Abstract
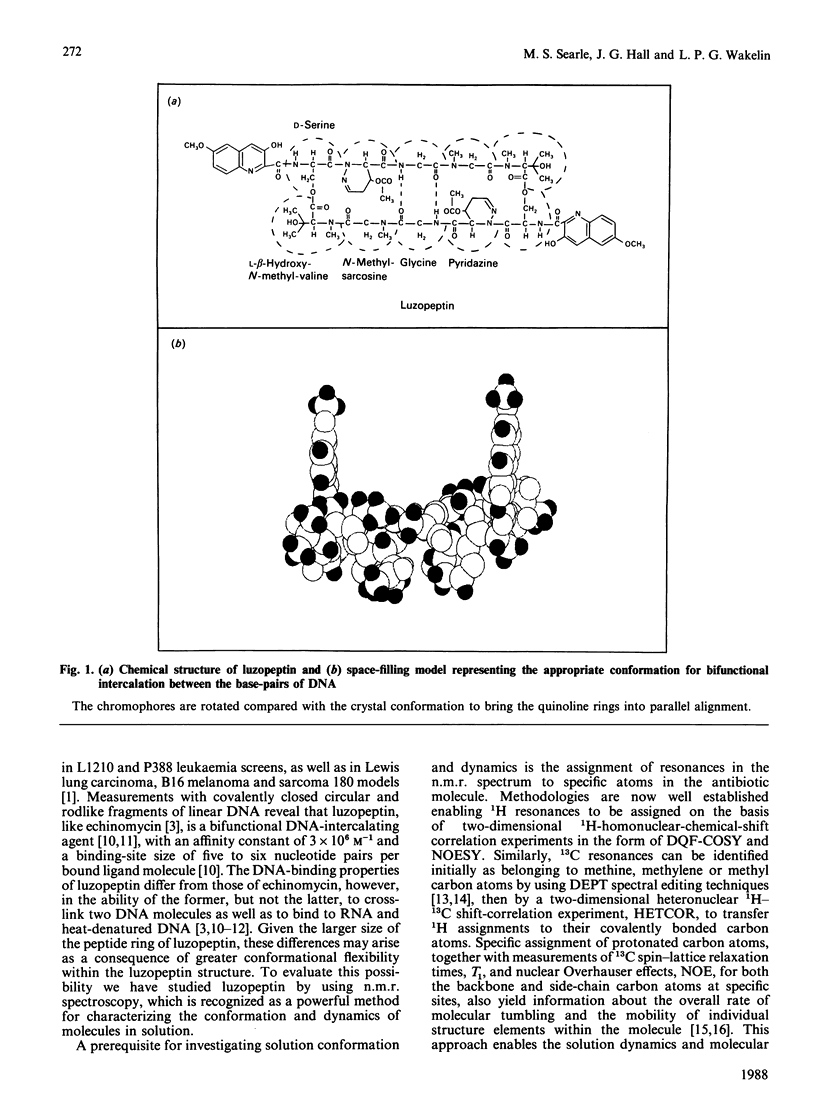
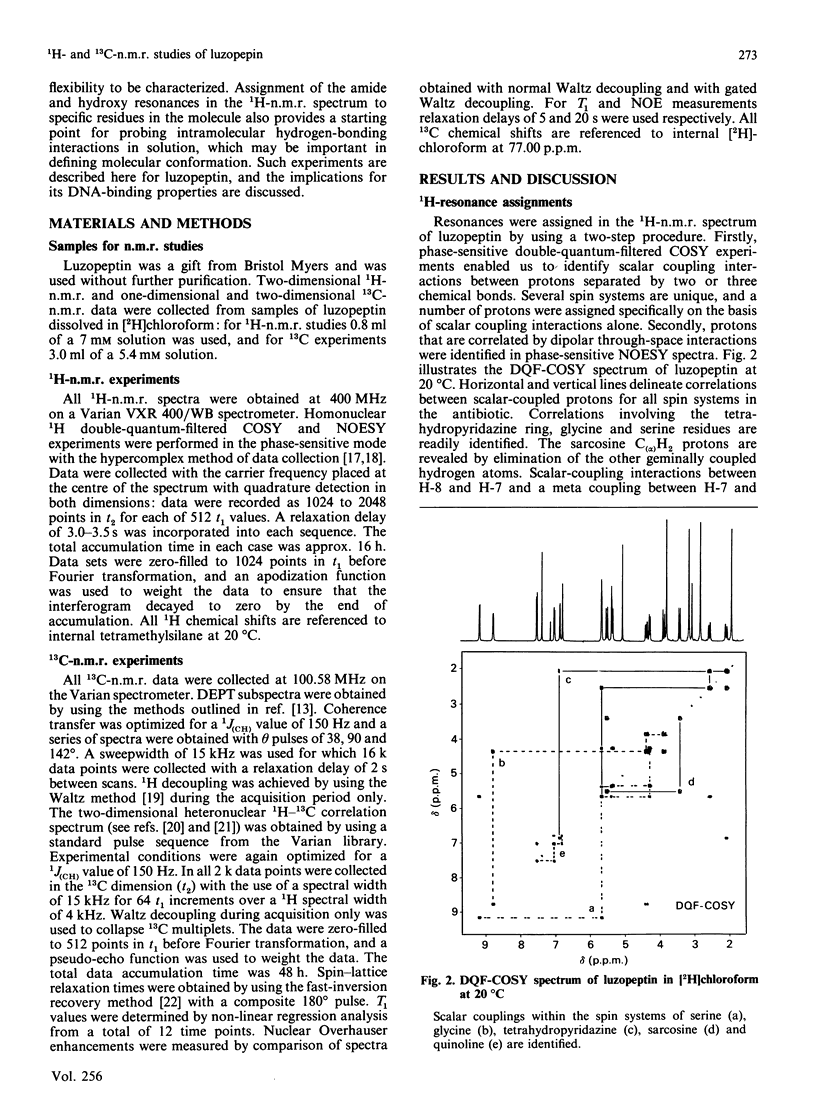
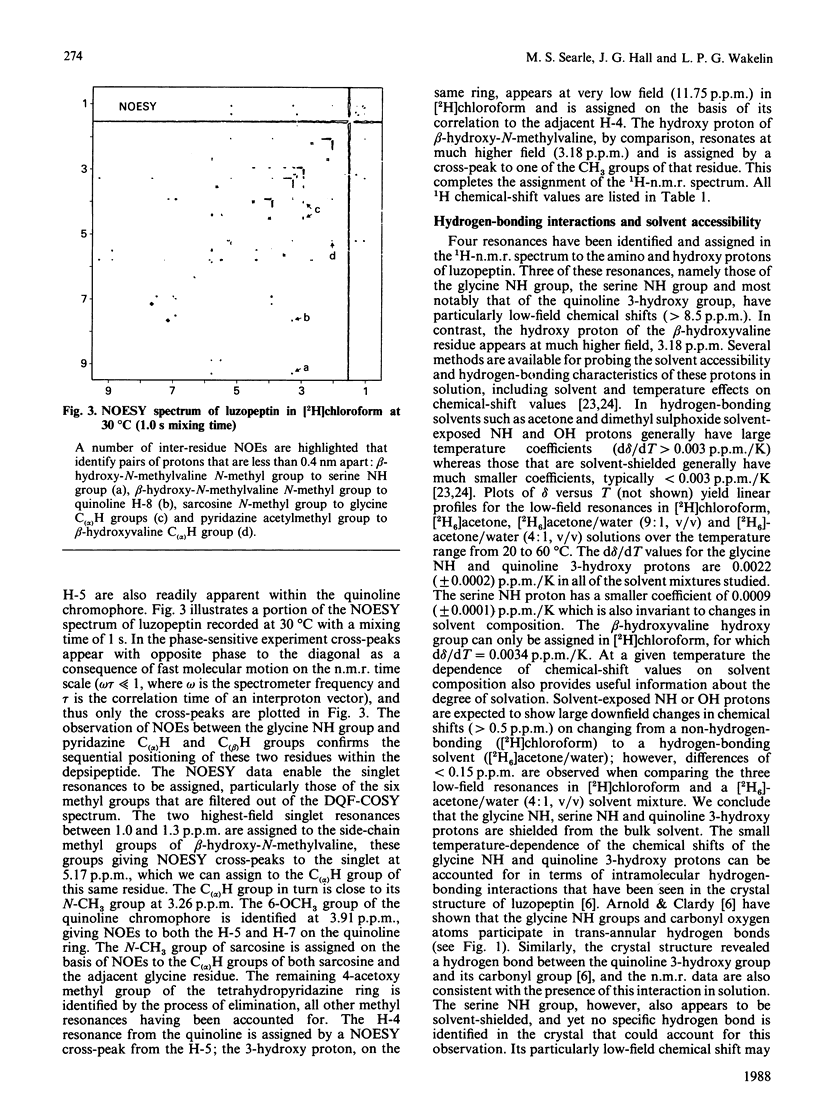
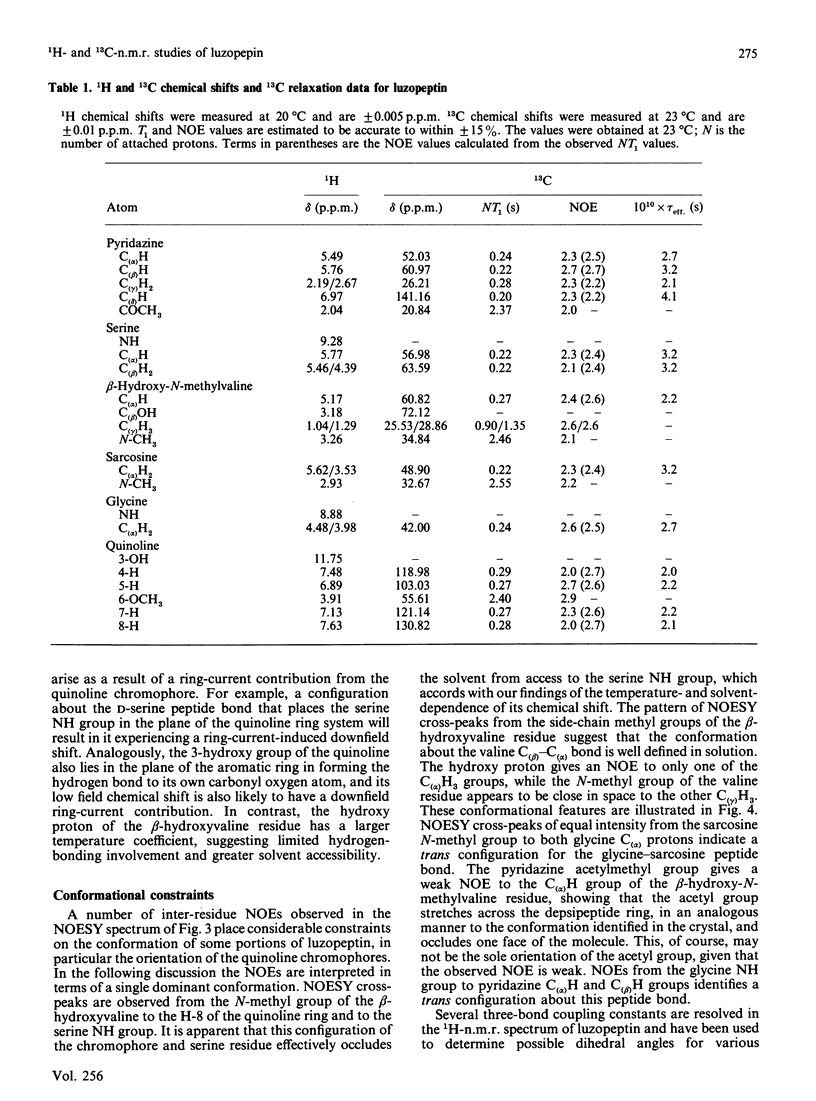
The depsipeptide DNA-intercalating antibiotic luzopeptin was studied in solution by n.m.r. methods. Two-dimensional 1H double-quantum-filtered correlation spectroscopy (DQF-COSY) and nuclear-Overhauser-effect spectroscopy (NOESY) confirm the primary structure and twofold symmetry of luzopeptin and provide details of its three-dimensional conformation in solution. Trans-annular hydrogen bonds between the glycine NH groups and carbonyl oxygen atoms have been identified in the crystalline state [Arnold & Clardy (1981) J. Am. Chem. Soc. 103, 1243-1244], and are important in maintaining an antiparallel beta-sheet conformation. The n.m.r. data indicate that the glycine NH protons are appreciably shielded from the solvent molecules, which suggests that these hydrogen bonds are maintained in solution. The orientation of the quinoline chromophores is defined by two-dimensional NOE cross-peaks that position the N-methyl group of the L-beta-hydroxyvaline residue close in space to both the quinoline H-8 and serine NH proton. This pattern of NOEs is in accord both with the chromophore configuration found in the crystal and one where the quinoline rings are aligned in a parallel manner at right-angles to the depsipeptide ring. The n.m.r. data are consistent with a hydrogen bond between the quinoline