Abstract
Rice sheath blight is a serious disease caused by Rhizoctonia solani that reduces rice yield. Currently, there is a lack of efficient and environmentally friendly control methods. In this study, we found that Bacillus velezensis (B. velezensis) Y6 could significantly inhibit the growth of mycelium in Rhizoctonia solani, and its control efficiency against rice sheath blight was 58.67% (p < 0.01) in a pot experiment. Lipopeptides play an important role in the control of rice sheath blight by B. velezensis Y6, among which iturin and fengycin are essential, and iturin W, a novel lipopeptide in B. velezensis, plays a major role in lipopeptide antagonism to Rhizoctonia solani. In the field, we also found that inoculation with B. velezensis Y6 can increase rice yield (dry weight) by 11.75%. Furthermore, the transcriptome profiling results of the rice roots revealed that there were a total of 1227 differential genes (DEGs) regulated when treated with Y6, of which 468 genes were up-regulated and 971 genes were down-regulated in rice roots compared with the control. Among them, the DEGs were mainly distributed in biological processes (BP) and were mainly enriched in response to stimulus (GO:0050896), response to stress (GO:0006950), and response to abiotic stimulus (GO:0009628). According to the KEGG pathway analysis, there were 338 DEGs classified into 87 KEGG functional pathway categories. Compared with the control, a large number of enriched genes were distributed in phenylpropanoid biosynthesis (map00940), glutathione metabolism (map00480), glycolysis/gluconeogenesis (map00010), and amino sugar and nucleotide sugar metabolism (map00520). In summary, this investigation provides a new perspective for studying the molecular mechanism of B. velezensis in controlling rice sheath blight.
Keywords: rice sheath blight, Bacillus velezensis, biocontrol, lipopeptides, transcriptome
1. Introduction
Rice sheath blight is one of China’s three major rice diseases [1]. The disease is caused by Rhizoctonia solani Kühn and has a wide disease area and a high frequency of incidence, which brings huge obstacles to the high and stable yield of rice [2]. At present, there are no cultivars with high resistance to rice sheath blight in rice cultivation, and agrochemical application is still the main means of controlling this disease [3,4]. However, the extensive use of pesticides in the field can lead to increased disease resistance and environmental pollution, and the search for safer and more effective methods of controlling rice sheath blight is now a mainstream trend. Biological control is widely recognized as a reliable, safe, and sustainable method of plant disease control [5,6].
Biological control mainly involves the use of microbial strains such as Bacillus, Harzianum Pseudomonas, Actinomycetes and Burkholderia to control plant diseases [7]. The use of Bacillus or its metabolites to control plant diseases is currently the focus of biocontrol research [8,9]. Bacillus can act as a biocontrol agent against fungal, bacterial and viral infections by inducing systemic resistance (ISR) in host plants [10,11]. B. velezensis YYC enhances the activity of defense-related enzymes (e.g., PAL, POD, and SOD), thereby increasing the basal immunity of the plant [12]. Bacillus cereus (Bce-2) prevents strawberry leaf spot disease caused by N. clavispora via increasing the expression of disease resistance genes in strawberry leaves [13]. Bacillus cereus AR156 can induce systemic resistance of Arabidopsis thaliana to Botrytis cinerea via JA/ET and NPR1-dependent signaling pathways [14]. In addition, Bacillus produces a variety of antibiotics, such as cyclic lipopeptides (CLPs), which can directly antagonize pathogens [15,16]. Surfactants secreted by Bacillus amyloliquefaciens Ba01 play an important role in the biological control of potato common scab [17]. Iturin A and Fengycin A secreted by Bacillus megaterium WL-3 have an inhibitory effect on P. infestans mycelium growth, as well as a good control effect on potato late blight in greenhouse experiments and field assays [18]. Although there are many studies on the biological control of plant diseases by Bacillus, the application of B. velezensis to the biological control of rice blight has not been reported.
In this study, we verified through pot experiments that B. velezensis Y6 could effectively control rice sheath blight. The main types and basic structures of lipopeptides secreted by B. velezensis Y6 were discovered by mass spectrometry; the lipopeptides essential for the antagonism of Rhizoctonia solani by B. velezensis Y6 were discovered by constructing mutant strains with deletions of genes related to lipopeptide synthesis; and the effect of B. velezensis Y6 on the expression of the rice transcriptome was revealed by RNAseq. This study provides new insights into how B. velezensis could be used to control rice sheath blight.
2. Materials and Methods
2.1. Strains and Growth Media
The strains used in this study are listed in Table S1. Mutants lacking lipopeptide synthesis were constructed as described previously [19]. B. velezensis Y6 and its derivative mutant strains were cultured on LB (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl, and distilled water 1 L at pH 7.0–7.2), or on solid LB medium supplemented with 1.5% agar. The corresponding antibiotics were added when culturing mutants. Rhizoctonia solani AG1-IA GD118 was cultured on PDA (potato 200 g/L, glucose 20 g/L, agar 20 g/L, and distilled water 1 L at pH 7.0–7.2) at 30 °C. The rice variety was Meixiangzhan 2.
2.2. B. velezensis Y6 Antagonizes Rhizoctonia solani
Rhizoctonia solani AG1-IA GD118 was cultured on PDA plates at 30 °C for 3 days. A 5 mm diameter mycelial agar block was cut and transferred to the center of a new PDA plate. After 1 day of culture, 30 µL of B. velezensis Y6 or its derivative mutants (OD600 ~0.4) grown in LB medium was added to a 5 mm diameter well 2.5 cm from the center of the mycelial agar block. Antifungal activity was evaluated by measuring the diameter of the inhibition zone after 2 days of culture at 30 °C.
2.3. Pot Experiment
After soaking rice seeds in warm water for 1 day, they were germinated at 30 °C for 2 days before sowing. When the rice had 3 leaves at 28 °C, it was transplanted. The B. velezensis Y6 solution with a concentration of 1 × 106 CFU/mL was inoculated on the rice root. After overnight cultivation, the rice was transplanted into a 6 L planting bucket (containing 5 kg of paddy soil). At 15 days after transplanting, the Y6 liquid bacterial agent with a concentration of 106 CFU/mL was sprayed on the rice until the rice leaves were fully moistened. The control group was sprayed with water, and the pesticide group was sprayed with 5% of Validamycin. At 24 h after the treatment, the Rhizoctonia solani grown on 2% agar PDA medium was cut into 5 mm pieces, fixed on the leaf sheath of the lowest rice leaf with plastic wrap, and kept moist with absorbent cotton. The disease severity data were collected from 15 days after the pathogen inoculation. The degree of disease severity was assigned as follows (Figure 6A): 0 = no lesions, 1 = appearance of water-soaked lesions, 2 = appearance of necrotic lesions, 3 = less than 50% necrosis on the leaf cross section, 4 = more than 50% necrosis on the leaf cross section, and 5 = necrosis across the entire leaf section resulting in leaf death [4]. The severity of the disease and the biocontrol efficiency were calculated according to the following formula.
2.4. Field Experiment
The field experiment was conducted on 1 April 2022, at the South China Agricultural Teaching and Research Base, Zengcheng District, Guangzhou City, Guangdong Province (113.633677° E, 23.243212° N, 22 m a.s.l.). The rice variety used was Meixiangzhan No. 2 (conventional rice), with a planting area of 30 m2 in each plot, a plant spacing of 15 cm, a row spacing of 30 cm, and a spacing of 50 cm between plots. Each treatment was repeated four times.
