Abstract
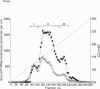
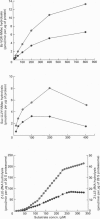
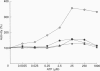
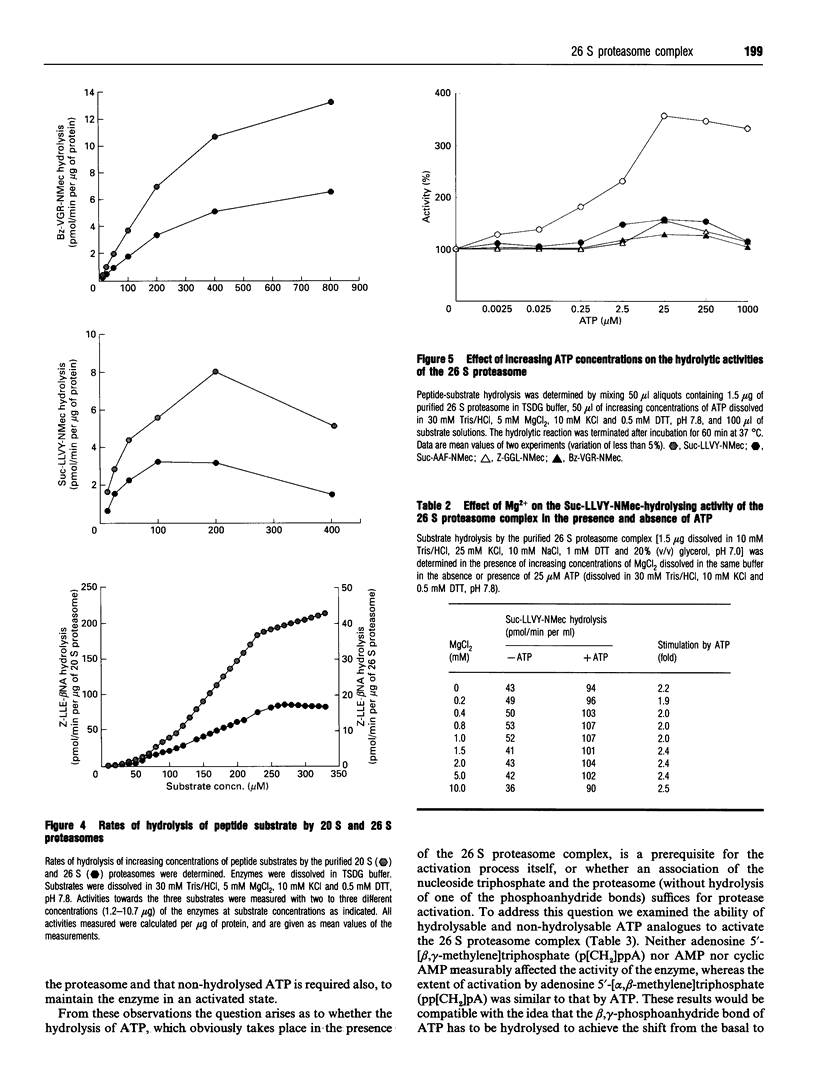
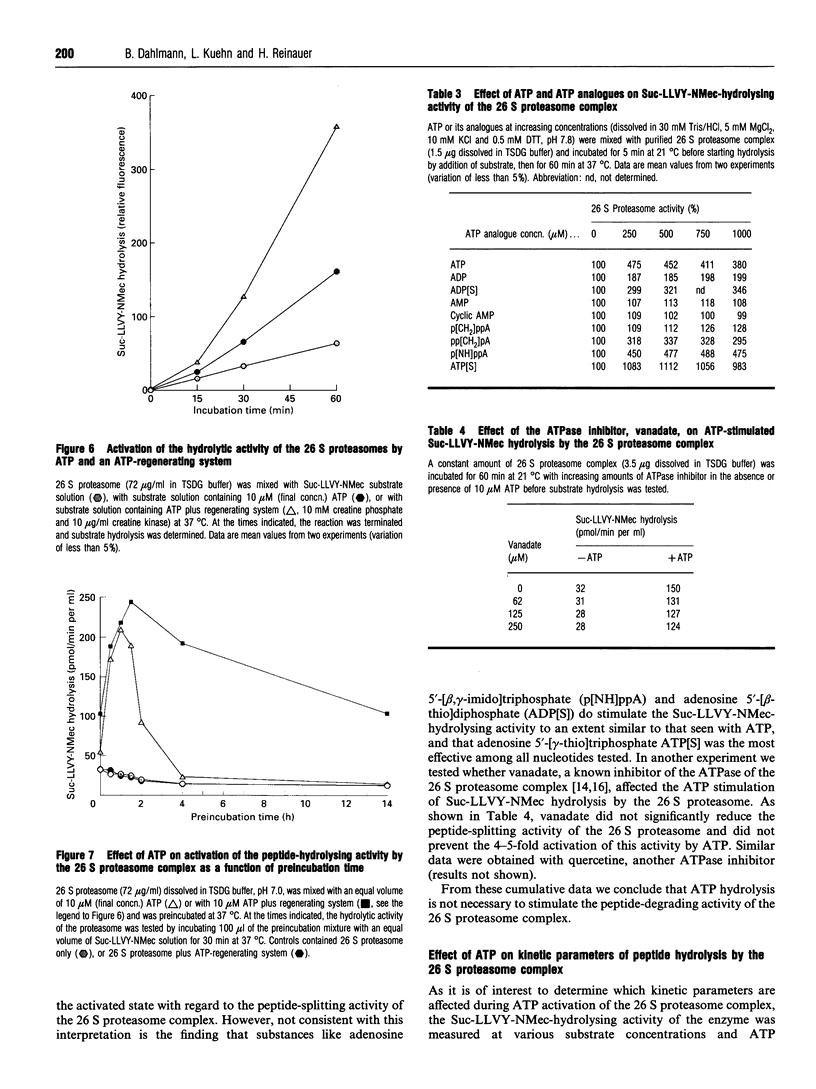
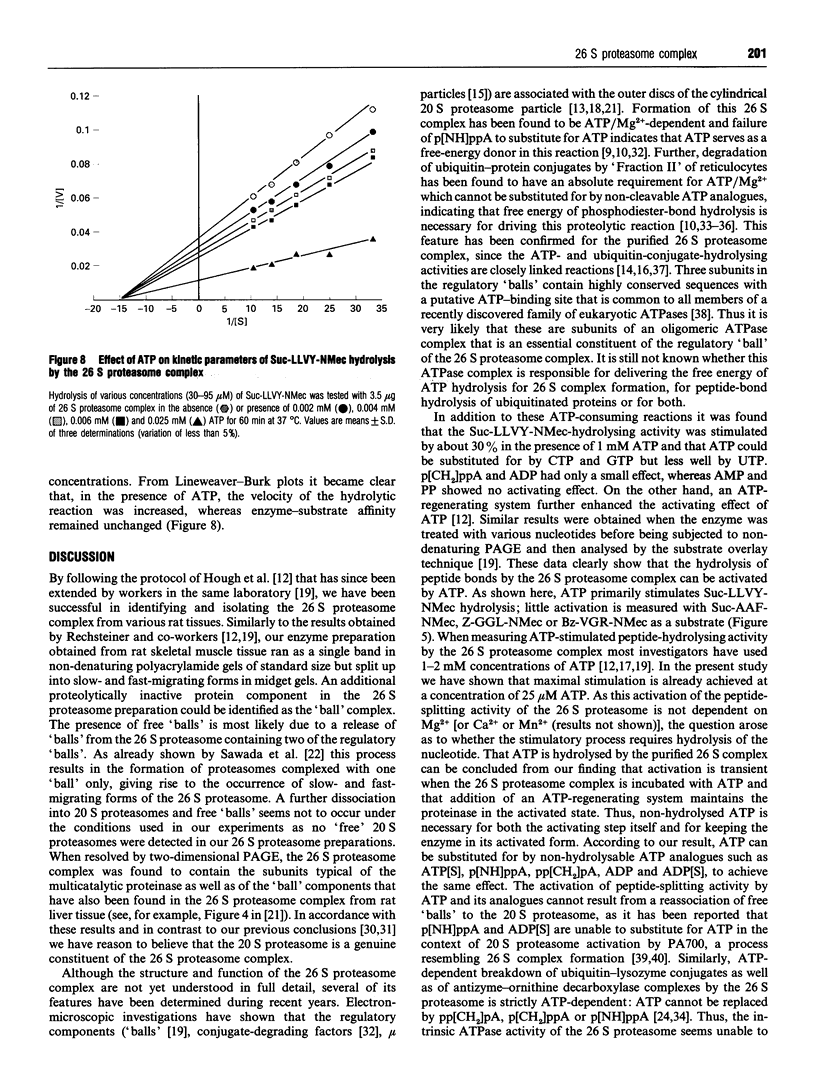
The 26 S proteasome complex is thought to catalyse the breakdown of ubiquitinated proteins within eukaryotic cells. In addition it has been found that the complex also degrades short-lived proteins such as ornithine decarboxylase in a ubiquitin-independent manner. Both proteolytic processes are paralleled by the hydrolysis of ATP. Here we show that ATP also affects the hydrolytic activity towards fluorigenic peptide substrates by the 26 S proteasome complex from rat skeletal muscle tissue. Low concentrations of ATP (about 25 microM) optimally activate the so-called chymotryptic and tryptic activity by increasing the rate of peptide hydrolysis but not peptidylglutamylpeptide hydrolysis. Activation of the enzyme by ATP is transient but this effect can be enhanced and prolonged by including in the assay an ATP-regenerating system, indicating that ATP is hydrolysed by the 26 S proteasome complex. Although ATP cannot be substituted for by adenosine 5'-[beta,gamma-methylene]triphosphate or AMP, hydrolysis of the phosphoanhydride bond of ATP seems not to be necessary for the activation process of the proteasome complex, a conclusion drawn from the findings that ATP analogues such as adenosine 5'-[beta,gamma-imido]triphosphate, adenosine 5'-O-[gamma-thio]triphosphate, adenosine 5'-O-[beta-thio]-diphosphate and adenosine 5'-[alpha,beta-methylene]triphosphate give the same effect as ATP, and vanadate does not prevent ATP activation. These effects are independent of the presence of Mg2+. Thus, ATP and other nucleotides may act as allosteric activators of peptide-hydrolysing activities of the 26 S proteasome complex as has also been found with the lon protease from Escherichia coli.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armon T., Ganoth D., Hershko A. Assembly of the 26 S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J Biol Chem. 1990 Dec 5;265(34):20723–20726. [PubMed] [Google Scholar]
