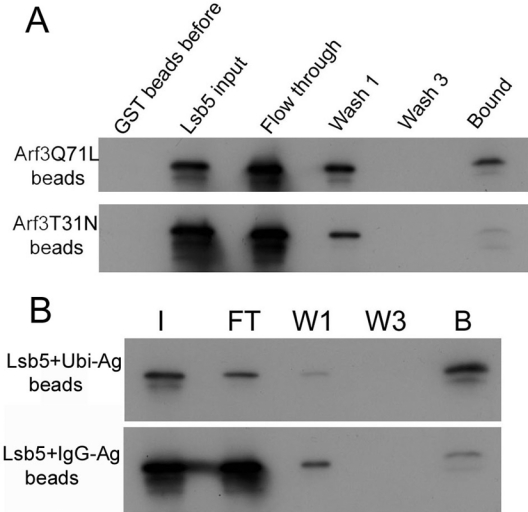
Figure 4. Interaction of Lsb5 with Arf3 and ubiquitin.
(A) Lsb5 was purified and incubated with GST–Arf3 beads with Arf3 in either the active (Q71L) or inactive (T31N) form as described in the Materials and methods section. Binding was observed only with the active Arf3p and Lsb5p. (B) Lsb5p was then incubated with ubiquitin–agarose or, as a control, IgG–agarose beads. After washing, binding was assessed by SDS/PAGE. Binding was detected with Lsb5 only on the ubiquitin-conjugated beads. I, input; FT, flow through; W1, first wash; W3, third wash; B, bound; Ubi, ubiquitin; Ag, agarose.
The GAT domain of mammalian GGA3 protein has been reported to interact both with Arf proteins and with ubiquitin. This latter interaction is postulated to be important for stabilizing and enhancing the interaction between Arfs and GGA proteins [26–28]. We expressed Lsb5p and incubated it with ubiquitin–agarose beads. As shown in Figure 4(B), there is a significant proportion of Lsb5p that is able to bind to the ubiquitin-bound beads compared with binding to an IgG–agarose bead control, suggesting that, like the GGA proteins, the Lsb5 protein is also able to bind to ubiquitin. The site of interaction is unlikely to be at the C-terminus, because a degradation product also shows binding to the beads.
In addition to a straightforward binding assay, we also considered whether the presence of ubiquitin might enhance binding of Lsb5p to Arf3p. This situation would be analogous to that observed for GGA3 protein interacting with Arf and ubiquitin. To do this, we first repeated the above assay with GST–Arf3, but added either Lsb5p alone or Lsb5p pre-incubated with His6-tagged ubiquitin. We also used GST–ubiquitin and added Lsb5p and then Arf3p, and looked for enhanced binding. Neither approach revealed any increase in binding of Arf3p to Lsb5p (results not shown), demonstrating that Arf3p and ubiquitin binding to Lsb5p are independent and not co-operative events.
Dependence of Arf3 and Lsb5 localization
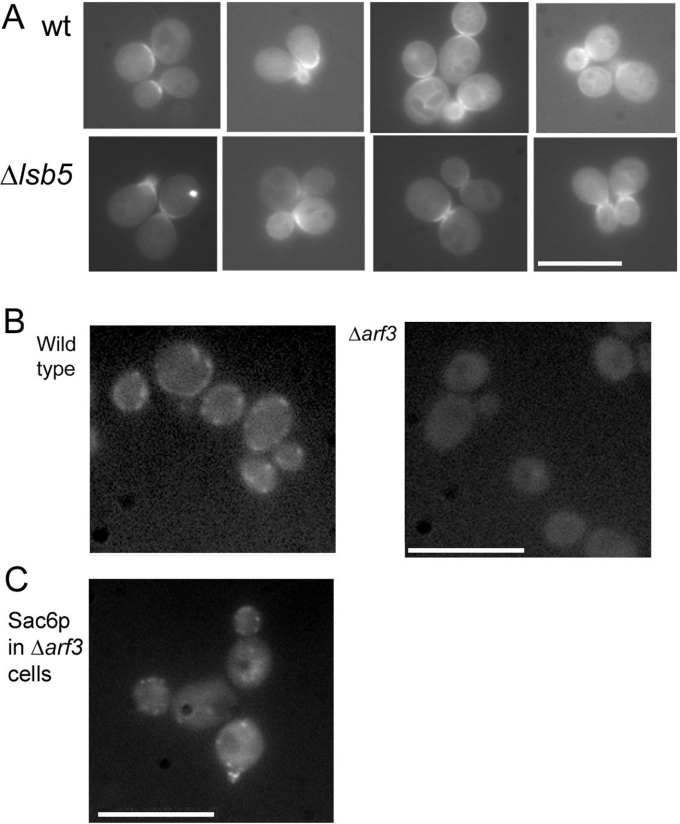
