Abstract
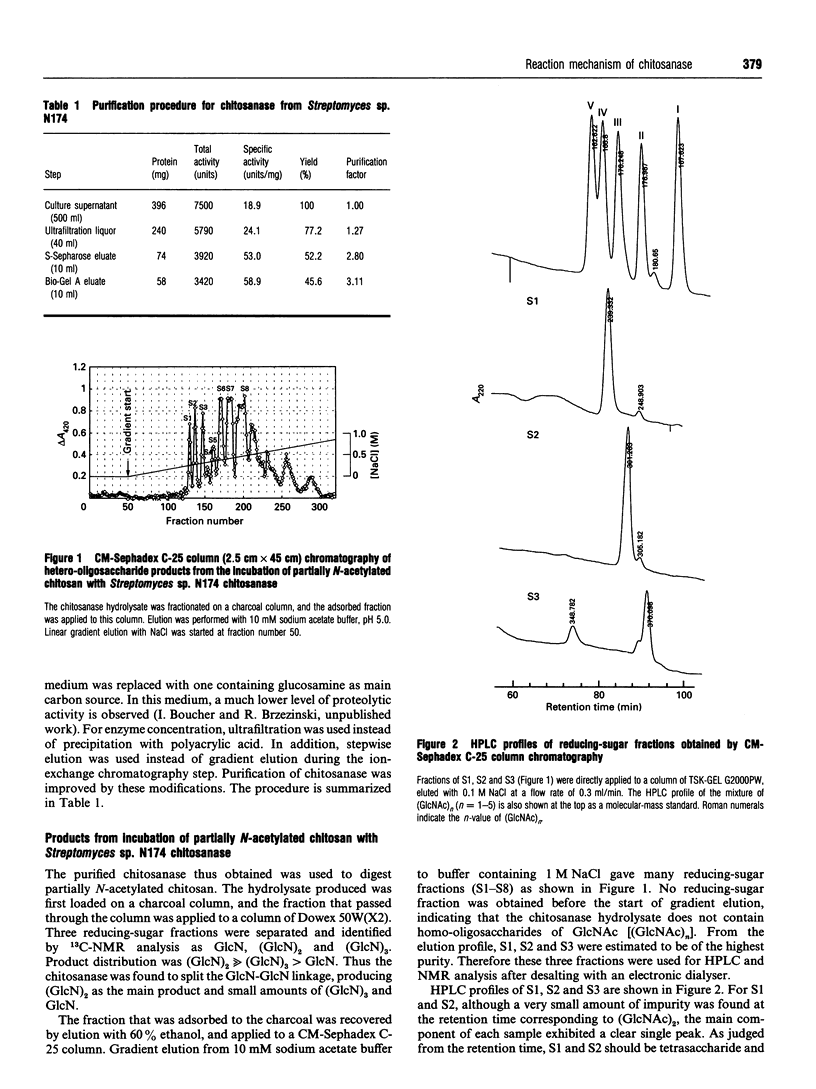
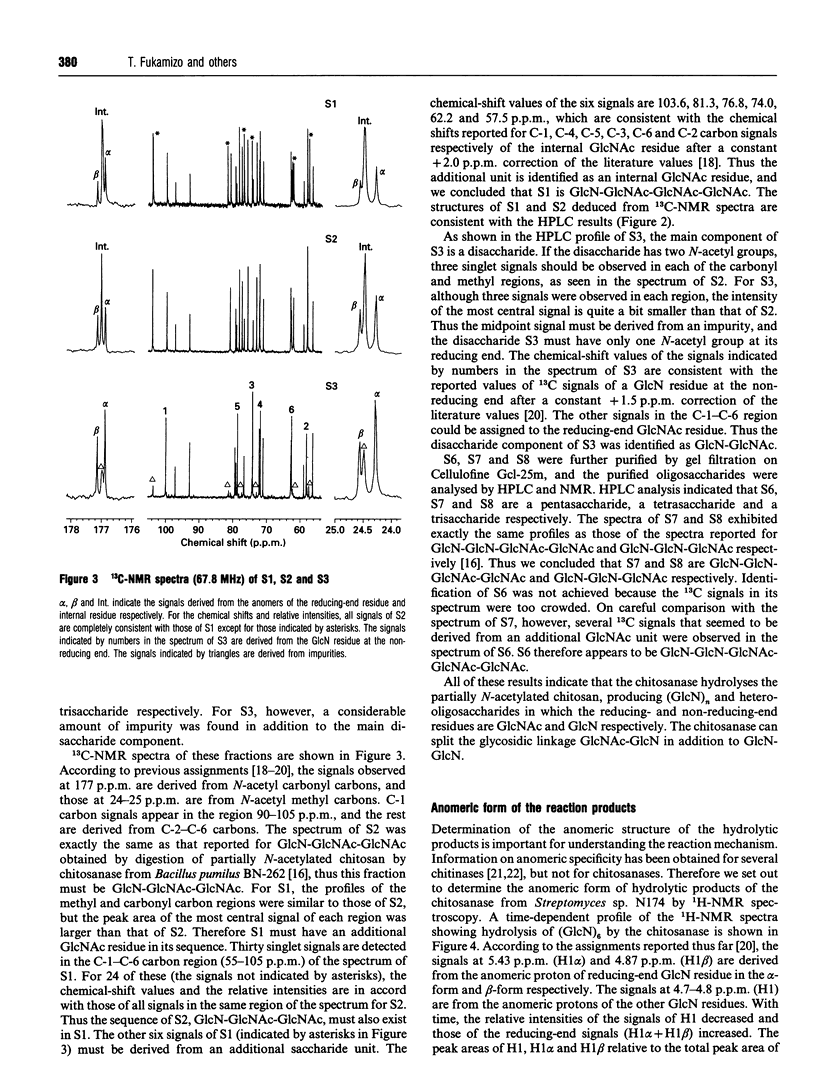
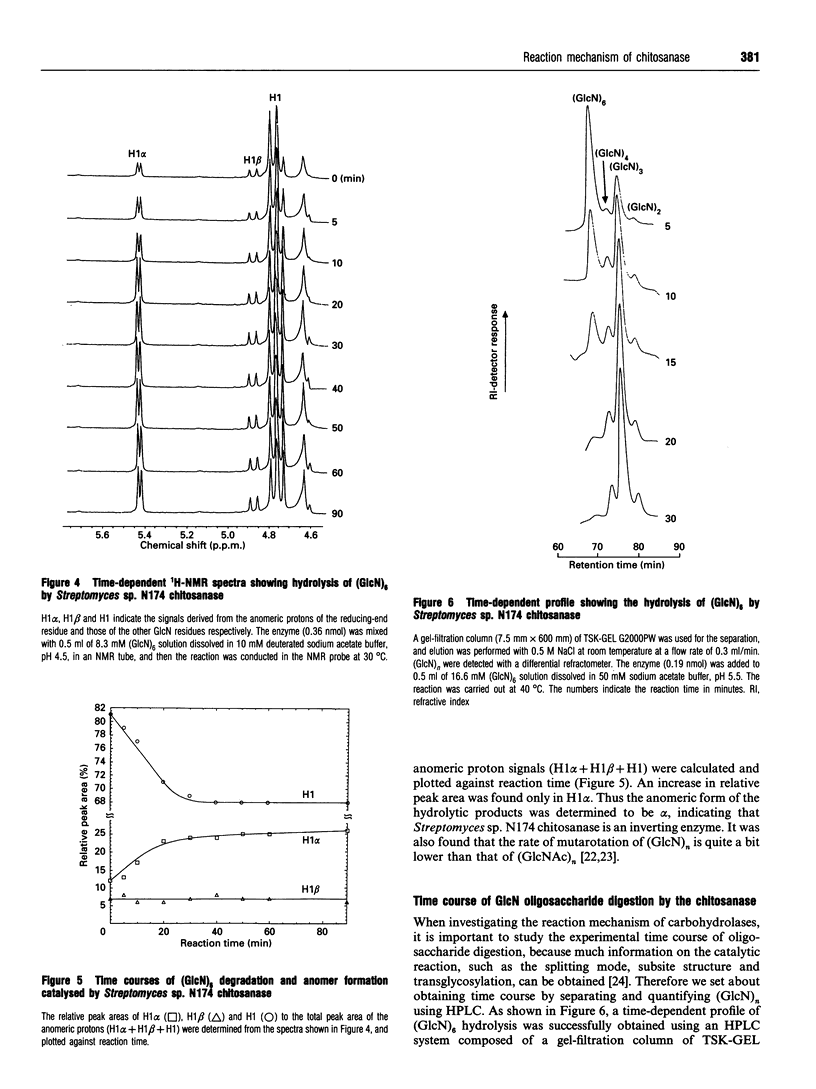
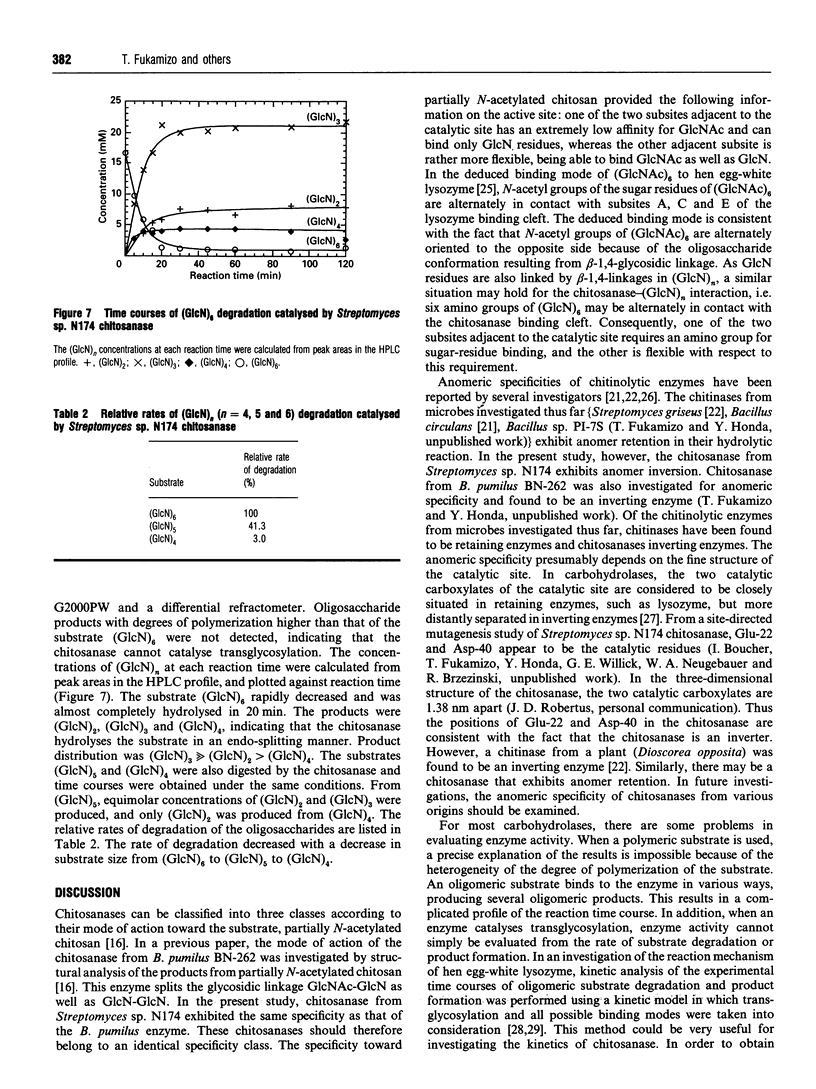
Chitosanase was produced by the strain of Streptomyces lividans TK24 bearing the csn gene from Streptomyces sp. N174, and purified by S-Sepharose and Bio-Gel A column chromatography. Partially (25-35%) N-acetylated chitosan was digested by the purified chitosanase, and structures of the products were analysed by NMR spectroscopy. The chitosanase produced heterooligosaccharides consisting of D-GlcN and GlcNAc in addition to glucosamine oligosaccharides [(GlcN)n, n = 1, 2 and 3]. The reducing- and non-reducing-end residues of the heterooligosaccharide products were GlcNAc and GlcN respectively, indicating that the chitosanase can split the GlcNAc-GlcN linkage in addition to that of GlcN-GlcN. Time-dependent 1H-NMR spectra showing hydrolysis of (GlcN)6 by the chitosanase were obtained in order to determine the anomeric form of the reaction products. The chitosanase was found to produce only the alpha-form; therefore it is an inverting enzyme. Separation and quantification of (GlcN)n was achieved by HPLC, and the time course of the reaction catalysed by the chitosanase was studied using (GlcN)n (n = 4, 5 and 6) as the substrate. The chitosanase hydrolysed (GlcN)6 in an endo-splitting