The rice seedlings were inoculated with B. velezensis Y6 solution at a concentration of 1 × 106 CFU/mL by root irrigation one day before transplanting. The field was managed with normal fertilizer and water, and B. velezensis Y6 solution with a concentration of 1 × 106 CFU/mL was sprayed on the leaves during the rice booting stage. After the rice matured, 20 rice plants were randomly selected from each plot to determine the number of effective panicles. The rice yield was determined by sampling and measuring the yield in the plot. The yield measurement area was 1 m2, and the measurement was repeated three times along the diagonal of the plot.
2.5. Extraction of Crude Lipopeptides
Crude lipopeptides were extracted by acid precipitation and methanol extraction. B. velezensis Y6 and its derivative mutant strains were cultured on LB liquid medium at 37 °C for 1 d. The culture was centrifuged at 12,000 rpm for 10 min to remove cells; the supernatant was adjusted to pH 2.0 with concentrated hydrochloric acid and allowed to precipitate overnight. It was then centrifuged at 12,000 rpm for 10 min and the supernatant was discarded. The precipitate was retained and extracted twice with 1/10 volume of methanol. It was allowed to stand at 4 °C for 2 h each time, and then the methanol extracts were combined.
2.6. Extraction of Lipopeptides (LP) from the Transparent Inhibition Zone
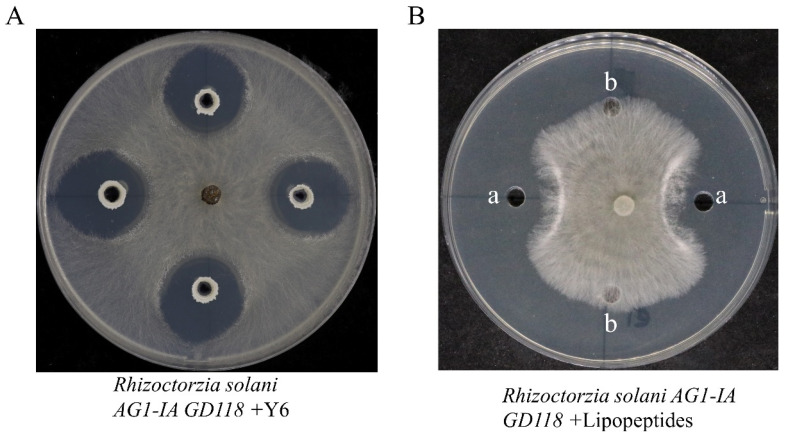
We extracted LPs from the clear zone (Figure 1A). A 600 mg agar sample was collected from the clear zone, minced and mixed with 2 mL of acetonitrile/water (1:1 v/v), shaken, sonicated twice for 30 s each and centrifuged, and then the supernatant was collected and filtered [19].
Figure 1.
Inhibitory effect of B. velezensis Y6 and lipopeptides against Rhizoctonia solani mycelium growth. (A) Photograph of the inhibitory effect of B. velezensis Y6 on the mycelial growth of Rhizoctonia solani; (B) photograph of the inhibitory effect of lipopeptides on the mycelial growth of Rhizoctonia solani, (a) crude lipopeptides, (b) methanol.
2.7. Identification of LPs by UPLC/Q-TOF-MS
The acetonitrile/water extracts were analyzed by reversed-phase Ultra-Performance Liquid Chromatography coupled with Time-of-Flight Mass Spectrometry (UPLC/Q-TOF-MS). The mass-to-charge ratio (m/z) was used to identify the lipopeptide compounds. Gradient elution was performed using (A) acetonitrile and (B) water (containing 0.2% formic acid), and the column temperature was maintained at 40 °C [20]. The detection conditions of the Agilent Ultra-Performance Liquid Chromatography coupled with Time-of-Flight Mass Spectrometry (UPLC/Q-TOF-MS) were as shown in Table 1:
Table 1.
UPLC/Q-TOF Time-of-Flight Mass Spectrometer detection mobile phase settings.
| Time (min) | Mobile Phase A% | Mobile Phase B% | Flow Rate |
|---|---|---|---|
| 4.5 | 80 | 20 | 0.3 mL/min |
| 5 | 80 | 20 | 0.3 mL/min |
| 7 | 95 | 5 | 0.3 mL/min |
| 9 | 95 | 5 | 0.3 mL/min |
| 10 | 40 | 60 | 0.3 mL/min |
| 15 | 40 | 60 | 0.3 mL/min |
2.8. Total RNA Extraction and Transcriptomic Analysis
To evaluate the effect of Y6 on the rice transcriptome, the following experiment was performed: 15 days after rice plants were transplanted, three samples were collected from the roots of rice plants in the control and Y6 treatments. The rice roots were washed twice with sterile water and wiped dry, then quickly frozen in liquid nitrogen, and finally the samples were stored in an ultra-low-temperature freezer at −80 °C. Total RNA was then extracted using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). The concentration, purity, and integrity of the RNA were then assessed using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA).
We selected a total amount of >=1 μg of RNA and used the NEBNext Ultra II RNA Library Prep Kit for Illumina to construct the library. We used the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and the Agilent High Sensitivity DNA Kit (Agilent, 5067-4626) to determine the library quality. We used Cutadapt (v1.15) software to filter the sequencing data and obtain high-quality sequences (clean data) for further analysis. The Oryza sativa genome was constructed using IRGSP-1.0 as the reference genome. A term was assigned to each gene in the Gene Ontology database, and the number of differentially enriched genes in each term was calculated. The hypergeometric distribution method was used to calculate the p value, and topGO (2.40.0) was used to perform GO enrichment analysis on the differentially expressed genes (the benchmark for significant enrichment was p value 0.05) and locate the GO of significant enrichment of the differentially expressed genes to determine the main biological functions of the differentially expressed genes. ClusterProfiler (3.16.1) software was used to perform enrichment analysis on the KEGG pathways of differentially expressed genes, with an emphasis on the significantly enriched pathways.
2.9. Statistical Analysis
Statistical analysis in the experiments was performed using the t test in IBM SPSS Statistics 20, and p < 0.05 indicated a significant difference. Tables were created using GraphPad Prism 8.
3. Results
3.1. B. velezensis Y6 Can Effectively Inhibit the Growth of Rhizoctonia solani
A plate antagonism test was used to investigate B. velezensis Y6’s antagonistic effect on the rice sheath blight pathogen. Rhizoctonia solani’s mycelium can be inhibited by B. velezensis Y6, as shown in Figure 1A, resulting in the formation of a clear, transparent region on the plate. The lipopeptides secreted by B. velezensis Y6 were extracted, and the plate dual culture method was used to study the effect of the lipopeptides secreted by B. velezensis Y6 on rice sheath blight pathogen. As shown in Figure 1B, B. velezensis Y6 can significantly restrict the growth of mycelium of Rhizoctonia solani. The above results show that B. velezensis Y6 can effectively antagonize rice sheath blight pathogens, and lipopeptides play an important role in this process.
