Abstract
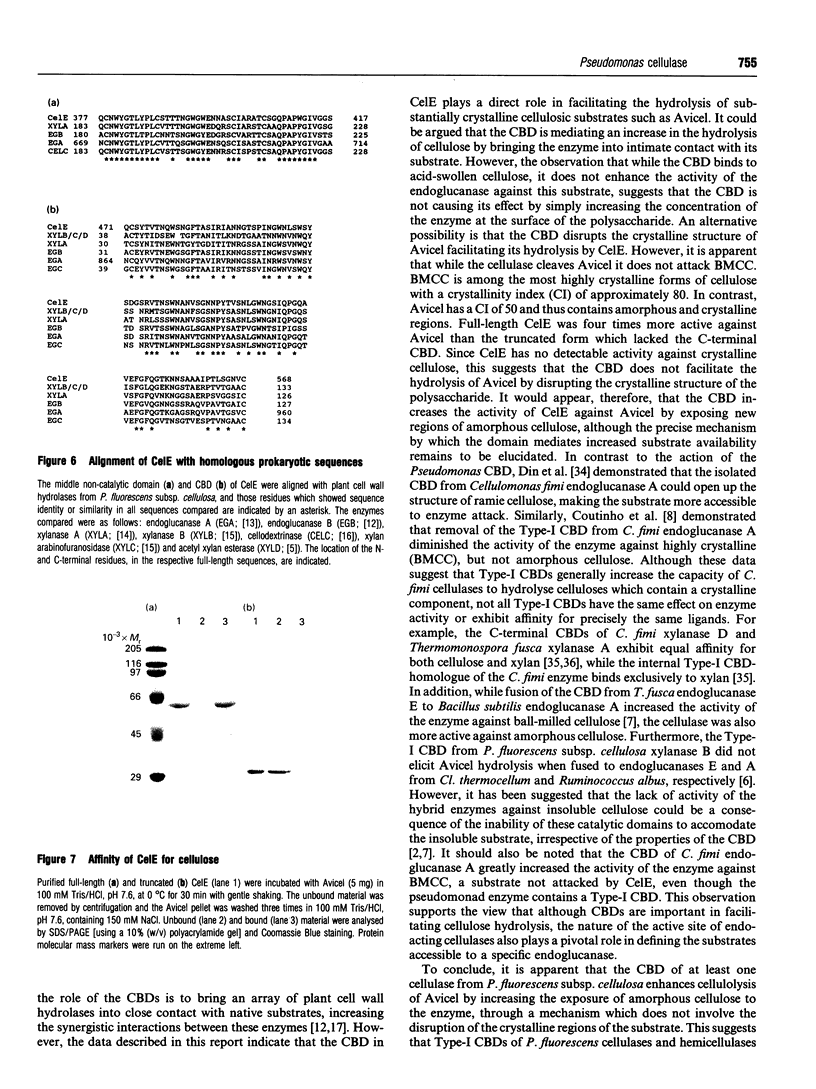
A genomic library of Pseudomonas fluorescens subsp. cellulosa DNA, constructed in lambda ZAPII, was screened for carboxymethyl-cellulase activity. The pseudomonad insert from a recombinant phage which displayed elevated cellulase activity in comparison with other cellulase-positive clones present in the library, was excised into pBluescript SK- to generate the plasmid pC48. The nucleotide sequence of the cellulase gene, designated celE, revealed a single open reading frame of 1710 bp that encoded a polypeptide, defined as endoglucanase E (CelE), of M(r) 59663. The deduced primary structure of CelE revealed an N-terminal signal peptide followed by a 300-amino-acid sequence that exhibited significant identity with the catalytic domains of cellulases belonging to glycosyl hydrolase Family 5. Adjacent to the catalytic domain was a 40-residue region that exhibited strong sequence identity to non-catalytic domains located in two other endoglucanases and a xylanase from P. fluorescens. The C-terminal 100 residues of CelE were similar to Type-I cellulose-binding domains (CBDs). The three domains of the cellulase were joined by linker sequences rich in serine residues. Analysis of the biochemical properties of full-length and truncated derivatives of CelE confirmed that the enzyme comprised an N-terminal catalytic domain and a C-terminal CBD. Analysis of purified CelE revealed that the enzyme had an M(r) of 56000 and an experimentally determined N-terminal sequence identical to residues 40-54 of the deduced primary structure of full-length CelE. The enzyme exhibited an endo mode of action in hydrolysing a range of cellulosic substrates including Avicel and acid-swollen cellulose, but did not attack xylan or any other hemicelluloses. A truncated form of the enzyme, which lacked the C-terminal CBD, displayed the same activity as full-length CelE against soluble cellulose and acid-swollen cellulose, but exhibited substantially lower activity than the full-length cellulase against Avicel. The significance of these data in relation to the role of the CBD is discussed.