manner producing (GlcN)2, (GlcN)3 and (GlcN)4, and did not catalyse transglycosylation. Product distribution was (GlcN)3 >> (GlcN)2 > (GlcN)4. Cleavage to (GlcN)3 + (GlcN)3 predominated over that to (GlcN)2 + (GlcN)4. Time courses showed a decrease in rate of substrate degradation from (GlcN)6 to (GlcN)5 to (GlcN)4. It is most likely that the substrate-binding cleft of the chitosanase can accommodate at least six GlcN residues, and that the cleavage point is located at the midpoint of the binding cleft.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armand S., Tomita H., Heyraud A., Gey C., Watanabe T., Henrissat B. Stereochemical course of the hydrolysis reaction catalyzed by chitinases A1 and D from Bacillus circulans WL-12. FEBS Lett. 1994 Apr 25;343(2):177–180. doi: 10.1016/0014-5793(94)80314-5. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Dahlquist F. W., Borders C. L., Jr, Jacobson G., Raftery M. A. The stereospecificity of human, hen, and papaya lysozymes. Biochemistry. 1969 Feb;8(2):694–700. doi: 10.1021/bi00830a035. [DOI] [PubMed] [Google Scholar]
- Denis F., Brzezinski R. A versatile shuttle cosmid vector for use in Escherichia coli and actinomycetes. Gene. 1992 Feb 1;111(1):115–118. doi: 10.1016/0378-1119(92)90611-r. [DOI] [PubMed] [Google Scholar]