3.2. Characterization of B. velezensis Y6-Secreted Lipopeptides by UPLC/Q-TOF-MS
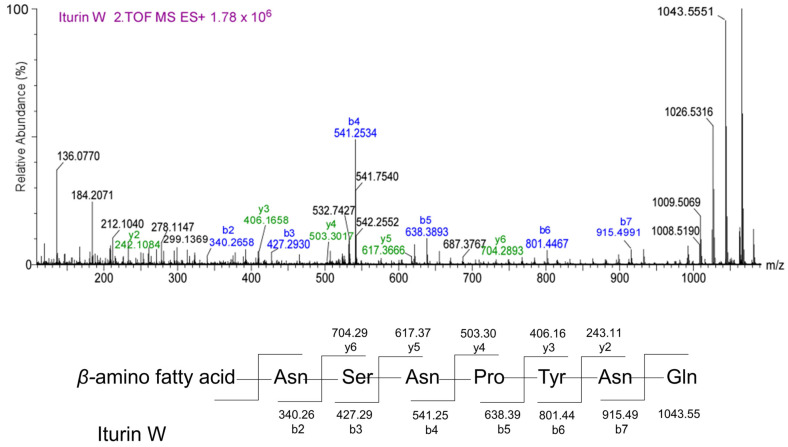
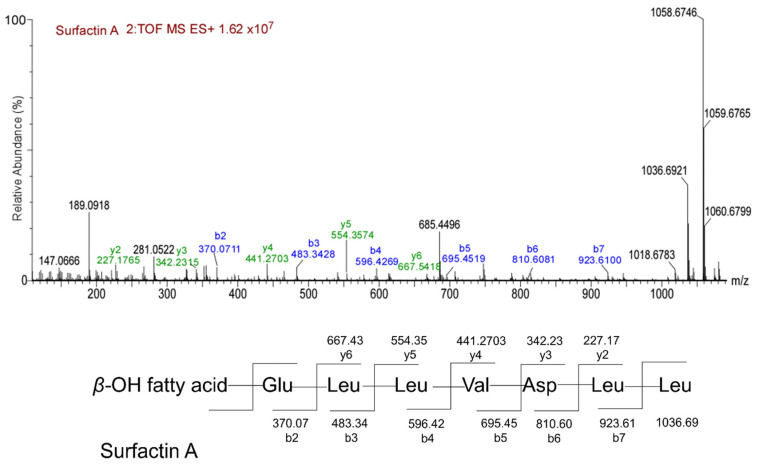
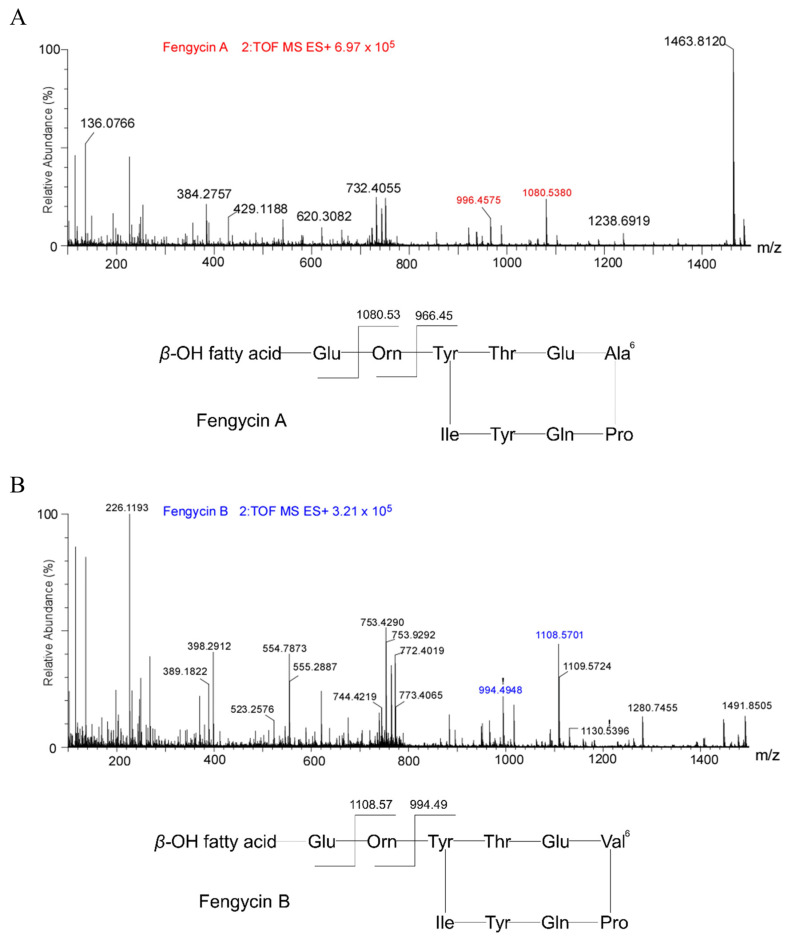
To clarify the types and structures of lipopeptides (LPs) produced by B. velezensis Y6, we extracted LPs from the transparent inhibition zone using acetonitrile/water (1:1) and then analyzed them by UPLC/Q-TOF-MS. According to the mass-to-charge ratio (m/z), we found three types of compounds in the extracted lipopeptides: iturin, fengycin, and surfactin (Figure S1). Each LP has multiple homologues, depending on the length of the fatty acid carbon chain. According to the chromatographic peak retention time, we found that iturin has four homologues (fatty acid carbon chain length C14 to C17), fengycin has four homologues (fatty acid carbon chain length C14 to C15), and surfactin has five homologues (fatty acid carbon chain length C12 to C16) (Figure S2). Iturin, fengycin, and surfactin all have a ring structure composed of amino acid chains and fatty acids. In order to determine the molecular structure of the compounds, the information from their secondary mass spectra was collected and analyzed. Figure 2 is an ion fragment diagram formed by the cleavage of iturin with a mass-to-charge ratio (m/z) of 1043.5551, which mainly includes b-type ion fragments generated from the cleavage of the N-terminus and y-type ion fragments generated from the cleavage of the C-terminus. Starting from the N-terminus, the b-ion fragments were 915.49 (b7), 801.44 (b6), 638.39 (b5), 541.25 (b4), 427.29 (b3), and 340.26 (b2). Assuming that the value of [M+H]+ is 1043.5551, the differences between these values were exactly the masses of Gln, Asn, Tyr, Pro, Asn, Ser, and Asn ion fragments. Starting from the C-terminus, the detectable y-ion fragments were 704.29 (y6), 617.37 (y5), 503.30 (y4), 406.16 (y3), and 243.11 (y2). The differences between their values corresponded to the masses of Ser, Asn, Pro, and Tyr ion fragments, which is consistent with the analysis results of the b-ion fragments. The preliminary amino acid sequence of iturin (1043.5551) was speculated to be β-amino fatty acid-Asn-Ser-Asn-Pro-Tyr-Asn-Gln from the N-terminus to the C-terminus, which belongs to Iturin W in the iturin family. Figure 3 is an ion fragment diagram formed by the cleavage of surfactin with a mass-to-charge ratio (m/z) of 1036.6921, which mainly includes b-type ion fragments generated from the cleavage of the N-terminus and y-type io1036.6921n fragments generated from the cleavage of the C-terminus. Starting from the N-terminus, the b-ion fragments were 923.61 (b7), 810.60 (b6), 695.45 (b5), 596.42 (b4), 483.34 (b3), and 370.07 (b2). Assuming that the value of [M+H]+ is 1036.6921, the differences between these values were exactly the masses of Leu, Leu, Asp, Val, Leu, Leu, and Glu ion fragments. Starting from the C-terminus, the detectable y-ion fragments were 667.43 (y6), 554.35 (y5), 441.27 (y4), 342.23 (y3), and 227.17 (y2). The differences between their values were the masses of Leu, Leu, Val, and Asp ion fragments, which is consistent with the analysis results of the b-ion fragments. The preliminary amino acid sequence of surfactin (1036.6921) was speculated to be β-amino fatty acid- Glu-Leu-Leu-Val-Asp-Leu-Leu from the N-terminus to the C-terminus, which belongs to Surfactin A in the surfactin family. Figure 4 is an ion fragment diagram formed by the cleavage of fengyrin with a mass-to-charge ratio (m/z) of 1463.8120 and 1491.8505. In the primary structure of fengycin, the main difference is the amino acid composition at the sixth position, which is usually Ala or Val. According to the characteristic ion fragment mass-to-charge ratio of 1080.5380 and 996.4575, it is judged that the sixth position of the amino acid chain is Ala, belonging to Fengycin A. According to the characteristic ion fragment mass-to-charge ratios of 1108.5701 and 994.4948, it is judged that the sixth position of the amino acid chain is Val, belonging to Fengycin B. These results show that Fengycin secreted by B. velezensis Y6 contains two structures: Fengycin A and Fengycin B.