In order to determine whether the interaction between Lsb5p and Arf3p is required for the localization of Arf3, we obtained a construct to allow the expression of GFP-tagged Arf3 (a gift from Dr Sean Munro, MRC Laboratory of Molecular Biology, Cambridge University, Cambridge, U.K.). Previous studies have shown that Arf3p has a highly polarized localization at the plasma membrane of buds in actively growing cells [25]. On transformation of the GFP–Arf3 plasmid into wild-type cells (KAY446), we also saw this localization pattern (Figure 5A). In addition to the localizations reported previously, we also were able to observe a lower level of staining around vacuoles, indicating possible trafficking through the endosomal pathway (Figure 5A). Cells lacking lsb5 were also studied, and these were found to contain a similar, but distinct, pattern of GFP–Arf3 localization. While there were no significant differences in levels of Arf3p at the membrane, subtle differences in localization were noted. Wild-type cells showed GFP–Arf3 localization all around the bud at the small-budded stage, while at the large-budded stage, staining was mostly on the bud side, rather than in the mother cell. However, in the Δlsb5 cells, Arf3p localization appeared to be more confined to the mother bud neck region and did not extend around the periphery of small-budded cells, and was also found on both mother and daughter sides of large-budded cells.
Figure 5. In vivo interaction between Lsb5p and Arf3p.
(A) GFP–Arf3 was expressed in wild-type cells (KAY446) and in cells lacking lsb5 (KAY583). As shown, Arf3p is still able to localize to the plasma membrane in the absence of Lsb5 expression. Scale bar, 10 μm. (B) GFP–Lsb5p was expressed in wild-type cells (KAY446) and in cells lacking Arf3 (KAY659). As shown, cells lacking Arf3 are no longer able to localize GFP–Lsb5p to the cell cortex, demonstrating a role for Arf3p in organization of the membrane-trafficking endocytic machinery. (C) Cells lacking Arf3 expression are still able to localize Sac6p to punctate cortical spots, demonstrating that the actin cytoskeleton is still organized in the ΔArf3 cells. Scale bar, 10 μm.
We also examined GFP–Lsb5 localization in cells lacking arf3. As shown in Figure 5(B), in the wild-type cells, we obtained the usual punctate and cortical patch staining of GFP–Lsb5p, whereas in the mutants, no cortical staining was obtained at all. These Δarf3 cells do, however, show a wild-type actin organization, as revealed by localization of Sac6p at appropriate punctate cortical spots (Figure 5C). This adds further to the evidence for an in vivo link between Lsb5p and Arf3p, and a role for Arf3p in endocytosis.