- Fukamizo T., Hayashi K. Separation and mutarotation of anomers of chitooligosaccharides. J Biochem. 1982 Feb;91(2):619–626. doi: 10.1093/oxfordjournals.jbchem.a133733. [DOI] [PubMed] [Google Scholar]
- Fukamizo T., Minematsu T., Yanase Y., Hayashi K., Goto S. Substrate size dependence of lysozyme-catalyzed reaction. Arch Biochem Biophys. 1986 Nov 1;250(2):312–321. doi: 10.1016/0003-9861(86)90732-0. [DOI] [PubMed] [Google Scholar]
- Fukamizo T., Ohkawa T., Ikeda Y., Goto S. Specificity of chitosanase from Bacillus pumilus. Biochim Biophys Acta. 1994 Apr 13;1205(2):183–188. doi: 10.1016/0167-4838(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Fukamizo T., Ohtakara A., Mitsutomi M., Goto S. NMR spectra of partially deacetylated chitotrisaccharides. Agric Biol Chem. 1991 Oct;55(10):2653–2655. [PubMed] [Google Scholar]
- Hart P. J., Monzingo A. F., Ready M. P., Ernst S. R., Robertus J. D. Crystal structure of an endochitinase from Hordeum vulgare L. seeds. J Mol Biol. 1993 Jan 5;229(1):189–193. doi: 10.1006/jmbi.1993.1017. [DOI] [PubMed] [Google Scholar]
- Izume M., Nagae S., Kawagishi H., Mitsutomi M., Ohtakara A. Action pattern of Bacillus sp. no. 7-M chitosanase on partially N-acetylated chitosan. Biosci Biotechnol Biochem. 1992 Mar;56(3):448–453. doi: 10.1271/bbb.56.448. [DOI] [PubMed] [Google Scholar]