hydroxy groups and the quinoline carbonyl oxygen atoms. The pyridazine acetylmethyl groups give NOEs to the C(alpha)H groups of the beta-hydroxy-N-methylvaline residues, showing that the acetyl groups, for at least some of the time, stretch over the depsipeptide ring, occluding one face of the molecule. Both of the latter features are also found in the crystal structure. Resonances in the 13C-n.m.r. spectrum of luzopeptin have been assigned by transferring 1H assignments to their covalently bonded carbon atoms via a heteronuclear shift-correlation experiment (HETCOR). The measurement of spin-lattice relaxation times and 1H-13C NOEs at specific sites in the molecule has led us to conclude that segmental motions within the depsipeptide ring are restricted and that the 13C relaxation data for luzopeptin's protonated carbon atoms are adequately described by isotropic tumbling in solution. Furthermore, relaxation data for the carbon atoms of the quinoline chromophores show that these rings exhibit similar motion to the depsipeptide ring and are not rotating rapidly with respect to it. Taken together all the data imply that luzopeptin is fairly rigid in solution, on the time scale of molecular tumbling, and has, or can readily attain, a staple-like structure suitable for bisintercalation.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allerhand A., Doddrell D., Glushko V., Cochran D. W., Wenkert E., Lawson P. J., Gurd F. R. Conformation and segmental motion of native and denatured ribonuclease A in solution. Application of natural-abundance carbon-13 partially relaxed Fourier transform nuclear magnetic resonance. J Am Chem Soc. 1971 Jan 27;93(2):544–546. doi: 10.1021/ja00731a053. [DOI] [PubMed] [Google Scholar]
- Allerhand A., Komoroski R. A. Study of internal rotations in gramicidin S by means of carbon-13 spin-lattice relaxation measurements. J Am Chem Soc. 1973 Dec 12;95(25):8228–8231. doi: 10.1021/ja00806a003. [DOI] [PubMed] [Google Scholar]
- Deslauriers R., Levy G. C., McGregor W. H., Sarantakis K., Smith I. C. Conformational flexibility of luteinizing hormone-releasing hormone in aqueous solution. A carbon-13 spin-lattice relaxation time study. Biochemistry. 1975 Sep 23;14(19):4335–4343. doi: 10.1021/bi00690a030. [DOI] [PubMed] [Google Scholar]
- Deslauriers R., Paiva A. C., Schaumburg K., Smith I. C. Conformational flexibility of angiotensin II. A carbon-13 spin-lattice relaxation study. Biochemistry. 1975 Mar 11;14(5):878–886. doi: 10.1021/bi00676a003. [DOI] [PubMed] [Google Scholar]
- Deslauriers R., Smith I. C., Walter R. Conformational mobility of the pyrrolidine ring of proline in peptides and peptide hormones as manifest in carbon 13 spin-lattice relaxation times. J Biol Chem. 1974 Nov 10;249(21):7006–7010. [PubMed] [Google Scholar]
- Huang C. H., Mirabelli C. K., Mong S., Crooke S. T. Intermolecular cross-linking of DNA through bifunctional intercalation of an antitumor antibiotic, luzopeptin A (BBM-928A). Cancer Res. 1983 Jun;43(6):2718–2724. [PubMed] [Google Scholar]