Lsb5p interacts with Arf6 and localizes to the midbody in mammalian cells
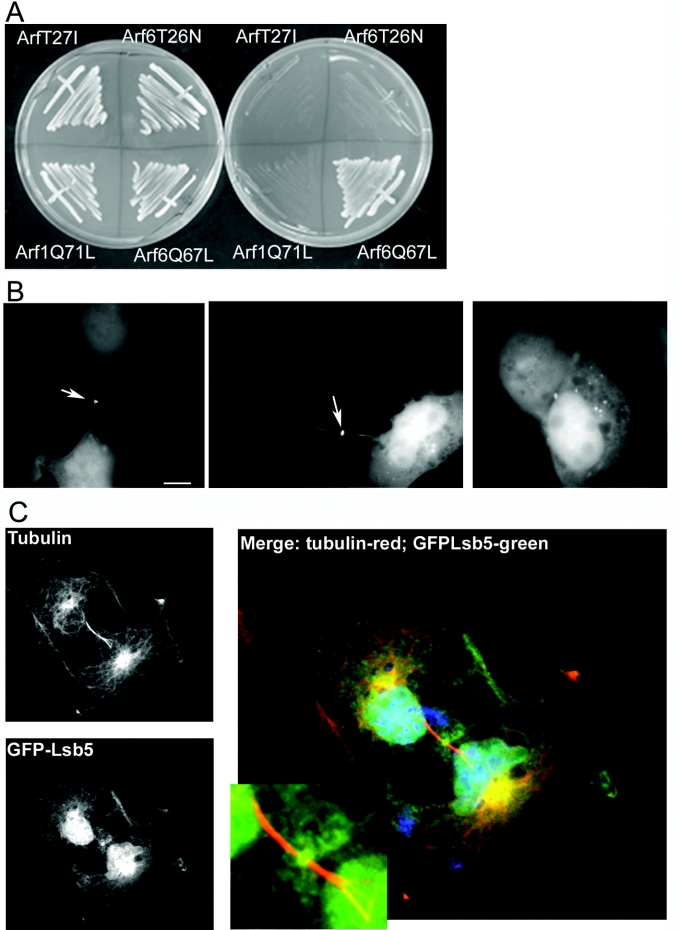
Despite the absence of a clear LSB5 homologue in mammalian cells, we postulated that, if yeast Lsb5p was able to interact with Arf3p, then it might also interact with its mammalian homologue Arf6 [30]. If this was the case, then it may be informative about possible conservation of Lsb5p-binding domains. Two approaches were taken. First, yeast two-hybrid analysis was performed, comparing growth on plates, of cells co-transformed with Lsb5p and Arf1Q71L (active), Arf1T27I (inactive), Arf6Q67L (active) and Arf6T26N (inactive). Only cells containing both Lsb5 and Arf6Q71L showed activation to permit growth on plates lacking uracil, leucine, histidine and tryptophan (Figure 6A).
Figure 6. Interaction between Lsb5p and mammalian Arf6.
(A) Yeast two-hybrid analysis in cells expressing Lsb5 bait plasmid and different Arf constructs fused to an activation domain sequence. Only co-expression of Lsb5 and activated Arf6Q67L gave a positive interaction on selection plates. (B) GFP–Lsb5 was transfected into COS7 cells as described. Localization was observed in two main localizations: in punctate spots dispersed in the cytoplasm (right-hand panel) and at the midbody, indicated by arrows (left-hand and central panels). (C) To demonstrate that the staining of Lsb5GFP is at the midbody, we used immunofluorescence microscopy to observe tubulin localization in the cells. Tubulin localizes to distinct structures either side of a midbody, but has a clear zone at the midbody itself. As depicted, GFP–Lsb5p localizes to the midbody, as defined by the tubulin structures on either side. Scale bar, 10 μm.
Secondly, LSB5 was cloned into a mammalian GFP expression vector and was transfected into COS7 cells. While many cells showed disperse or vesicular staining, up to 20% showed a clear staining to the midbody (Figures 6B and 6C). This localization was verified by co-localization with tubulin, which allows clear identification of the midbody position (Figure 6C). In cells transfected with GFP alone, no midbody staining was seen at all. We also observed that, in cells transfected with GFP–Lsb5, the presence of Lsb5 appeared to have a detrimental effect such that cytokinesis appeared to be blocked and binucleate cells could be observed. The proportion of binucleate cells in the population increased from 2% in untransfected cells to 12% in cells transfected with GFP alone, and to 28% in cells transfected with GFP–Lsb5p. It is known that Arf6 localizes to the midbody [29,30], and our data indicate that Lsb5p is likely to be interacting with Arf6 in this location possibly disrupting its normal function.
DISCUSSION
Studies linking endocytosis to the actin cytoskeleton have identified many proteins that are involved in this process, and it seems clear that a number of sequential complexes are formed during invagination, scission and movement of the vesicle into the cell ([4,8,9], and reviewed in [31]). One suggestion is that different actin organizations are required for each step, and so proteins that can be regulated to stimulate actin remodelling are central to the endocytic mechanism. In previous studies, we have shown that the Sla1 protein plays a central role in linking regulators of the actin cytoskeleton to the endocytic machinery [8]. Through a yeast two-hybrid screen, we identified Lsb5p as an Sla1p interactor [14]. Others also identified Lsb5p as a protein that binds to the Arp2/3 regulator Las17p [15]. Thus, from two studies, this protein shows links to the actin-regulatory machinery. Deletion of lsb5 alone does not confer any observable phenotype on cells. However, when lsb5 is deleted in a background already lacking the actin-associated protein gene ysc84, there is a severe phenotype with aberrant actin organization and an almost complete loss of fluid-phase endocytosis. Because neither single deletion has a distinct phenotype, we reasoned that the double deletion highlights the existence of two endocytic pathways that are able to function in at least a partially overlapping fashion [14].