- Marcotte E., Hart P. J., Boucher I., Brzezinski R., Robertus J. D. Crystallization of a chitosanase from Streptomyces N174. J Mol Biol. 1993 Aug 5;232(3):995–996. doi: 10.1006/jmbi.1993.1447. [DOI] [PubMed] [Google Scholar]
- Masaki A., Fukamizo T., Otakara A., Torikata T., Hayashi K., Imoto T. Estimation of rate constants in lysozyme-catalyzed reaction of chitooligosaccharides. J Biochem. 1981 Oct;90(4):1167–1175. doi: 10.1093/oxfordjournals.jbchem.a133569. [DOI] [PubMed] [Google Scholar]
- Masaki A., Fukamizo T., Otakara A., Torikata T., Hayashi K., Imoto T. Lysozyme-catalyzed reaction of chitooligosaccharides. J Biochem. 1981 Aug;90(2):527–533. doi: 10.1093/oxfordjournals.jbchem.a133501. [DOI] [PubMed] [Google Scholar]
- Masson J. Y., Denis F., Brzezinski R. Primary sequence of the chitosanase from Streptomyces sp. strain N174 and comparison with other endoglycosidases. Gene. 1994 Mar 11;140(1):103–107. doi: 10.1016/0378-1119(94)90738-2. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Tews I., Dauter Z., Oppenheim A. B., Chet I., Wilson K. S., Vorgias C. E. Crystal structure of a bacterial chitinase at 2.3 A resolution. Structure. 1994 Dec 15;2(12):1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- Sakai K., Katsumi R., Isobe A., Nanjo F. Purification and hydrolytic action of a chitosanase from Nocardia orientalis. Biochim Biophys Acta. 1991 Aug 9;1079(1):65–72. doi: 10.1016/0167-4838(91)90025-u. [DOI] [PubMed] [Google Scholar]