- Huang C. H., Mong S., Crooke S. T. Interactions of a new antitumor antibiotic BBM-928A with deoxyribonucleic acid. Bifunctional intercalative binding studied by fluorometry and viscometry. Biochemistry. 1980 Nov 25;19(24):5537–5542. doi: 10.1021/bi00565a012. [DOI] [PubMed] [Google Scholar]
- Huang C. H., Prestayko A. W., Crooke S. T. Bifunctional intercalation of antitumor antibiotics BBM-928A and echinomycin with deoxyribonucleic acid. Effects of intercalation on deoxyribonucleic acid degradative activity of bleomycin and phleomycin. Biochemistry. 1982 Jul 20;21(15):3704–3710. doi: 10.1021/bi00258a028. [DOI] [PubMed] [Google Scholar]
- Konishi M., Ohkuma H., Sakai F., Tsuno T., Koshiyama H., Naito T., Kawaguchi H. BBM-928, a new antitumor antibiotic complex. III. Structure determination of BBM-928 A, B and C. J Antibiot (Tokyo) 1981 Feb;34(2):148–159. doi: 10.7164/antibiotics.34.148. [DOI] [PubMed] [Google Scholar]
- Led J. J., Grant D. M., Horton W. J., Sundby F., Vihelmsen K. Carbon-13 magnetic resonance study of structural and dynamical features in carbamylated insulins. J Am Chem Soc. 1975 Oct 15;97(21):5997–6008. doi: 10.1021/ja00854a008. [DOI] [PubMed] [Google Scholar]
- Ohkuma H., Sakai F., Nishiyama Y., Ohbayashi M., Imanishi H., Konishi M., Miyaki T., Koshiyama H., Kawaguchi H. BBM-928, a new antitumor antibiotic complex. I. Production, isolation, characterization and antitumor activity. J Antibiot (Tokyo) 1980 Oct;33(10):1087–1097. doi: 10.7164/antibiotics.33.1087. [DOI] [PubMed] [Google Scholar]