Figure 2.
MS/MS analysis of iturin. MS/MS spectra of the iturin produced by B. velezensis Y6 and possible b- and y-type fragments generated from iturin W by collision.
Figure 3.
MS/MS analysis of surfactin. MS/MS spectra of the surfactin produced by B. velezensis Y6 and possible b- and y-type fragments generated from surfactin A by collision.
Figure 4.
MS/MS analysis of fengycin. (A) MS/MS spectra of the peak at m/z of 1463.8120 fengycin produced by B. velezensis Y6 and possible basic structures of Fengycin A; (B) MS/MS spectra of the peak at m/z of 1491.8505 fengycin produced by B. velezensis Y6 and possible basic structures of Fengycin B.
3.3. Antibacterial Activity of B. velezensis Y6 and Its Derived Mutants against Rhizoctonia solani
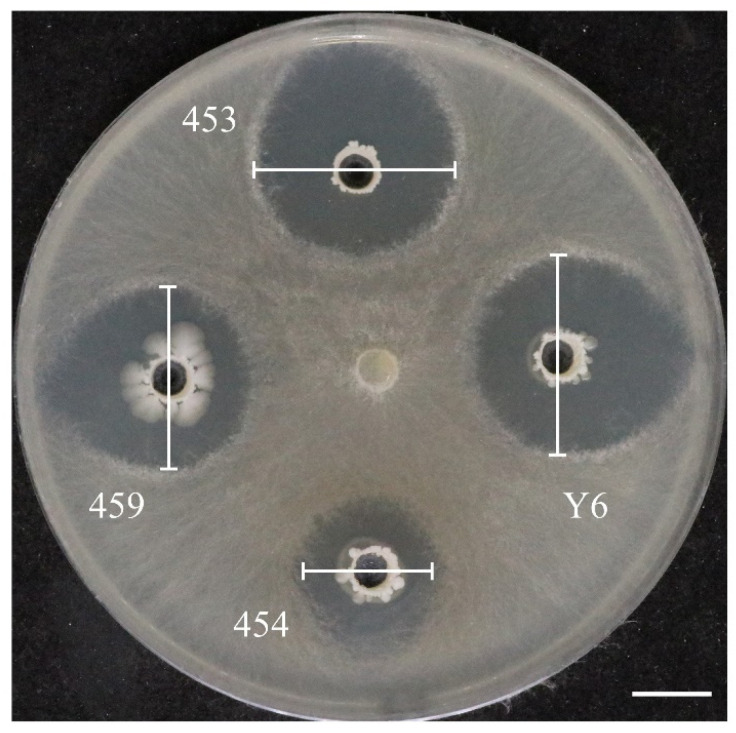
In order to clarify the type of lipopeptides that play a major role in the antagonism of B. velezensis Y6 to rice sheath blight, the effects of the wild-type and three mutants (453 (srfAA::mls), 454 (ituA::mls), and 459 (fenC::spc)) of B. velezensis Y6 on the mycelial growth of Rhizoctonia solani were studied by the plate dual-culture method. The results in Figure 5 show that there were obvious differences in the antibacterial activity of the wild type and mutant strains of B. velezensis Y6 against the Rhizoctonia solani. The diameter of the inhibition zone of the wild type of B. velezensis Y6 was 24.88 ± 0.22 mm. Compared with the wild type of B. velezensis Y6, the inhibition zone diameter of mutant strain 453 (srfAA::mls) was 24.50 ± 0.50 mm, with no significant difference; the inhibition zone diameter of mutant 454 (ituA::mls) was 17.13 ± 0.74 mm, which was reduced by 31.15%, with a significant difference (p < 0.01); the inhibition zone diameter of mutant 459 (fenC::spc) was 21.88 ± 0.65 mm, which was reduced by 12.06%, with a significant difference (p < 0.01) (Table 2). These results indicate that fengycin and iturin are essential for the antagonism of B. velezensis Y6 against the Rhizoctonia solani, among which iturin plays a major role in the antagonism of B. velezensis Y6 against the Rhizoctonia solani.
Figure 5.
The antagonistic activity of strain Y6 and its mutants (453 (srfAA::mls), 454 (ituA::mls), and 459 (fenC::spc)). The white line represents the size of the inhibition circle, which was evaluated after 24 h of incubation at 30 °C. The bar symbolizes 10 mm.
Table 2.
The effects of B. velezensis Y6 and its derived mutants on the mycelial growth of Rhizoctonia solani.
| Strain | Diameter of the Inhibition Zone (mm) |
|---|---|
| WT (Y6) | 24.88 ± 0.22 a |
| 453 (srfAA::mls) | 24.50 ± 0.50 a |
| 454 (ituA::mls) | 17.13 ± 0.74 c |
| 459 (fenC::spc)) | 21.88 ± 0.65 b |
Note: Different letters indicate significant differences.
3.4. Biocontrol Effects of Y6 on Rice Plant Tissues in Pot Experiment
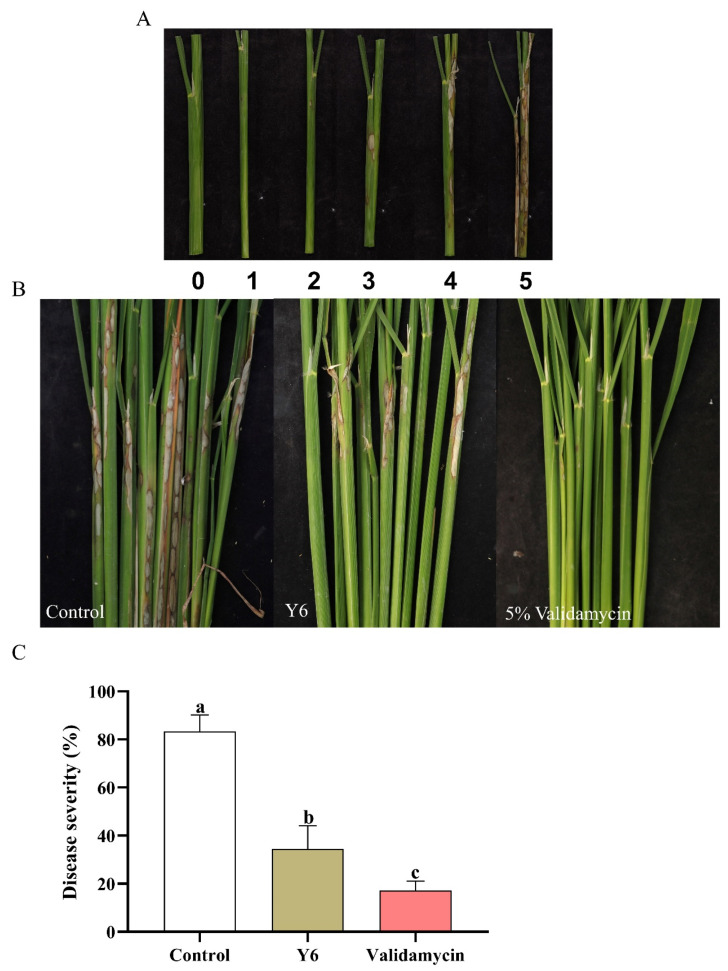
We found that B. velezensis Y6 can effectively inhibit the growth of Rhizoctonia solani. In order to evaluate the effect of B. velezensis Y6 on the biological control of rice sheath blight, we conducted a pot experiment. As shown in Figure 6, the disease severity of rice sheath blight in the control group was 83.33 ± 5.98%, and the disease severity of rice sheath blight in the group treated with 5% Validamycin was 17.22 ± 3.29%. Compared with the control group, the disease severity of rice sheath blight in the group inoculated with B. velezensis Y6 was 34.44 ± 8.39%, and the control efficiency was 58.67% (p < 0.01). The above results show that B. velezensis Y6 can be an effective biological control of rice sheath blight.
