Abstract
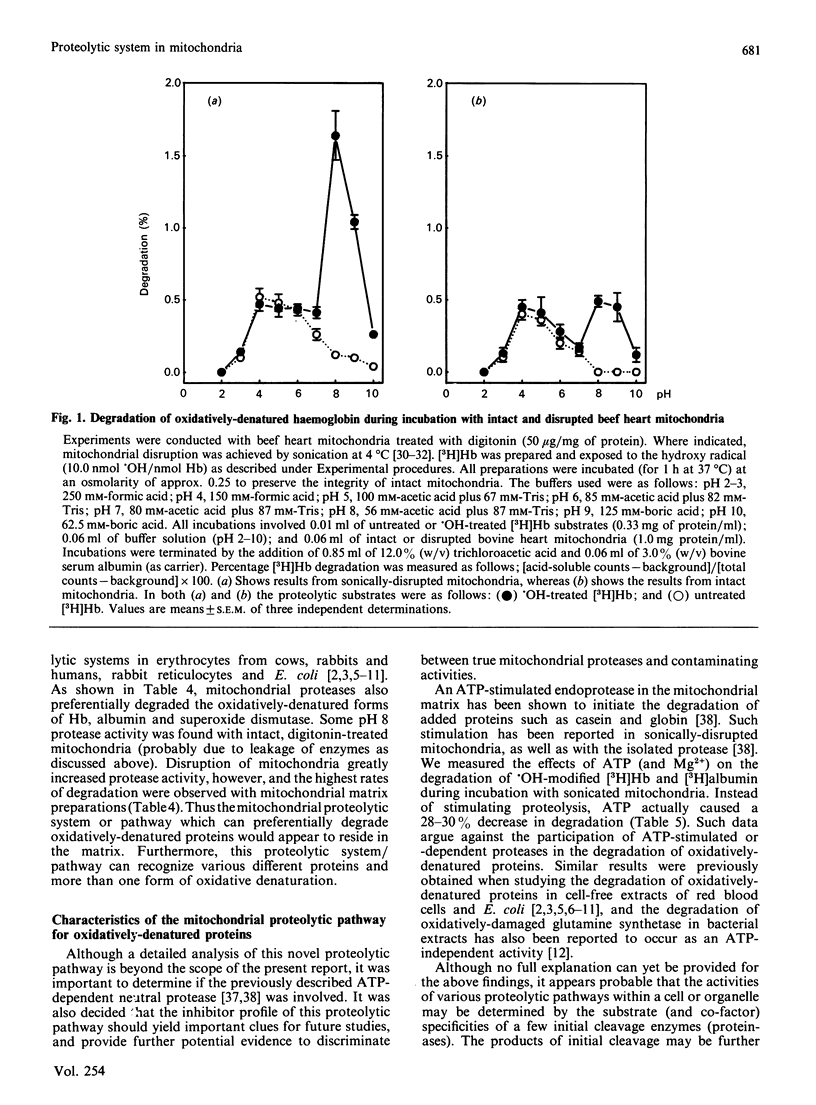
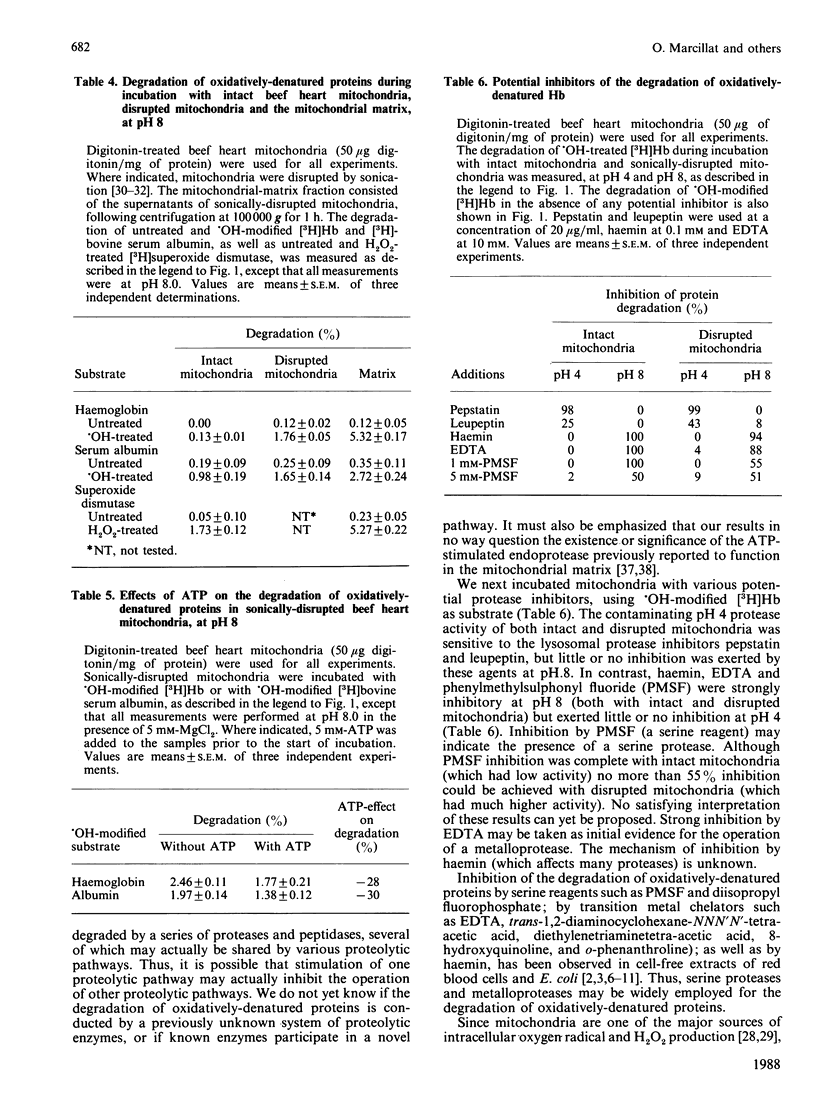
When incubated with mitochondria in an air atmosphere, menadione and doxorubicin (which redox cycle with the respiratory chain to produce oxygen radicals), as well as xanthine oxidase plus xanthine (which generate superoxide and H2O2), stimulated the degradation of newly-synthesized [( 3H]leucine-labelled) mitochondrial polypeptides. No stimulation was observed in an N2 atmosphere, ATP was not required, and xanthine oxidase was not effective without xanthine. Various forms of oxidative stress induced varying degrees of protein cross-linking, protein fragmentation and proteolysis, as judged by gel electrophoresis and amino acid analysis. To learn more about the proteolytic enzymes involved in degradation, we undertook studies with purified protein substrates which had been exposed to oxidative stress (OH or H2O2) in vitro. Despite mitochondrial contamination with acid proteases of lysosomal (and other) origin, pH profiles revealed distinct proteolytic activities at both pH 4 and pH 8. The pH 8 activity preferentially degraded the oxidatively-denatured forms of haemoglobin, albumin and superoxide dismutase; was unaffected by digitonin; and exhibited a several-fold increase in activity upon mitochondrial disruption (highest activity being found in the matrix). In contrast, the pH 4 activity was dramatically decreased by digitonin treatment (to reduce lysosomal contamination); was unaffected by mitochondrial disruption; and showed no preference for oxidatively-denatured proteins. The pH 8 activity was not stimulated by ATP, but was inhibited by EDTA, haemin and phenylmethylsulphonyl fluoride. In contrast, the contaminating pH 4 activity was only inhibited by pepstatin and leupeptin. Thus, our experiments reveal a distinct mitochondrial (matrix) proteolytic pathway which can preferentially degrade oxidatively-denatured proteins.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blech D. M., Borders C. L., Jr Hydroperoxide anion, HO-2, is an affinity reagent for the inactivation of yeast Cu,Zn superoxide dismutase: modification of one histidine per subunit. Arch Biochem Biophys. 1983 Jul 15;224(2):579–586. doi: 10.1016/0003-9861(83)90245-x. [DOI] [PubMed] [Google Scholar]
- Chance B., Sies H., Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979 Jul;59(3):527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- Davies K. J., Delsignore M. E., Lin S. W. Protein damage and degradation by oxygen radicals. II. Modification of amino acids. J Biol Chem. 1987 Jul 15;262(20):9902–9907. [PubMed] [Google Scholar]
- Davies K. J., Delsignore M. E. Protein damage and degradation by oxygen radicals. III. Modification of secondary and tertiary structure. J Biol Chem. 1987 Jul 15;262(20):9908–9913. [PubMed] [Google Scholar]