- Sahal D., Balaram P. Peptide models of electrostatic interactions in proteins: NMR studies on two beta-turn tetrapeptides containing Asp-His and Asp-Lys salt bridges. Biochemistry. 1986 Oct 7;25(20):6004–6013. doi: 10.1021/bi00368a026. [DOI] [PubMed] [Google Scholar]
- Tomita K., Hoshino Y., Sasahira T., Kawaguchi H. BBM-928, a new antitumor antibiotic complex. II. Taxonomic studies on the producing organism. J Antibiot (Tokyo) 1980 Oct;33(10):1098–1102. doi: 10.7164/antibiotics.33.1098. [DOI] [PubMed] [Google Scholar]
- Ughetto G., Wang A. H., Quigley G. J., van der Marel G. A., van Boom J. H., Rich A. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 1985 Apr 11;13(7):2305–2323. doi: 10.1093/nar/13.7.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswamitra M. A., Kennard O., Cruse W. B., Egert E., Sheldrick G. M., Jones P. G., Waring M. J., Wakelin L. P., Olsen R. K. Structure of TANDEM and its implication for bifunctional intercalation into DNA. Nature. 1981 Feb 26;289(5800):817–819. doi: 10.1038/289817a0. [DOI] [PubMed] [Google Scholar]
- Wakelin S. P., Waring M. J. The binding of echinomycin to deoxyribonucleic acid. Biochem J. 1976 Sep 1;157(3):721–740. doi: 10.1042/bj1570721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974 Dec 20;252(5485):653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]