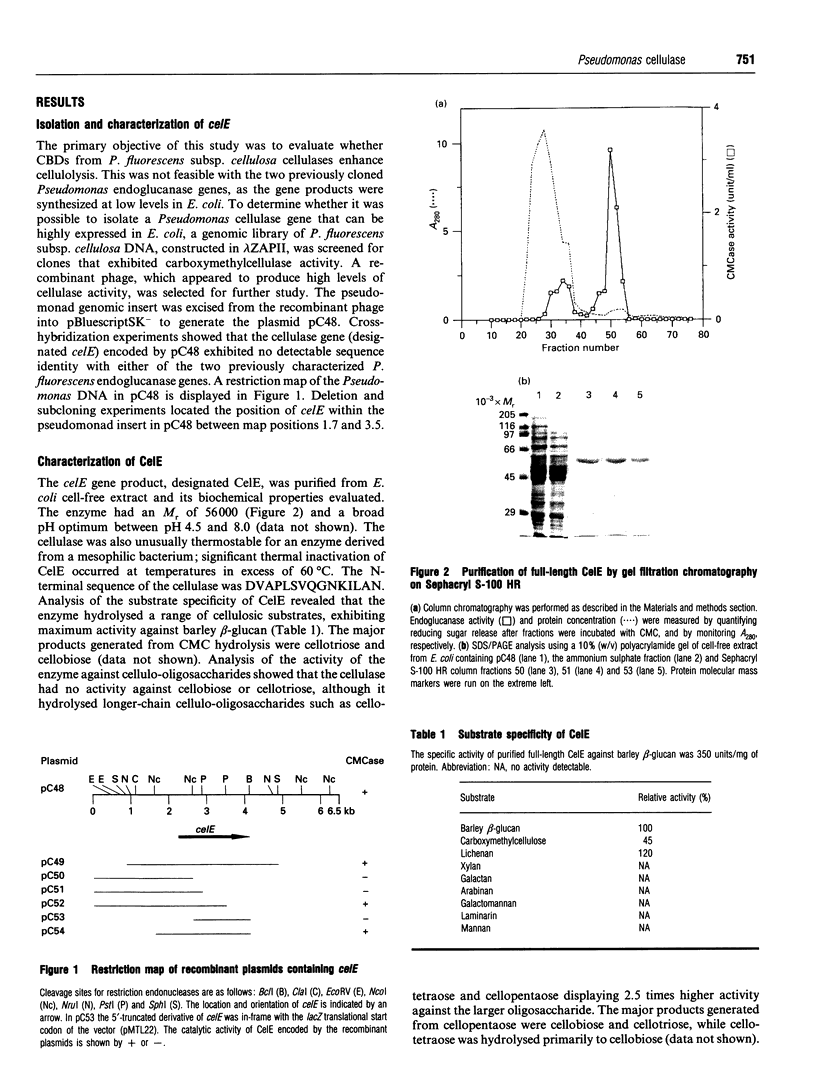
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black G. W., Hazlewood G. P., Millward-Sadler S. J., Laurie J. I., Gilbert H. J. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem J. 1995 Apr 1;307(Pt 1):191–195. doi: 10.1042/bj3070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite K. L., Black G. W., Hazlewood G. P., Ali B. R., Gilbert H. J. A non-modular endo-beta-1,4-mannanase from Pseudomonas fluorescens subspecies cellulosa. Biochem J. 1995 Feb 1;305(Pt 3):1005–1010. doi: 10.1042/bj3051005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun C., Meinke A., Ziser L., Withers S. G. Simultaneous high-performance liquid chromatographic determination of both the cleavage pattern and the stereochemical outcome of the hydrolysis reactions catalyzed by various glycosidases. Anal Biochem. 1993 Jul;212(1):259–262. doi: 10.1006/abio.1993.1320. [DOI] [PubMed] [Google Scholar]
- Béguin P., Aubert J. P. The biological degradation of cellulose. FEMS Microbiol Rev. 1994 Jan;13(1):25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- Chambers S. P., Prior S. E., Barstow D. A., Minton N. P. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988 Aug 15;68(1):139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Ferreira L. M., Hazlewood G. P., Barker P. J., Gilbert H. J. The cellodextrinase from Pseudomonas fluorescens subsp. cellulosa consists of multiple functional domains. Biochem J. 1991 Nov 1;279(Pt 3):793–799. doi: 10.1042/bj2790793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. M., Wood T. M., Williamson G., Faulds C., Hazlewood G. P., Black G. W., Gilbert H. J. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem J. 1993 Sep 1;294(Pt 2):349–355. doi: 10.1042/bj2940349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes C. M., Hall J., Hirst B. H., Hazlewood G. P., Gilbert H. J. The resistance of cellulases and xylanases to proteolytic inactivation. Appl Microbiol Biotechnol. 1995 Apr;43(1):52–57. doi: 10.1007/BF00170622. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Hall J., Hazlewood G. P., Ferreira L. M. The N-terminal region of an endoglucanase from Pseudomonas fluorescens subspecies cellulosa constitutes a cellulose-binding domain that is distinct from the catalytic centre. Mol Microbiol. 1990 May;4(5):759–767. doi: 10.1111/j.1365-2958.1990.tb00646.x. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Jenkins G., Sullivan D. A., Hall J. Evidence for multiple carboxymethylcellulase genes in Pseudomonas fluorescens subsp. cellulosa. Mol Gen Genet. 1987 Dec;210(3):551–556. doi: 10.1007/BF00327211. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Sullivan D. A., Jenkins G., Kellett L. E., Minton N. P., Hall J. Molecular cloning of multiple xylanase genes from Pseudomonas fluorescens subsp. cellulosa. J Gen Microbiol. 1988 Dec;134(12):3239–3247. doi: 10.1099/00221287-134-12-3239. [DOI] [PubMed] [Google Scholar]