- Chu-Ping M., Vu J. H., Proske R. J., Slaughter C. A., DeMartino G. N. Identification, purification, and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20 S proteasome. J Biol Chem. 1994 Feb 4;269(5):3539–3547. [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A. L. The ubiquitin-mediated proteolytic pathway: mechanisms of recognition of the proteolytic substrate and involvement in the degradation of native cellular proteins. FASEB J. 1994 Feb;8(2):182–191. doi: 10.1096/fasebj.8.2.8119489. [DOI] [PubMed] [Google Scholar]
- Dahlmann B., Kopp F., Kuehn L., Niedel B., Pfeifer G., Hegerl R., Baumeister W. The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Lett. 1989 Jul 17;251(1-2):125–131. doi: 10.1016/0014-5793(89)81441-3. [DOI] [PubMed] [Google Scholar]
- Dahlmann B., Kuehn L., Heinrich P. C., Kirschke H., Wiederanders B. ATP-activated, high-molecular-mass proteinase-I from rat skeletal muscle is a cysteine proteinase-alpha 1-macroglobulin complex. Biochim Biophys Acta. 1989 May 31;991(2):253–262. doi: 10.1016/0304-4165(89)90113-x. [DOI] [PubMed] [Google Scholar]
- Dahlmann B., Kuehn L., Rutschmann M., Reinauer H. Purification and characterization of a multicatalytic high-molecular-mass proteinase from rat skeletal muscle. Biochem J. 1985 May 15;228(1):161–170. doi: 10.1042/bj2280161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino G. N., McCullough M. L., Reckelhoff J. F., Croall D. E., Ciechanover A., McGuire M. J. ATP-stimulated degradation of endogenous proteins in cell-free extracts of BHK 21/C13 fibroblasts. A key role for the proteinase, macropain, in the ubiquitin-dependent degradation of short-lived proteins. Biochim Biophys Acta. 1991 Mar 4;1073(2):299–308. doi: 10.1016/0304-4165(91)90135-4. [DOI] [PubMed] [Google Scholar]
- DeMartino G. N., Moomaw C. R., Zagnitko O. P., Proske R. J., Chu-Ping M., Afendis S. J., Swaffield J. C., Slaughter C. A. PA700, an ATP-dependent activator of the 20 S proteasome, is an ATPase containing multiple members of a nucleotide-binding protein family. J Biol Chem. 1994 Aug 19;269(33):20878–20884. [PubMed] [Google Scholar]
- Driscoll J., Goldberg A. L. The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J Biol Chem. 1990 Mar 25;265(9):4789–4792. [PubMed] [Google Scholar]
- Dubiel W., Ferrell K., Rechsteiner M. Tat-binding protein 7 is a subunit of the 26S protease. Biol Chem Hoppe Seyler. 1994 Apr;375(4):237–240. doi: 10.1515/bchm3.1994.375.4.237. [DOI] [PubMed] [Google Scholar]