- Davies K. J., Doroshow J. H., Hochstein P. Mitochondrial NADH dehydrogenase-catalyzed oxygen radical production by adriamycin, and the relative inactivity of 5-iminodaunorubicin. FEBS Lett. 1983 Mar 7;153(1):227–230. doi: 10.1016/0014-5793(83)80153-7. [DOI] [PubMed] [Google Scholar]
- Davies K. J., Doroshow J. H. Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J Biol Chem. 1986 Mar 5;261(7):3060–3067. [PubMed] [Google Scholar]
- Davies K. J., Goldberg A. L. Oxygen radicals stimulate intracellular proteolysis and lipid peroxidation by independent mechanisms in erythrocytes. J Biol Chem. 1987 Jun 15;262(17):8220–8226. [PubMed] [Google Scholar]
- Davies K. J., Goldberg A. L. Proteins damaged by oxygen radicals are rapidly degraded in extracts of red blood cells. J Biol Chem. 1987 Jun 15;262(17):8227–8234. [PubMed] [Google Scholar]
- Davies K. J., Lin S. W. Degradation of oxidatively denatured proteins in Escherichia coli. Free Radic Biol Med. 1988;5(4):215–223. doi: 10.1016/0891-5849(88)90015-9. [DOI] [PubMed] [Google Scholar]
- Davies K. J., Lin S. W. Oxidatively denatured proteins are degraded by an ATP-independent proteolytic pathway in Escherichia coli. Free Radic Biol Med. 1988;5(4):225–236. doi: 10.1016/0891-5849(88)90016-0. [DOI] [PubMed] [Google Scholar]
- Davies K. J., Lin S. W., Pacifici R. E. Protein damage and degradation by oxygen radicals. IV. Degradation of denatured protein. J Biol Chem. 1987 Jul 15;262(20):9914–9920. [PubMed] [Google Scholar]
- Davies K. J. Protein damage and degradation by oxygen radicals. I. general aspects. J Biol Chem. 1987 Jul 15;262(20):9895–9901. [PubMed] [Google Scholar]
- Dean B. Proteolysis in mitochondrial preparations and in lysosomal preparations derived from rat liver. Arch Biochem Biophys. 1983 Nov;227(1):154–163. doi: 10.1016/0003-9861(83)90358-2. [DOI] [PubMed] [Google Scholar]
- Dean R. T., Pollak J. K. Endogenous free radical generation may influence proteolysis in mitochondria. Biochem Biophys Res Commun. 1985 Feb 15;126(3):1082–1089. doi: 10.1016/0006-291x(85)90296-7. [DOI] [PubMed] [Google Scholar]
- Dean R. T., Thomas S. M., Garner A. Free-radical-mediated fragmentation of monoamine oxidase in the mitochondrial membrane. Roles for lipid radicals. Biochem J. 1986 Dec 1;240(2):489–494. doi: 10.1042/bj2400489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desautels M., Goldberg A. L. Demonstration of an ATP-dependent, vanadate-sensitive endoprotease in the matrix of rat liver mitochondria. J Biol Chem. 1982 Oct 10;257(19):11673–11679. [PubMed] [Google Scholar]
- Desautels M., Goldberg A. L. Liver mitochondria contain an ATP-dependent, vanadate-sensitive pathway for the degradation of proteins. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1869–1873. doi: 10.1073/pnas.79.6.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroshow J. H., Davies K. J. Redox cycling of anthracyclines by cardiac mitochondria. II. Formation of superoxide anion, hydrogen peroxide, and hydroxyl radical. J Biol Chem. 1986 Mar 5;261(7):3068–3074. [PubMed] [Google Scholar]