Figure 6.
B. velezensis Y6 can reduce the disease severity of rice sheath blight in pot experiments. (A) Photographs of rice sheath blight severity rating values. (B) Photographs of rice sheath blight under different treatments. (C) Disease severity of rice sheath blight. Different letters indicate significant differences.
3.5. B. velezensis Y6 Can Increase Rice Yield in the Field
In general, B. velezensis has a growth-promoting effect on plants. In order to verify the effect of B. velezensis Y6 on rice yield, we conducted pot experiments and field experiments. In the rice pot experiment (Figure S5), we found that after inoculation with B. velezensis Y6, the thousand-grain weight, seed-setting rate, and effective panicle number of rice increased by 5.4%, 22.18% (p < 0.01), and 21.23% (p < 0.01), respectively (Table S2). Through field experiments, we confirmed the effect of B. velezensis Y6 on rice yield (Figure S5). The results showed that, compared with the control group, the panicle length, 1000-grain weight, and rice yield (dry weight) of rice increased by 7.7% (p < 0.05), 5.4%, and 11.75%, respectively, after inoculation with B. velezensis Y6 (Table S3). The above results show that inoculation with B. velezensis Y6 can increase rice field yield.
3.6. Transcriptome Profiling Effects of Strain Y6 on Rice
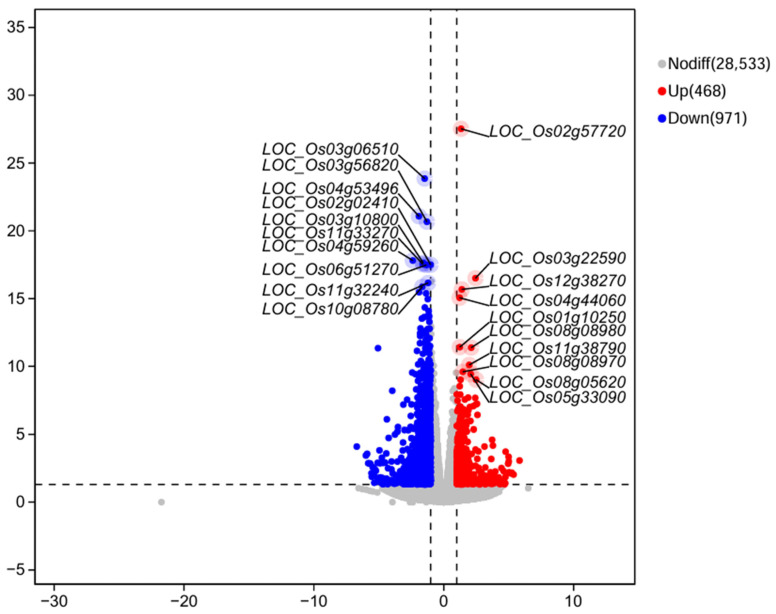
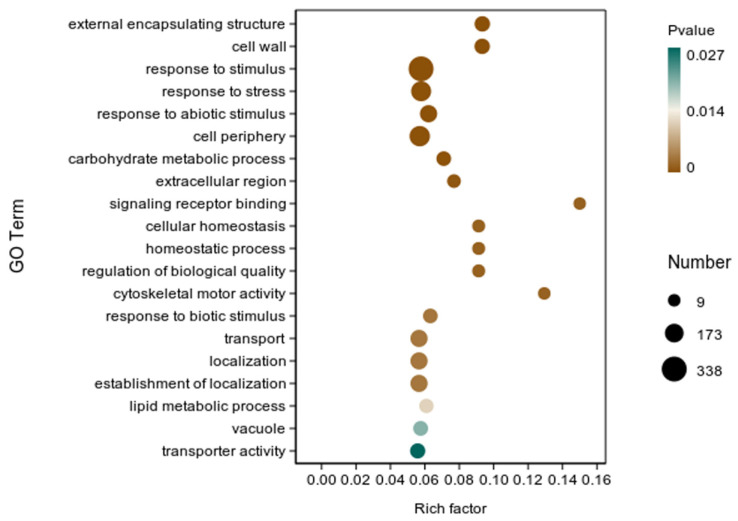
We analyzed the transcriptome of rice roots inoculated with strain Y6. The results showed that 468 genes were up-regulated and 971 genes were down-regulated in rice roots compared with the control (Figure 7). In addition, we also performed enrichment analysis of DEGs in rice using the GO database. A total of 1439 genes were enriched in 167 categories of three major categories in the GO database: molecular function (MF), cellular component (CC), and biological process (BP). The top 20 GO entries with the most significant enrichment were selected in the three broad categories of CC, MF, and BP (Figure S7). The DEGs in CC were mainly enriched in the external encapsulating structure (GO:0030312), cell wall (GO:0005618), cell periphery (GO:0071944), extracellular region (GO:0005576), and vacuole (GO:0005773) categories. In MF, they were mainly enriched in the signaling receptor binding (GO:0005102), cytoskeletal motor activity (GO:0003774), and transporter activity (GO:0005215) categories. In BP, they were mainly enriched in the response to stimulus (GO:0050896), response to stress (GO:0006950), response to abiotic stimulus (GO:0009628), carbohydrate metabolic process (GO:0005975), cellular homeostasis (GO:0019725), homeostatic process (GO:0042592), regulation of biological quality (GO:0065008), response to biotic stimulus (GO:0009607), transport (GO:0006810), localization (GO:0051179), establishment of localization (GO:0051234), and lipid metabolic process (GO:0006629) categories. While in the GO factor graph, DEGs were mainly distributed in BP and were mainly enriched in the response to stimulus (GO:0050896), response to stress (GO:0006950), and response to abiotic stimulus (GO:0009628) categories (Figure 8).
Figure 7.
Volcano plot of differentially expressed genes (DEGs) in the transcriptome of rice inoculated with B. velezensis Y6 and control. Red dots indicate up-regulated genes, and blue dots indicate down-regulated genes.
Figure 8.
GO enrichment analysis of differentially expressed genes (DEGs) in the transcriptome of rice inoculated with strain Y6 and control. The X-axis represents the enrichment factor. The Y-axis represents the GO term name.