- Etlinger J. D., Goldberg A. L. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci U S A. 1977 Jan;74(1):54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eytan E., Ganoth D., Armon T., Hershko A. ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7751–7755. doi: 10.1073/pnas.86.20.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoth D., Leshinsky E., Eytan E., Hershko A. A multicomponent system that degrades proteins conjugated to ubiquitin. Resolution of factors and evidence for ATP-dependent complex formation. J Biol Chem. 1988 Sep 5;263(25):12412–12419. [PubMed] [Google Scholar]
- Goldberg A. L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992 Jan 15;203(1-2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., Waxman L. The role of ATP hydrolysis in the breakdown of proteins and peptides by protease La from Escherichia coli. J Biol Chem. 1985 Oct 5;260(22):12029–12034. [PubMed] [Google Scholar]
- Haas A. L., Rose I. A. Hemin inhibits ATP-dependent ubiquitin-dependent proteolysis: role of hemin in regulating ubiquitin conjugate degradation. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6845–6848. doi: 10.1073/pnas.78.11.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Heller H., Haas A. L., Rose I. A. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hershko A., Leshinsky E., Ganoth D., Heller H. ATP-dependent degradation of ubiquitin-protein conjugates. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1619–1623. doi: 10.1073/pnas.81.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L., Pratt G., Rechsteiner M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J Biol Chem. 1992 Nov 5;267(31):22362–22368. [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987 Jun 15;262(17):8303–8313. [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J Biol Chem. 1986 Feb 15;261(5):2400–2408. [PubMed] [Google Scholar]
- Ikai A., Nishigai M., Tanaka K., Ichihara A. Electron microscopy of 26 S complex containing 20 S proteasome. FEBS Lett. 1991 Nov 4;292(1-2):21–24. doi: 10.1016/0014-5793(91)80824-m. [DOI] [PubMed] [Google Scholar]