- Duque-Magalhaes M. C., Regnier P. Study on the localization of proteases of mitochondrial origin. Biochimie. 1982 Oct;64(10):907–913. doi: 10.1016/s0300-9084(82)80353-2. [DOI] [PubMed] [Google Scholar]
- Duque-Magalhães M. C., Gualberto J. M. Regulation of mitochondrial proteolysis. Selective degradation of inner membrane polypeptides. FEBS Lett. 1987 Jan 5;210(2):142–146. doi: 10.1016/0014-5793(87)81324-8. [DOI] [PubMed] [Google Scholar]
- Duque-Magalhães M. C., Régnier P. Discrimination of distinct proteinases at the four structural levels of rat liver mitochondria. Biochem J. 1986 Jan 1;233(1):283–286. doi: 10.1042/bj2330283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan J. M., Waxman L., Goldberg A. L. Red blood cells contain a pathway for the degradation of oxidant-damaged hemoglobin that does not require ATP or ubiquitin. J Biol Chem. 1986 May 5;261(13):5705–5713. [PubMed] [Google Scholar]
- Font B., Vial C., Goldschmidt D., Eichenberger D., Gautheron D. C. Heart mitochondrial creatine kinase solubilization. Effect of mitochondrial swelling and SH group reagents. Arch Biochem Biophys. 1981 Nov;212(1):195–203. doi: 10.1016/0003-9861(81)90359-3. [DOI] [PubMed] [Google Scholar]
- Fucci L., Oliver C. N., Coon M. J., Stadtman E. R. Inactivation of key metabolic enzymes by mixed-function oxidation reactions: possible implication in protein turnover and ageing. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1521–1525. doi: 10.1073/pnas.80.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas R., Heinrich P. C. A novel SH-type carboxypeptidase in the inner membrane of rat-liver mitochondria. Eur J Biochem. 1979 May 2;96(1):9–15. doi: 10.1111/j.1432-1033.1979.tb13007.x. [DOI] [PubMed] [Google Scholar]
- Hare J. F. A novel proteinase associated with mitochondrial membranes. Biochem Biophys Res Commun. 1978 Aug 14;83(3):1206–1215. doi: 10.1016/0006-291x(78)91523-1. [DOI] [PubMed] [Google Scholar]
- Hodgson E. K., Fridovich I. The interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide: inactivation of the enzyme. Biochemistry. 1975 Dec 2;14(24):5294–5299. doi: 10.1021/bi00695a010. [DOI] [PubMed] [Google Scholar]
- Kalnov S. L., Novikova L. A., Zubatov A. S., Luzikov V. N. Proteolysis of the products of mitochondrial protein synthesis in yeast mitochondria and submitochondrial particles. Biochem J. 1979 Jul 15;182(1):195–202. doi: 10.1042/bj1820195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine R. L., Oliver C. N., Fulks R. M., Stadtman E. R. Turnover of bacterial glutamine synthetase: oxidative inactivation precedes proteolysis. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2120–2124. doi: 10.1073/pnas.78.4.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R. L. Oxidative modification of glutamine synthetase. I. Inactivation is due to loss of one histidine residue. J Biol Chem. 1983 Oct 10;258(19):11823–11827. [PubMed] [Google Scholar]
