Abstract
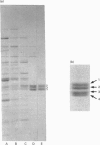
Bovine NAD(+)-dependent isocitrate dehydrogenase was shown previously to contain four subunits of approx. 40 kDa (subunits 1-4) possessing different peptide maps and electrophoretic properties [Rushbrook and Harvey (1978) Biochemistry 17, 5339-5346]. In this study the heterogeneity is confirmed using enzyme purified by updated methods and from single animals, ruling out allelic variability. Subunits 1 and 2 were differentiated from each other and from subunits 3 and 4 by N-terminal amino acid sequencing. Subunits 3 and 4 (subunits 3/4) were identical in sequence over 30 residues. The N-terminal residues of subunits 1 and 2 were homologous but not identical with the beta- and gamma-subunits respectively of the comparable pig heart enzyme. Subunits 3/4 were identical over 30 residues with the N-terminus of the pig heart alpha-subunit. Full-length sequence, including that for mitochondrial import, is presented for a protein with the processed N-terminus of subunits 3/4, deduced from cloned cDNA obtained utilizing the N-terminal sequence information. The derived amino acid sequence for the mature protein contains 339 amino acids and has a molecular mass of 36,685 Da. Complete identity with N-terminal and Cys-containing peptides totalling 92 residues from the alpha-subunit of the pig heart enzyme [Huang and Colman (1990) Biochemistry 29, 8266-8273] suggests that maintenance of a particular three-dimensional structure in this subunit is crucial to the function of the enzyme. An electrophoretic heterogeneity within the pig heart alpha-subunit, similar to that shown by bovine subunits 3/4, was demonstrated. One reordering of the Cys-containing peptides of the pig heart alpha-subunit is indicated. Sequence comparison with the distantly related NADP(+)-dependent enzyme from Escherichia coli, for which the three-dimensional structure is known [Stoddard, Dean and Koshland (1993) Biochemistry 32, 9310-9316] shows strong conservation of residues binding isocitrate, Mg2+ and the NAD+ moiety of NADP+, consistent with a catalytic function.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes L. D., Kuehn G. D., Atkinson D. E. Yeast diphosphopyridine nucleotide specific isocitrate dehydrogenase. Purification and some properties. Biochemistry. 1971 Oct 12;10(21):3939–3944. doi: 10.1021/bi00797a022. [DOI] [PubMed] [Google Scholar]
- CHEN R. F., PLAUT G. W. ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES. Biochemistry. 1963 Sep-Oct;2:1023–1032. doi: 10.1021/bi00905a020. [DOI] [PubMed] [Google Scholar]
- Cohen P. F., Colman R. F. Purification of NAD-specific isocitrate dehydrogenase from porcine heart. Biochim Biophys Acta. 1971 Aug 20;242(2):325–330. doi: 10.1016/0005-2744(71)90224-5. [DOI] [PubMed] [Google Scholar]
- Cupp J. R., McAlister-Henn L. Cloning and characterization of the gene encoding the IDH1 subunit of NAD(+)-dependent isocitrate dehydrogenase from Saccharomyces cerevisiae. J Biol Chem. 1992 Aug 15;267(23):16417–16423. [PubMed] [Google Scholar]
- Cupp J. R., McAlister-Henn L. Kinetic analysis of NAD(+)-isocitrate dehydrogenase with altered isocitrate binding sites: contribution of IDH1 and IDH2 subunits to regulation and catalysis. Biochemistry. 1993 Sep 14;32(36):9323–9328. doi: 10.1021/bi00087a010. [DOI] [PubMed] [Google Scholar]
- Cupp J. R., McAlister-Henn L. NAD(+)-dependent isocitrate dehydrogenase. Cloning, nucleotide sequence, and disruption of the IDH2 gene from Saccharomyces cerevisiae. J Biol Chem. 1991 Nov 25;266(33):22199–22205. [PubMed] [Google Scholar]