- Johnston N. L., Cohen R. E. Uncoupling ubiquitin-protein conjugation from ubiquitin-dependent proteolysis by use of beta, gamma-nonhydrolyzable ATP analogues. Biochemistry. 1991 Jul 30;30(30):7514–7522. doi: 10.1021/bi00244a021. [DOI] [PubMed] [Google Scholar]
- Kanayama H. O., Tamura T., Ugai S., Kagawa S., Tanahashi N., Yoshimura T., Tanaka K., Ichihara A. Demonstration that a human 26S proteolytic complex consists of a proteasome and multiple associated protein components and hydrolyzes ATP and ubiquitin-ligated proteins by closely linked mechanisms. Eur J Biochem. 1992 Jun 1;206(2):567–578. doi: 10.1111/j.1432-1033.1992.tb16961.x. [DOI] [PubMed] [Google Scholar]
- Kuehn L., Dahlmann B., Reinauer H. Evidence indicating that the multicatalytic proteinase of rabbit reticulocytes is not incorporated as a core enzyme into a 26 S proteinase complex. Arch Biochem Biophys. 1992 May 15;295(1):55–60. doi: 10.1016/0003-9861(92)90487-h. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee D. H., Kim S. S., Kim K. I., Ahn J. Y., Shim K. S., Nishigai M., Ikai A., Tamura T., Tanaka K., Ichihara A. Structure and properties of the 26S protease complex from chick skeletal muscle. Biochem Mol Biol Int. 1993 May;30(1):121–130. [PubMed] [Google Scholar]
- Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992 Dec 10;360(6404):597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Peters J. M., Cejka Z., Harris J. R., Kleinschmidt J. A., Baumeister W. Structural features of the 26 S proteasome complex. J Mol Biol. 1993 Dec 20;234(4):932–937. doi: 10.1006/jmbi.1993.1646. [DOI] [PubMed] [Google Scholar]
- Peters J. M., Harris J. R., Kleinschmidt J. A. Ultrastructure of the approximately 26S complex containing the approximately 20S cylinder particle (multicatalytic proteinase/proteasome). Eur J Cell Biol. 1991 Dec;56(2):422–432. [PubMed] [Google Scholar]
- Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993 Mar 25;268(9):6065–6068. [PubMed] [Google Scholar]
- SIMPSON M. V. The release of labeled amino acids from the proteins of rat liver slices. J Biol Chem. 1953 Mar;201(1):143–154. [PubMed] [Google Scholar]
- STEINBERG D., VAUGHAN M. Observations on intracellular protein catabolism studied in vitro. Arch Biochem Biophys. 1956 Nov;65(1):93–105. doi: 10.1016/0003-9861(56)90180-1. [DOI] [PubMed] [Google Scholar]