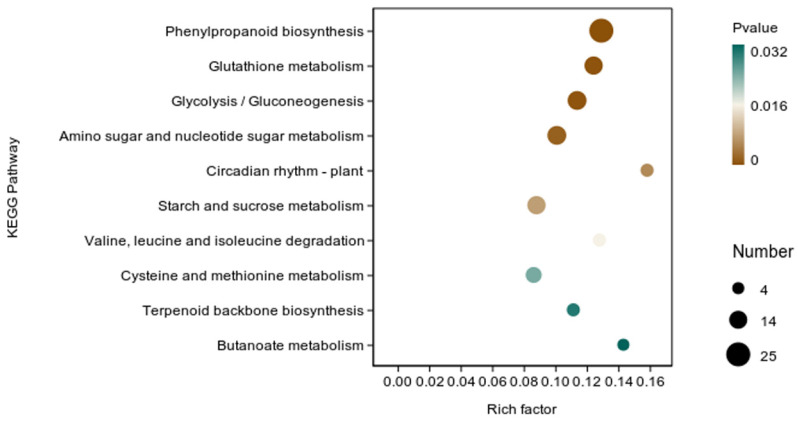
In addition, we performed enrichment analysis of DEGs in rice using the KEGG database. There were 338 DEGs classified into 87 KEGG functional pathway categories. The 10 most significant pathways mainly involved in plant metabolic pathways were phenylpropanoid biosynthesis (map00940), glutathione metabolism (map00480), glycolysis/gluconeogenesis (map00010), amino sugar and nucleotide sugar metabolism (map00520), starch and sucrose metabolism (map00500), valine, leucine and isoleucine degradation (map00280), cysteine and methionine metabolism (map00270), terpenoid backbone biosynthesis (map00900), butanoate metabolism (map00650), and circadian rhythm—plant (map04712) (Figure S8). In addition, a large number of enriched genes were distributed in phenylpropanoid biosynthesis (map00940), glutathione metabolism (map00480), glycolysis/gluconeogenesis (map00010), amino sugar and nucleotide sugar metabolism (map00520) (Figure 9).
Figure 9.
KEGG enrichment analysis of differentially expressed genes (DEGs) in the transcriptome of rice inoculated with B. velezensis Y6 and control. The X-axis represents the enrichment factor. The Y-axis represents the pathway name.
4. Discussion
This study aimed to elucidate the mechanism of B. velezensis Y6 in controlling rice sheath blight. Studies have found that strain Y6 can antagonize rice sheath blight pathogens, in which the lipopeptides play an important role. Pot experiment results show that inoculation with strain Y6 can significantly reduce the disease severity of rice sheath blight.
The antibacterial activity of Bacillus is related to the lipopeptides it produces [21,22]. Bacillus strains can create one or more types of lipopeptides, each of which usually includes numerous homologues with identical amino acid residues and various lengths of fatty acid chains [23,24]. Lipopeptides belonging to B. velezensis Y6 were characterized by UPLC/Q-TOF-MS analysis, and three subfamilies, Surfactin A (C12–C16), Iturin W (C14–C17), Fengycin A and Fengycin B (C16–C17) subfamilies, were found. Zhou et al. reported that Iturin W, a novel lipopeptide produced by the deep-sea bacterium Bacillus sp. strain wsm-1, has stronger antibacterial activity than the common iturin A and has good prospects for biological control of rice blast disease caused by M. grisea [25]. This is the first time that iturin W has been reported in B. velezensis and it plays an important role in the biological control of rice sheath blight by B. velezensis Y6. Surfactin, iturin, and fengycin generally have strong antagonistic activities against pathogens; however, their activities against Rhizoctonia solani were still unclear. By constructing lipopeptide synthesis-related gene deletion mutants (453 (srfAA::mls), 454 (ituA::mls), and 459 (fenC::spc)), we found that fengycin and iturin were required for strain Y6 to antagonize the rice sheath blight pathogen. This was consistent with previous studies reporting that surfactin exhibits significant antibacterial activity and that fengycin and iturin have activities against fungal pathogens [26,27,28]. Our results showed that iturin had stronger antagonistic activity than fengycin against Rhizoctonia solani. Zhang et al. reported that iturins produced by B. velezensis Jt84 play a key role in rice blast disease biocontrol [29].
Transcriptome analysis of the root system of rice pot trials was used in this experiment to further elucidate the biological control mechanism of B. velezensis Y6 against rice sheath blight. According to GO analysis, DEGs in rice roots after inoculation with B. velezensis Y6 mainly involved BP aspects of response to stimulus (GO:0050896), response to stress (GO:0006950), and response to abiotic stimulus (GO:0009628). Hao et al. reported that the DEGs in the maize were enriched in eight biological process terms under drought stress in maize, which were mainly related to stress response (e.g., “response to stimulus”, “response to stress”, “response to abiotic stimulus”, “response to biotic stimulus”, “response to endogenous stimulus”, and “response to external stimulus”) [30]. Bollier et al. reported that most DEGs in tomato response to heat stress belonged to GO classes linked to response to stimulus (GO:0050896), response to stress (GO:0006950) and response to abiotic stimulus (GO:0009628) [31]. Liang et al. reported that the DEGs in rice were enriched in the categories of response to temperature stimulus (GO:0009266), response to abiotic stimulus (GO:0009628), response to heat (GO:0009408), and response to stimulus (GO:0050896) under mild field drought stress during grain-filling stage [32]. Yan et al. reported that the DEGs in tomato treated with B. velezensis YYC were enriched in the categories of defense response (53 DEGs), response to fungus (20 DEGs) and defense response to fungus (19 DEGs) [12]. Usually, stresses such as high temperature, drought, and pathogens will affect the growth of rice and cause a decrease in rice yield [33,34,35]. Our results showed that inoculation with strain Y6 could increase rice yield in the field. The results of this study are consistent with current research, indicating that inoculation with strain Y6 can improve rice resistance to stress.
It has been reported that the phenylpropanoid pathway was crucial in the defense against plant diseases. When the cell wall is invaded by pathogens, the phenylpropanoid pathway is greatly activated, providing lignin-building monolignols, thus forming the first line of defense for plants against diseases [36,37]. Vranic et al. reported that activation of the wheat phenylpropanoid pathway can prevent fungal invasion and spread through the spike, thereby enhancing wheat plant resistance to Fusarium head blight [38]. According to the study by Liu et al., strain YC0136 can enhance systemic resistance in tobacco. After inoculation with strain YC0136, the expression of certain genes involved in the phenylpropanoid metabolic pathway in tobacco roots showed varying degrees of increase compared with tobacco roots that were not inoculated with strain YC0136 [39]. Graham et al. reported that glutathione is a key regulator of plant defense and a crucial player in governing the outcome of biotic stresses [40]. In this study, we found that the DEGs were involved in phenylpropanoid biosynthesis (map00940), glutathione metabolism (map00480), glycolysis/gluconeogenesis (map00010), and amino sugar and nucleotide sugar metabolism (map00520), which indicates that inoculation with B. velezensis Y6 can enhance the metabolic pathways related to plant defense in rice.