- Dean A. M., Lee M. H., Koshland D. E., Jr Phosphorylation inactivates Escherichia coli isocitrate dehydrogenase by preventing isocitrate binding. J Biol Chem. 1989 Dec 5;264(34):20482–20486. [PubMed] [Google Scholar]
- Dekker P. J., Stuurman J., van Oosterum K., Grivell L. A. Determinants for binding of a 40 kDa protein to the leaders of yeast mitochondrial mRNAs. Nucleic Acids Res. 1992 Jun 11;20(11):2647–2655. doi: 10.1093/nar/20.11.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. B., Delort J., Mallet J. Oligodeoxyribonucleotide ligation to single-stranded cDNAs: a new tool for cloning 5' ends of mRNAs and for constructing cDNA libraries by in vitro amplification. Nucleic Acids Res. 1991 Oct 11;19(19):5227–5232. doi: 10.1093/nar/19.19.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich R. S., Colman R. F. Binding of ligands to half of subunits of NAD-dependent isocitrate dehydrogenase from pig heart. Binding of manganous ion, isocitrate, ADP and NAD. J Biol Chem. 1981 Feb 10;256(3):1276–1282. [PubMed] [Google Scholar]
- Ehrlich R. S., Colman R. F. Interrelationships among nucleotide binding sites of pig heart NAD-dependent isocitrate dehydrogenase. J Biol Chem. 1982 May 10;257(9):4769–4774. [PubMed] [Google Scholar]
- Ehrlich R. S., Colman R. F. Separation, recombination, and characterization of dissimilar subunits of the DPN-dependent isocitrate dehydrogenase from pig heart. J Biol Chem. 1983 Jun 10;258(11):7079–7086. [PubMed] [Google Scholar]
- Ehrlich R. S., Colman R. F. The role of dissimilar subunits of NAD-specific isocitrate dehydrogenase from pig heart. Evaluation using affinity labeling. J Biol Chem. 1984 Oct 10;259(19):11936–11942. [PubMed] [Google Scholar]
- Ehrlich R. S., Hayman S., Ramachandran N., Colman R. F. Re-evaluation of molecular weight of pig heart NAD-specific isocitrate dehydrogenase. J Biol Chem. 1981 Oct 25;256(20):10560–10564. [PubMed] [Google Scholar]
- Elzinga S. D., Bednarz A. L., van Oosterum K., Dekker P. J., Grivell L. A. Yeast mitochondrial NAD(+)-dependent isocitrate dehydrogenase is an RNA-binding protein. Nucleic Acids Res. 1993 Nov 25;21(23):5328–5331. doi: 10.1093/nar/21.23.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C. C., Lin J. P., Plaut G. W. Effects of temperature on diphosphopyridine nucleotide-linked isocitrate dehydrogenase from bovine heart. Aspects of the kinetics, stability, and quarternary structure of the enzyme. J Biol Chem. 1975 Mar 25;250(6):2022–2027. [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio N. A., Jr, Yip A. T., Fleming J., Plaut G. W. Diphosphopyridine nucleotide-linked isocitrate dehydrogenase from bovine heart. Polymeric forms and subunits. J Biol Chem. 1970 Oct 25;245(20):5469–5477. [PubMed] [Google Scholar]
- Hentze M. W. Enzymes as RNA-binding proteins: a role for (di)nucleotide-binding domains? Trends Biochem Sci. 1994 Mar;19(3):101–103. doi: 10.1016/0968-0004(94)90198-8. [DOI] [PubMed] [Google Scholar]
- Huang Y. C., Colman R. F. Subunit location and sequences of the cysteinyl peptides of pig heart NAD-dependent isocitrate dehydrogenase. Biochemistry. 1990 Sep 11;29(36):8266–8273. doi: 10.1021/bi00488a010. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Dean A. M., Koshland D. E., Jr, Stroud R. M. Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes. Biochemistry. 1991 Sep 3;30(35):8671–8678. doi: 10.1021/bi00099a026. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Thorsness P. E., Ramalingam V., Helmers N. H., Koshland D. E., Jr, Stroud R. M. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth J. A. Purification of yeast isocitrate dehydrogenase. Biochem J. 1972 Oct;129(5):1119–1124. doi: 10.1042/bj1291119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada K., Sato M., Tanaka N., Katsube Y., Matsuura Y., Oshima T. Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution. J Mol Biol. 1991 Dec 5;222(3):725–738. doi: 10.1016/0022-2836(91)90508-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte D. C., Koshland D. E., Jr Phosphorylation of isocitrate dehydrogenase as a demonstration of enhanced sensitivity in covalent regulation. Nature. 1983 Sep 22;305(5932):286–290. doi: 10.1038/305286a0. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Nichols B. J., Hall L., Perry A. C., Denton R. M. Molecular cloning and deduced amino acid sequences of the gamma-subunits of rat and monkey NAD(+)-isocitrate dehydrogenases. Biochem J. 1993 Oct 15;295(Pt 2):347–350. doi: 10.1042/bj2950347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou B., Dekker P., Blom J., Grivell L. A. A 40 kd protein binds specifically to the 5'-untranslated regions of yeast mitochondrial mRNAs. EMBO J. 1990 Dec;9(12):4135–4143. doi: 10.1002/j.1460-2075.1990.tb07636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaut G. W., Schramm V. L., Aogaichi T. Action of magnesium ion on diphosphopyridine nucleotide-linked isocitrate dehydrogenase from bovine heart. Characterization of the forms of the substrate and the modifier of the reaction. J Biol Chem. 1974 Mar 25;249(6):1848–1856. [PubMed] [Google Scholar]
- Ramachandran N., Colman R. F. Chemical characterization of distinct subunits of pig heart DPN-specific isocitrate dehydrogenase. J Biol Chem. 1980 Sep 25;255(18):8859–8864. [PubMed] [Google Scholar]
- Ramachandran N., Colman R. F. Evidence for the presence of two nonidentical subunits in NAD-dependent isocitrate dehydrogenase of pig heart. Proc Natl Acad Sci U S A. 1978 Jan;75(1):252–255. doi: 10.1073/pnas.75.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushbrook J. I., Harvey R. A. Nicotinamide adenine dinucleotide dependent isocitrate dehydrogenase from beef heart: subunit heterogeneity and enzyme dissociation. Biochemistry. 1978 Dec 12;17(25):5339–5346. doi: 10.1021/bi00618a003. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Shen W. C., Mauck L., Colman R. F. Physicochemical properties of the diphosphopyridine nucleotide-specific isocitrate dehydrogenase of pig heart. J Biol Chem. 1974 Dec 25;249(24):7942–7949. [PubMed] [Google Scholar]
- Stoddard B. L., Dean A., Koshland D. E., Jr Structure of isocitrate dehydrogenase with isocitrate, nicotinamide adenine dinucleotide phosphate, and calcium at 2.5-A resolution: a pseudo-Michaelis ternary complex. Biochemistry. 1993 Sep 14;32(36):9310–9316. doi: 10.1021/bi00087a008. [DOI] [PubMed] [Google Scholar]
- Stoddard B. L., Koshland D. E., Jr Structure of isocitrate dehydrogenase with alpha-ketoglutarate at 2.7-A resolution: conformational changes induced by decarboxylation of isocitrate. Biochemistry. 1993 Sep 14;32(36):9317–9322. doi: 10.1021/bi00087a009. [DOI] [PubMed] [Google Scholar]
- Thorsness P. E., Koshland D. E., Jr Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987 Aug 5;262(22):10422–10425. [PubMed] [Google Scholar]
- Zasloff M., Ginder G. D., Felsenfeld G. A new method for the purification and identification of covalently closed circular DNA molcules. Nucleic Acids Res. 1978 Apr;5(4):1139–1152. doi: 10.1093/nar/5.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]