- Sawada H., Muto K., Fujimuro M., Akaishi T., Sawada M. T., Yokosawa H., Goldberg A. L. Different ratios in 20 S proteasomes and regulatory subunit complexes in two isoforms of the 26 S proteasome purified from rabbit skeletal muscle. FEBS Lett. 1993 Dec 6;335(2):207–212. doi: 10.1016/0014-5793(93)80731-9. [DOI] [PubMed] [Google Scholar]
- Seelig A., Kloetzel P. M., Kuehn L., Dahlmann B. Molecular interaction of the proteasome (multicatalytic proteinase). Evidence that the proteasome is not a constituent of the '26 S' multienzyme complex. Biochem J. 1991 Nov 15;280(Pt 1):225–232. doi: 10.1042/bj2800225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A. Purification and characterization of a multiprotein component of the Drosophila 26 S (1500 kDa) proteolytic complex. J Biol Chem. 1993 Apr 25;268(12):9055–9062. [PubMed] [Google Scholar]
- Ugai S., Tamura T., Tanahashi N., Takai S., Komi N., Chung C. H., Tanaka K., Ichihara A. Purification and characterization of the 26S proteasome complex catalyzing ATP-dependent breakdown of ubiquitin-ligated proteins from rat liver. J Biochem. 1993 Jun;113(6):754–768. doi: 10.1093/oxfordjournals.jbchem.a124116. [DOI] [PubMed] [Google Scholar]
- Umaña C. R. Protein degradation at neutral pH. Possible enzymic and control mechanisms. Proc Soc Exp Biol Med. 1971 Oct;138(1):31–38. [PubMed] [Google Scholar]
- Waxman L., Goldberg A. L. Selectivity of intracellular proteolysis: protein substrates activate the ATP-dependent protease (La). Science. 1986 Apr 25;232(4749):500–503. doi: 10.1126/science.2938257. [DOI] [PubMed] [Google Scholar]
- Wilkinson K. D., Urban M. K., Haas A. L. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J Biol Chem. 1980 Aug 25;255(16):7529–7532. [PubMed] [Google Scholar]
- Woo K. M., Chung W. J., Ha D. B., Goldberg A. L., Chung C. H. Protease Ti from Escherichia coli requires ATP hydrolysis for protein breakdown but not for hydrolysis of small peptides. J Biol Chem. 1989 Feb 5;264(4):2088–2091. [PubMed] [Google Scholar]
- Yoshimura T., Kameyama K., Takagi T., Ikai A., Tokunaga F., Koide T., Tanahashi N., Tamura T., Cejka Z., Baumeister W. Molecular characterization of the "26S" proteasome complex from rat liver. J Struct Biol. 1993 Nov-Dec;111(3):200–211. doi: 10.1006/jsbi.1993.1050. [DOI] [PubMed] [Google Scholar]