Analysis of the domain structure of Lsb5p provides some interesting ideas for how Lsb5p may function in endocytosis. Its structure is reminiscent of the GGA proteins, having an N-terminal VHS domain and a central GAT domain. It does not, however, have the γ-adaptin ear motif or the clathrin-binding sequence, but it does have the C-terminal NPF motif. In the present study, we have furthered our understanding of the Lsb5 protein, and have shown that the NPF motif is the likely interaction site with the HD1 domain of Sla1. This is the first non-integral membrane protein with an NPF motif that has been demonstrated to have a specific interaction role. As HD1 also interacts with the NPF motifs of other proteins, e.g. Ste2p, Ste3p and Kex2p [23], it seems likely that these interactions are mutually exclusive. It will be interesting now to determine whether the interactions of Sla1p with receptors and Lsb5p occurs sequentially or whether there is an equilibrium with interactions occurring at the same time, but with different Sla1 molecules. The interaction between Lsb5p and Sla1p was stronger when the GAT domain was present, indicating that the presence of other binding partners, such as Arf3p or ubiquitin, might stabilize this interaction.
The other central findings of the present study were the demonstration that Lsb5p is able to interact with Arf3 and, importantly, that Arf3p is necessary for Lsb5p localization. In previous studies, we showed that Sla1 deletion appears to decrease Lsb5p localization at the membrane. However, there was still a distinct, low level of Lsb5p localization to cortical patches in the ΔSla1 cells [14]. This might suggest that, while Arf3p localizes Lsb5p to the cortex, binding is then transferred to Sla1p (and possibly to Las17p). In the absence of the latter proteins, there would be expected to be some binding, while deletion of Arf3 might lead to a much more severe reduction in Lsb5p localization. This outcome was what was observed. A schematic model depicting Lsb5 interactions is shown in Figure 7.
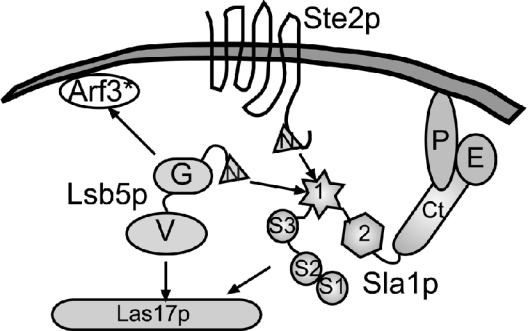
Figure 7. Schematic diagram depicting interactions of Lsb5 with other proteins involved in actin remodelling and endocytosis.
In the present study and in previous work [14], we have identified interaction sites between Lsb5p and Sla1p. This interaction occurs between the HD1 (1) domain of Sla1 and most likely the NPF motif (N) of Lsb5p. The interaction appears to be stabilized by the presence of the GAT domain (G). The GAT domain of Lsb5 is most likely to interact with Arf3. Our studies, and those of Madania et al. [15], indicate that the VHS domain of Lsb5 is the domain likely to interact with Las17p. Other domains and proteins depicted: S1, S2, S3, Src homology 3 domains 1, 2 and 3 of Sla1; 2, HD2 of Sla1; Ct, C-terminal repeats region of Sla1; P, Pan1p; E, End3p. Arf3*, active Arf3.