- Levine R. L. Oxidative modification of glutamine synthetase. II. Characterization of the ascorbate model system. J Biol Chem. 1983 Oct 10;258(19):11828–11833. [PubMed] [Google Scholar]
- Nagano T., Fridovich I. Superoxide radical from xanthine oxidase acting upon lumazine. J Free Radic Biol Med. 1985;1(1):39–42. doi: 10.1016/0748-5514(85)90027-3. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Stadtman E. R. Oxidative inactivation of glutamine synthetase subunits. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2011–2015. doi: 10.1073/pnas.81.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen P. L., Greenawalt J. W., Reynafarje B., Hullihen J., Decker G. L., Soper J. W., Bustamente E. Preparation and characterization of mitochondria and submitochondrial particles of rat liver and liver-derived tissues. Methods Cell Biol. 1978;20:411–481. doi: 10.1016/s0091-679x(08)62030-0. [DOI] [PubMed] [Google Scholar]
- Quintanilha A. T., Packer L., Davies J. M., Racanelli T. L., Davies K. J. Membrane effects of vitamin E deficiency: bioenergetic and surface charge density studies of skeletal muscle and liver mitochondria. Ann N Y Acad Sci. 1982;393:32–47. doi: 10.1111/j.1749-6632.1982.tb31230.x. [DOI] [PubMed] [Google Scholar]
- Rapoport S. M., Schewe T., Wiesner R., Halangk W., Ludwig P., Janicke-Höhne M., Tannert C., Hiebsch C., Klatt D. The lipoxygenase of reticulocytes. Purification, characterization and biological dynamics of the lipoxygenase; its identity with the respiratory inhibitors of the reticulocyte. Eur J Biochem. 1979 Jun 1;96(3):545–561. doi: 10.1111/j.1432-1033.1979.tb13068.x. [DOI] [PubMed] [Google Scholar]
- Rice R. H., Means G. E. Radioactive labeling of proteins in vitro. J Biol Chem. 1971 Feb 10;246(3):831–832. [PubMed] [Google Scholar]
- Rivett A. J. Preferential degradation of the oxidatively modified form of glutamine synthetase by intracellular mammalian proteases. J Biol Chem. 1985 Jan 10;260(1):300–305. [PubMed] [Google Scholar]
- Roseman J. E., Levine R. L. Purification of a protease from Escherichia coli with specificity for oxidized glutamine synthetase. J Biol Chem. 1987 Feb 15;262(5):2101–2110. [PubMed] [Google Scholar]
- Stadtman E. R., Wittenberger M. E. Inactivation of Escherichia coli glutamine synthetase by xanthine oxidase, nicotinate hydroxylase, horseradish peroxidase, or glucose oxidase: effects of ferredoxin, putidaredoxin, and menadione. Arch Biochem Biophys. 1985 Jun;239(2):379–387. doi: 10.1016/0003-9861(85)90703-9. [DOI] [PubMed] [Google Scholar]
- Wolff S. P., Dean R. T. Fragmentation of proteins by free radicals and its effect on their susceptibility to enzymic hydrolysis. Biochem J. 1986 Mar 1;234(2):399–403. doi: 10.1042/bj2340399. [DOI] [PMC free article] [PubMed] [Google Scholar]