5. Conclusions
In this study, we found that B. velezensis Y6 could significantly inhibit the growth of mycelium in Rhizoctonia solani. Lipopeptides play an important role in the control of rice sheath blight by B. velezensis Y6, among which iturin and fengycin are essential, and iturinW, a novel lipopeptide in B. velezensis Y6, plays a major role in lipopeptide antagonism to Rhizoctonia solani. We also found that inoculation with B. velezensis Y6 can increase rice yield in the field. In addition, transcriptome analysis showed that strain Y6 may improve rice resistance to Rhizoctonia solani, mainly by upregulating genes related to response to stress, response to abiotic stimulus, phenolylpropanoid biosynthesis, and glutathione metabolism. This study helps to reveal the molecular mechanism of B. velezensis Y6 in the biological control of rice sheath blight and provides a theoretical basis for the application of B. velezensis Y6 in the biological control of rice sheath blight.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12081694/s1, Table S1: Strains used in this study; Table S2: The effect of B. velezensis Y6 on rice growth; Table S3: Effects of B. velezensis Y6 on rice yield in field; Figure S1: Representative chromatograms of the LPs (iturin, fengycin, and surfactin) from B. velezensis Y6 using UPLC/Q-TOF-MS analysis. Figure S2: UPLC/Q-TOF analysis of crude extract lipopeptide of B. velezensis Y6. [M+H]+ ions of iturin at the m/z of 1043.55088, 1057.56695, 1071.57828, and 1085.59657. Figure S3: UPLC/Q-TOF analysis of crude extract lipopeptide of B. velezensis Y6. [M+H]+ ions of fengycin at the m/z of 1463.79237, 1477.81396, 1491.82280, and 1505.83657. Figure S4: UPLC/Q-TOF analysis of crude extract lipopeptide of B. velezensis Y6. [M+H]+ ions of surfactin at the m/z of 994.63888, 1008.65622, 1022.67069, 1036.68825, and 1050.70099. Figure S5: Photographs of a pot experiment on the effect of B. velezensis Y6 on rice growth. Figure S6: Photographs of field experiment on the effect of B. velezensis Y6 on rice yield. Figure S7: Top 20 pathways with the most significant enrichment in GO term in the transcriptome of rice inoculated with B. velezensis Y6 and control. The X-axis is GO term, indicating the number and percentage of DEGs under each functional classification; the Y-axis is GO term enrichment −log10 (p value), indicating the enriched GO functional classification, which is divided into three categories: cellular components (CC), molecular functions (MF), and biological processes (BP). Figure S8: Top 10 pathways with the most significant enrichment in KEGG pathway in the transcriptome of rice inoculated with B. velezensis Y6 and control. The X-axis is pathway, indicating the number and percentage of DEGs under each functional classification; the Y-axis is pathway enrichment −log10 (p value), indicating the enriched KEGG pathway functional classification, which is divided into metabolism and organismal systems.
Author Contributions
Conceptualization, H.T. and A.C.; methodology, H.T. and Y.C.; software, X.L.; validation, H.T. and X.L.; formal analysis, H.T., X.L. and H.H.; investigation, H.T., X.L. and H.H.; resources, H.T. and X.L; data curation, H.T., X.L. and H.H.; writing—original draft preparation, H.T.; writing—review and editing, A.C. and Y.C.; visualization, X.L.; supervision, X.L. and H.H.; project administration, A.C. and Y.C.; funding acquisition, A.C. and Y.C. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1136222.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This study was supported by the National Natural Science Foundation of China (41977035) and the Fundamental Research Fund of Guangxi Institute of Botany (GZY22001).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Bernardes-de-Assis J., Storari M., Zala M., Wang W., Jiang D., Shidong L., Jin M., McDonald B.A., Ceresini P.C. Genetic structure of populations of the rice-infecting pathogen Rhizoctonia solani AG-1 IA from China. Phytopathology. 2009;99:1090–1099. doi: 10.1094/PHYTO-99-9-1090. [DOI] [PubMed] [Google Scholar]
- 2.Yu Y.Y., Jiang C.H., Wang C., Chen L.J., Li H.Y., Xu Q., Guo J.H. An improved strategy for stable biocontrol agents selecting to control rice sheath blight caused by Rhizoctonia solani. Microbiol. Res. 2017;203:1–9. doi: 10.1016/j.micres.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Deng S., Guo Q., Gao Y., Li J., Xu Z. Induced resistance to rice sheath blight (Rhizoctonia solani Kuhn) by beta-amino-butyric acid conjugate of phenazine-1-carboxylic acid. Pestic Biochem. Physiol. 2023;194:105502. doi: 10.1016/j.pestbp.2023.105502. [DOI] [PubMed] [Google Scholar]
- 4.Park D.S., Sayler R.J., Hong Y.G., Nam M.H., Yang Y. A Method for Inoculation and Evaluation of Rice Sheath Blight Disease. Plant Dis. 2008;92:25–29. doi: 10.1094/PDIS-92-1-0025. [DOI] [PubMed] [Google Scholar]
- 5.Senapati M., Tiwari A., Sharma N., Chandra P., Bashyal B.M., Ellur R.K., Bhowmick P.K., Bollinedi H., Vinod K.K., Singh A.K., et al. Rhizoctonia solani Kuhn Pathophysiology: Status and Prospects of Sheath Blight Disease Management in Rice. Front. Plant Sci. 2022;13:881116. doi: 10.3389/fpls.2022.881116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akash Datta A.D., Vurukonda S.S.K.P. Rice Sheath Blight: A Review of The Unsung Fatal Disease. Trends Biosci. 2017;10:9216–9219. [Google Scholar]
- 7.Singh R., Singh B., Singh A., Kureel R. Management of sheath blight in rice through application of Validamycin, Trichoderma harzianum and Pseudomonas fluorescence. J. Appl. Nat. Sci. 2010;2:121–125. doi: 10.31018/jans.v2i1.110. [DOI] [Google Scholar]
- 8.Meena K.R., Kanwar S.S. Lipopeptides as the antifungal and antibacterial agents: Applications in food safety and therapeutics. Biomed Res. Int. 2015;2015:473050. doi: 10.1155/2015/473050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veras F.F., Silveira R.D., Welke J.E. Bacillus spp. as a strategy to control fungi and mycotoxins in food. Curr. Opin. Food Sci. 2023;52:101068. doi: 10.1016/j.cofs.2023.101068. [DOI] [Google Scholar]
- 10.Li B., Li Q., Xu Z., Zhang N., Shen Q., Zhang R. Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production. Front. Microbiol. 2014;5:636. doi: 10.3389/fmicb.2014.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etesami H., Jeong B.R., Glick B.R. Biocontrol of plant diseases by Bacillus spp. Physiol. Mol. Plant Pathol. 2023;126:102048. doi: 10.1016/j.pmpp.2023.102048. [DOI] [Google Scholar]
- 12.Yan Y., Xu W., Hu Y., Tian R., Wang Z. Bacillus velezensis YYC promotes tomato growth and induces resistance against bacterial wilt. Biol. Control. 2022;172:104977. doi: 10.1016/j.biocontrol.2022.104977. [DOI] [Google Scholar]
- 13.Zhang S., Wu J., Chen J., Jun S., Yuan Y., Dai X., Wang F., Ma Y. The biological control effect of Bacillus cereus on strawberry leaf spot disease caused by Neopestalotiopsis clavispora. Sci. Hortic. 2024;327:112841. doi: 10.1016/j.scienta.2024.112841. [DOI] [Google Scholar]
- 14.Nie P., Li X., Wang S., Guo J., Zhao H., Niu D. Induced Systemic Resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-Dependent Signaling Pathway and Activates PAMP-Triggered Immunity in Arabidopsis. Front. Plant Sci. 2017;8:238. doi: 10.3389/fpls.2017.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad B., Sharma D., Kumar P., Chandra Dubey R. Biocontrol potential of Bacillus spp. for resilient and sustainable agricultural systems. Physiol. Mol. Plant Pathol. 2023;128:102173. doi: 10.1016/j.pmpp.2023.102173. [DOI] [Google Scholar]