The possible evolutionary importance of the Lsb5p interactions with Arf3p was highlighted in the observed interaction of Lsb5p with active mammalian Arf6 and in the localization of Lsb5p to the midbody of mammalian cells, a recognized site of Arf6 function [29,30]. This conserved interaction then leads to the question of whether there is an Lsb5p homologue in mammalian cells. Many of the proteins involved in the endocytic process, other than those that directly bind to and regulate actin, do not appear to be highly conserved. Indeed, many potential homologues, such as amphiphysin and Rvs167, and epsin and Ent1/2, share a very low level of direct sequence identity, but their domain structure is very similar, and, for that reason, they have been proposed to carry out related functions. Searching databases on the basis of sequence identity does not reveal any clear Lsb5p homologues other than in all fungal species listed. However, analysis of proteins carrying a similar domain structure reveals some interesting candidates, the best characterized of which is the mammalian protein Tom1. Current data on Tom1 indicate that it is a protein that localizes to the endosomal network and that it is involved in the recruitment of ubiquitin-conjugated proteins to early endosomes [32]. Given the localization of Lsb5p, mostly to the cell cortex and probably to early endosomes, and that we are able to show interactions with ubiquitin, the similarities between the two proteins indicate that Lsb5p and Tom1 might perform the same function in yeast and mammalian cells respectively. Tom1 itself has not been studied intensively, but additional interacting proteins have been identified which are involved in localizing it to the endosomes (endofin) and which form a complex involved in the binding of ubiquitinylated proteins (Tol1-IP) [32,33]. Investigating whether homologues of these are found in yeast and also whether they interact with Lsb5p will strengthen the evidence for the functional similarity of these proteins. Conversely, determination of whether Tom1 is able to interact with Arf6 and with actin regulatory proteins may shed light on the role of this protein, and in particular highlight whether Tom1 is also able to act to couple actin regulators to the endocytic process.
Acknowledgments
We thank Campbell Gourlay (Sheffield) and Steve Winder (Sheffield) for critical reading of the manuscript, Sean Munro and Alison Gillingham (Laboratory of Molecular Biology, Cambridge) for Arf3 plasmids, Nia Bryant (Glasgow) for the CUP–GFP plasmid, Gwyn Gould (Glasgow) for the mammalian Arf plasmids, and Steve Winder for assistance with mammalian cell transfection. This work was supported by an MRC (Medical Research Council) Senior Research Fellowship to K.R.A. (G117/394).
References
- 1.Engvist-Goldstein A. E., Drubin D. G. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 2.Merrifield C. J., Feldman M. E., Wan L., Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 3.Lee E., De Camilli P. Dynamin at actin tails. Proc. Natl. Acad. Sci. U.S.A. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 5.Holtzman D. A., Yang S., Drubin D. G. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayscough K. R., Eby J. J., Lila T., Dewar H., Kozminski K. G., Drubin D. G. Sla1p is a functionally modular component of the yeast cortical actin cytoskeleton required for correct localization of both Rho1p-GTPase and Sla2p, a protein with talin homology. Mol. Biol. Cell. 1999;10:1061–1075. doi: 10.1091/mbc.10.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang H. Y., Xu J., Cai M. J. Pan1p, End3p, and Sla1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren D. T., Andrews P. D., Gourlay C. G., Ayscough K. R. Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J. Cell Sci. 2002;115:1703–1715. doi: 10.1242/jcs.115.8.1703. [DOI] [PubMed] [Google Scholar]
- 9.Gourlay C. W., Dewar H., Warren D. T., Costa R., Satish N., Ayscough K. R. An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J. Cell Sci. 2003;116:2551–2564. doi: 10.1242/jcs.00454. [DOI] [PubMed] [Google Scholar]
- 10.Brett T., Traub L., Fremont D. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure. 2002;10:797–809. doi: 10.1016/s0969-2126(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 11.Lehtonen S., Zhao F., Lehtonen E. CD2-associated protein directly interacts with the actin cytoskeleton. Am. J. Physiol. Renal Physiol. 2002;283:F734–F743. doi: 10.1152/ajprenal.00312.2001. [DOI] [PubMed] [Google Scholar]