- Hall J., Gilbert H. J. The nucleotide sequence of a carboxymethylcellulase gene from Pseudomonas fluorescens subsp. cellulosa. Mol Gen Genet. 1988 Jul;213(1):112–117. doi: 10.1007/BF00333406. [DOI] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Huskisson N. S., Durrant A. J., Gilbert H. J. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol Microbiol. 1989 Sep;3(9):1211–1219. doi: 10.1111/j.1365-2958.1989.tb00271.x. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991 Dec 1;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993 Aug 1;293(Pt 3):781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hewick R. M., Dreyer W. J., Hood L. E. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 1983;91:399–413. doi: 10.1016/s0076-6879(83)91038-8. [DOI] [PubMed] [Google Scholar]
- Irwin D., Jung E. D., Wilson D. B. Characterization and sequence of a Thermomonospora fusca xylanase. Appl Environ Microbiol. 1994 Mar;60(3):763–770. doi: 10.1128/aem.60.3.763-770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellett L. E., Poole D. M., Ferreira L. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maglione G., Matsushita O., Russell J. B., Wilson D. B. Properties of a genetically reconstructed Prevotella ruminicola endoglucanase. Appl Environ Microbiol. 1992 Nov;58(11):3593–3597. doi: 10.1128/aem.58.11.3593-3597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward-Sadler S. J., Poole D. M., Henrissat B., Hazlewood G. P., Clarke J. H., Gilbert H. J. Evidence for a general role for high-affinity non-catalytic cellulose binding domains in microbial plant cell wall hydrolases. Mol Microbiol. 1994 Jan;11(2):375–382. doi: 10.1111/j.1365-2958.1994.tb00317.x. [DOI] [PubMed] [Google Scholar]
- Murphy G., Kavanagh T. Speeding-up the sequencing of double-stranded DNA. Nucleic Acids Res. 1988 Jun 10;16(11):5198–5198. doi: 10.1093/nar/16.11.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole D. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Characterization of hybrid proteins consisting of the catalytic domains of Clostridium and Ruminococcus endoglucanases, fused to Pseudomonas non-catalytic cellulose-binding domains. Biochem J. 1991 Nov 1;279(Pt 3):787–792. doi: 10.1042/bj2790787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sedmak J. J., Grossberg S. E. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal Biochem. 1977 May 1;79(1-2):544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Wood P. J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982 Apr;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988 Jan 4;170(3):575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- Wood T. M. The cellulase of Fusarium solani. Purification and specificity of the -(1-4)-glucanase and the -D-glucosidase components. Biochem J. 1971 Feb;121(3):353–362. doi: 10.1042/bj1210353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Xue G. P., Orpin C. G., Black G. W., Gilbert H. J., Hazlewood G. P. Intronless celB from the anaerobic fungus Neocallimastix patriciarum encodes a modular family A endoglucanase. Biochem J. 1994 Jan 15;297(Pt 2):359–364. doi: 10.1042/bj2970359. [DOI] [PMC free article] [PubMed] [Google Scholar]