- 16.Han V.-C., Michael P.J., Swift B., Bennett S.J. Biological control of Sclerotinia sclerotiorum: Modes of action of biocontrol agents, soil organic amendments, and soil microbiome manipulation. Biol. Control. 2023;186:105346. doi: 10.1016/j.biocontrol.2023.105346. [DOI] [Google Scholar]
- 17.Feng R.Y., Chen Y.H., Lin C., Tsai C.H., Yang Y.L., Chen Y.L. Surfactin secreted by Bacillus amyloliquefaciens Ba01 is required to combat Streptomyces scabies causing potato common scab. Front. Plant Sci. 2022;13:998707. doi: 10.3389/fpls.2022.998707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Liang J., Zhang C., Wang L., Gao W., Jiang J. Bacillus megaterium WL-3 Lipopeptides Collaborate Against Phytophthora infestans to Control Potato Late Blight and Promote Potato Plant Growth. Front. Microbiol. 2020;11:1602. doi: 10.3389/fmicb.2020.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y., Pi H., Chandrangsu P., Li Y., Wang Y., Zhou H., Xiong H., Helmann J.D., Cai Y. Antagonism of Two Plant-Growth Promoting Bacillus velezensis Isolates Against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018;8:4360. doi: 10.1038/s41598-018-22782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao H., Wang S., Li X., Li X., Cai J., Zhao L., Wang J., Zeng J., Qin Y., Xiong X., et al. Biological control of potato common scab and growth promotion of potato by Bacillus velezensis Y6. Front. Microbiol. 2023;14:1295107. doi: 10.3389/fmicb.2023.1295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H., Shao D., Jiang C., Shi J., Li Q., Huang Q., Rajoka M.S.R., Yang H., Jin M. Biological activity of lipopeptides from Bacillus. Appl. Microbiol. Biotechnol. 2017;101:5951–5960. doi: 10.1007/s00253-017-8396-0. [DOI] [PubMed] [Google Scholar]
- 22.Ongena M., Jacques P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Roongsawang N., Washio K., Morikawa M. Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int. J. Mol. Sci. 2010;12:141–172. doi: 10.3390/ijms12010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sani A., Qin W.Q., Li J.Y., Liu Y.F., Zhou L., Yang S.Z., Mu B.Z. Structural diversity and applications of lipopeptide biosurfactants as biocontrol agents against phytopathogens: A review. Microbiol. Res. 2024;278:127518. doi: 10.1016/j.micres.2023.127518. [DOI] [PubMed] [Google Scholar]
- 25.Zhou S., Liu G., Zheng R., Sun C., Wu S. Structural and Functional Insights into Iturin W, a Novel Lipopeptide Produced by the Deep-Sea Bacterium Bacillus sp. Strain wsm-1. Appl. Environ. Microbiol. 2020;86:e01597-20. doi: 10.1128/AEM.01597-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeriouh H., de Vicente A., Perez-Garcia A., Romero D. Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 2014;16:2196–2211. doi: 10.1111/1462-2920.12271. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L., Sun C. Fengycins, Cyclic Lipopeptides from Marine Bacillus subtilis Strains, Kill the Plant-Pathogenic Fungus Magnaporthe grisea by Inducing Reactive Oxygen Species Production and Chromatin Condensation. Appl. Environ. Microbiol. 2018;84:e00445-18. doi: 10.1128/AEM.00445-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H., Li X., Li X., Yu H., Shen Z. Identification of lipopeptide isoforms by MALDI-TOF-MS/MS based on the simultaneous purification of iturin, fengycin, and surfactin by RP-HPLC. Anal. Bioanal. Chem. 2015;407:2529–2542. doi: 10.1007/s00216-015-8486-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhang R.-S., Wang F.-G., Qi Z.-Q., Qiao J.-Q., Du Y., Yu J.-J., Yu M.-N., Liang D., Song T.-Q., Yan P.-X., et al. Iturins produced by Bacillus velezensis Jt84 play a key role in the biocontrol of rice blast disease. Biol. Control. 2022;174:105001. doi: 10.1016/j.biocontrol.2022.105001. [DOI] [Google Scholar]
- 30.Hao L.-y., Liu X.-y., Zhang X.-j., Sun B.-c., Liu C., Zhang D.-f., Tang H.-j., Li C.-h., Li Y.-x., Shi Y.-s., et al. Genome-wide identification and comparative analysis of drought related genes in roots of two maize inbred lines with contrasting drought tolerance by RNA sequencing. J. Integr. Agric. 2020;19:449–464. doi: 10.1016/S2095-3119(19)62660-2. [DOI] [Google Scholar]
- 31.Bollier N., Micol-Ponce R., Dakdaki A., Maza E., Zouine M., Djari A., Bouzayen M., Chevalier C., Delmas F., Gonzalez N., et al. Various tomato cultivars display contrasting morphological and molecular responses to a chronic heat stress. Front. Plant Sci. 2023;14:1278608. doi: 10.3389/fpls.2023.1278608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Y., Tabien R.E., Tarpley L., Mohammed A.R., Septiningsih E.M. Transcriptome profiling of two rice genotypes under mild field drought stress during grain-filling stage. AoB Plants. 2021;13:plab043. doi: 10.1093/aobpla/plab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaturvedi A.K., Bahuguna R.N., Shah D., Pal M., Jagadish S.V.K. High temperature stress during flowering and grain filling offsets beneficial impact of elevated CO2 on assimilate partitioning and sink-strength in rice. Sci. Rep. 2017;7:8227. doi: 10.1038/s41598-017-07464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nahar L., Aycan M., Hanamata S., Baslam M., Mitsui T. Impact of Single and Combined Salinity and High-Temperature Stresses on Agro-Physiological, Biochemical, and Transcriptional Responses in Rice and Stress-Release. Plants. 2022;11:501. doi: 10.3390/plants11040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngalimat M.S., Mohd Hata E., Zulperi D., Ismail S.I., Ismail M.R., Mohd Zainudin N.A.I., Saidi N.B., Yusof M.T. Plant Growth-Promoting Bacteria as an Emerging Tool to Manage Bacterial Rice Pathogens. Microorganisms. 2021;9:682. doi: 10.3390/microorganisms9040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yadav V., Wang Z., Wei C., Amo A., Ahmed B., Yang X., Zhang X. Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense. Pathogens. 2020;9:312. doi: 10.3390/pathogens9040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen O., Deng L., Ruan C., Yi L., Zeng K. Pichia galeiformis Induces Resistance in Postharvest Citrus by Activating the Phenylpropanoid Biosynthesis Pathway. J. Agric. Food Chem. 2021;69:2619–2631. doi: 10.1021/acs.jafc.0c06283. [DOI] [PubMed] [Google Scholar]
- 38.Vranic M., Perochon A., Doohan F.M. Transcriptional Profiling Reveals the Wheat Defences against Fusarium Head Blight Disease Regulated by a NAC Transcription Factor. Plants. 2023;12:2708. doi: 10.3390/plants12142708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H., Wang J., Sun H., Han X., Peng Y., Liu J., Liu K., Ding Y., Wang C., Du B. Transcriptome Profiles Reveal the Growth-Promoting Mechanisms of Paenibacillus polymyxa YC0136 on Tobacco (Nicotiana tabacum L.) Front. Microbiol. 2020;11:584174. doi: 10.3389/fmicb.2020.584174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noctor G., Cohen M., Trémulot L., Châtel-Innocenti G., Van Breusegem F., Mhamdi A. Glutathione: A key modulator of plant defence and metabolism through multiple mechanisms. J. Exp. Bot. 2024;75:4549–4572. doi: 10.1093/jxb/erae194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1136222.