- 12.Stamenova S., Dunn R., Adler A., Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85–endophilin complex, Sla1–Rvs167. J. Biol. Chem. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- 13.Kowanetz K., Szymkiewicz I., Haglund K., Kowanetz M., Husnjak K., Taylor J., Soubeyran P., Engstrom U., Ladbury J., Dikic I. Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J. Biol. Chem. 2003;278:39735–39746. doi: 10.1074/jbc.M304541200. [DOI] [PubMed] [Google Scholar]
- 14.Dewar H., Warren D. T., Gardiner F. C., Gourlay C. G., Satish N., Richardson M. R., Andrews P. D., Ayscough K. R. Novel proteins linking the actin cytoskeleton to the endocytic machinery in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:3646–3661. doi: 10.1091/mbc.E02-05-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madania A., Dumoulin P., Grava S., Kitamoto H., Scharer-Brodbeck C., Soulard A., Moreau V., Winsor B. The Saccharomyces cerevisiae homologue of human Wiskott–Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell. 1999;10:3521–3538. doi: 10.1091/mbc.10.10.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Confalonieri S., Di Fiore P. The Eps15 homology (EH) domain. FEBS Lett. 2002;513:24–29. doi: 10.1016/s0014-5793(01)03241-0. [DOI] [PubMed] [Google Scholar]
- 17.Boman A. L. GGA proteins: new players in the sorting game. J. Cell Sci. 2001;114:3413–3418. doi: 10.1242/jcs.114.19.3413. [DOI] [PubMed] [Google Scholar]
- 18.Bonifacino J. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 19.Hirst J., Lindsay M. R., Robinson M. S. GGAs: roles of the different domains and comparison with AP-1 and clathrin. Mol. Biol. Cell. 2001;12:3573–3588. doi: 10.1091/mbc.12.11.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhdankina O., Strand N. L., Redmond J. M., Boman A. L. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast. 2001;18:1–18. doi: 10.1002/1097-0061(200101)18:1<1::AID-YEA644>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser C., Michaelis S., Mitchell A. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1994. Methods in Yeast Genetics: a Laboratory Course Manual. [Google Scholar]
- 22.Howard J. P., Hutton J. L., Olson J. M., Payne G. S. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'shea E. K. Global analysis of protein localization in budding yeast. Nature (London) 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 24.Collins B., Watson P., Owen D. The structure of the GGA1-GAT domain reveals the molecular basis for ARF binding and membrane association of GGAs. Dev. Cell. 2003;4:321–332. doi: 10.1016/s1534-5807(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 25.Huang C. F., Liu Y. W., Tung L., Lin C. H., Lee F. J. S. Role for Arf3p in development of polarity, but not endocytosis, in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:3834–3847. doi: 10.1091/mbc.E03-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puertollano R., Bonifacino J. S. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 27.Scott P. M., Bilodeau P. S., Zhdankina O., Winistorfer S. C., Hauglund M. J., Allaman M. M., Kearney W. R., Robertson A. D., Boman A. L., Piper R. C. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 2004;6:252–259. doi: 10.1038/ncb1107. [DOI] [PubMed] [Google Scholar]
- 28.Shiba Y., Katoh Y., Shiba T., Yoshino K., Takatsu H., Kobayashi H., Shin H., Wakatsuki S., Nakayama K. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J. Biol. Chem. 2004;279:7105–7111. doi: 10.1074/jbc.M311702200. [DOI] [PubMed] [Google Scholar]
- 29.Schweitzer J., D'souza-Schorey C. Localization and activation of the ARF6 GTPase during cleavage furrow ingression and cytokinesis. J. Biol. Chem. 2002;277:27210–27216. doi: 10.1074/jbc.M201569200. [DOI] [PubMed] [Google Scholar]
- 30.Donaldson J. G. Multiple Roles for Arf6: sorting, structuring and signaling at the plasma membrane. J. Biol. Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- 31.Ayscough K. R. Endocytosis: actin in the driving seat. Curr. Biol. 2004;14:R124–R126. [PubMed] [Google Scholar]
- 32.Katoh Y., Shiba Y., Mitsuhashi H., Yanagida Y., Takatsu H., Nakayama K. Tollip and Tom1 form a complex and recruit ubiquitin-conjugated proteins onto early endosomes. J. Biol. Chem. 2004;279:24435–24443. doi: 10.1074/jbc.M400059200. [DOI] [PubMed] [Google Scholar]
- 33.Seet L. F., Liu N. S., Hanson B. J., Hong W. J. Endofin recruits TOM1 to endosomes. J. Biol. Chem. 2004;279:4670–4679. doi: 10.1074/jbc.M311228200. [DOI] [PubMed] [Google Scholar]
- 34.James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]