Abstract
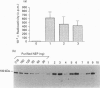
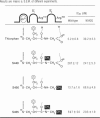
Neprilysin (EC 3.4.24.11) is a Zn2+ metallopeptidase involved in the degradation of biologically active peptides, e.g. enkephalins and atrial natriuretic peptide. The substrate specificity and catalytic activity of neprilysin resemble those of thermolysin, a crystallized bacterial Zn2+ metalloprotease. Despite little overall homology between the primary structures of thermolysin and neprilysin, many of the amino acid residues involved in catalysis, as well as Zn2+ and substrate binding, are highly conserved. Most of the active-site residues of neprilysin have their homologues in thermolysin and have been characterized by site-directed mutagenesis. Furthermore, hydrophobic cluster analysis has revealed some other analogies between the neprilysin and thermolysin sequences [Benchetrit, Bissery, Mornon, Devault, Crine and Roques (1988) Biochemistry 27, 592-596]. According to this analysis the role of Asn542 in the neprilysin active site is analogous to that of Asn112 of thermolysin, which is to bind the substrate. Site-directed mutagenesis was used to change Asn542 to Gly or Gln residues. The effect of these mutations on substrate catalysis and inhibitor binding was examined with a series of thiorphan-like compounds containing various degrees of methylation at the P2' residue. For both mutated enzymes, determination of kinetic parameters with [D-Ala2,Leu5]enkephalin as substrate showed that the large decrease in activity was attributable to an increase in Km (14-16-fold) whereas kcat values were only slightly affected (2-3-fold decrease). This is in agreement with Asn542 being involved in substrate binding rather than directly in catalysis. Finally, the IC50 values for thiorphan and substituted thiorphans strongly suggest that Asn542 of neprilysin binds the substrate on the amino side of the P2' residue by formation of a unique hydrogen bond.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubry M., Zollinger M., Fortin S., Vénien C., LeGrimellec C., Crine P. Monoclonal antibodies as probes for the transmembrane structure of neutral endopeptidase 24.11 ('enkephalinase'). Biochim Biophys Acta. 1988 Oct 13;967(1):56–64. doi: 10.1016/0304-4165(88)90188-2. [DOI] [PubMed] [Google Scholar]
- Bateman R. C., Jr, Jackson D., Slaughter C. A., Unnithan S., Chai Y. G., Moomaw C., Hersh L. B. Identification of the active-site arginine in rat neutral endopeptidase 24.11 (enkephalinase) as arginine 102 and analysis of a glutamine 102 mutant. J Biol Chem. 1989 Apr 15;264(11):6151–6157. [PubMed] [Google Scholar]
- Bateman R. C., Jr, Kim Y. A., Slaughter C., Hersh L. B. N-bromoacetyl-D-leucylglycine. An affinity label for neutral endopeptidase 24.11. J Biol Chem. 1990 May 25;265(15):8365–8368. [PubMed] [Google Scholar]
- Beaumont A., Barbe B., Le Moual H., Boileau G., Crine P., Fournié-Zaluski M. C., Roques B. P. Charge polarity reversal inverses the specificity of neutral endopeptidase-24.11. J Biol Chem. 1992 Feb 5;267(4):2138–2141. [PubMed] [Google Scholar]
- Beaumont A., Le Moual H., Boileau G., Crine P., Roques B. P. Evidence that both arginine 102 and arginine 747 are involved in substrate binding to neutral endopeptidase (EC 3.4.24.11). J Biol Chem. 1991 Jan 5;266(1):214–220. [PubMed] [Google Scholar]
- Benchetrit T., Bissery V., Mornon J. P., Devault A., Crine P., Roques B. P. Primary structure homologies between two zinc metallopeptidases, the neutral endopeptidase 24.11 ("enkephalinase") and thermolysin, through clustering analysis. Biochemistry. 1988 Jan 26;27(2):592–596. doi: 10.1021/bi00402a014. [DOI] [PubMed] [Google Scholar]
- Benchetrit T., Fournié-Zaluski M. C., Roques B. P. Relationship between the inhibitory potencies of thiorphan and retrothiorphan enantiomers on thermolysin and neutral endopeptidase 24.11 and their interactions with the thermolysin active site by computer modelling. Biochem Biophys Res Commun. 1987 Sep 30;147(3):1034–1040. doi: 10.1016/s0006-291x(87)80174-2. [DOI] [PubMed] [Google Scholar]
- Blundell T. L. Metalloproteinase superfamilies and drug design. Nat Struct Biol. 1994 Feb;1(2):73–75. doi: 10.1038/nsb0294-73. [DOI] [PubMed] [Google Scholar]
- Devault A., Lazure C., Nault C., Le Moual H., Seidah N. G., Chrétien M., Kahn P., Powell J., Mallet J., Beaumont A. Amino acid sequence of rabbit kidney neutral endopeptidase 24.11 (enkephalinase) deduced from a complementary DNA. EMBO J. 1987 May;6(5):1317–1322. doi: 10.1002/j.1460-2075.1987.tb02370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devault A., Nault C., Zollinger M., Fournie-Zaluski M. C., Roques B. P., Crine P., Boileau G. Expression of neutral endopeptidase (enkephalinase) in heterologous COS-1 cells. Characterization of the recombinant enzyme and evidence for a glutamic acid residue at the active site. J Biol Chem. 1988 Mar 15;263(8):4033–4040. [PubMed] [Google Scholar]
- Devault A., Sales V., Nault C., Beaumont A., Roques B., Crine P., Boileau G. Exploration of the catalytic site of endopeptidase 24.11 by site-directed mutagenesis. Histidine residues 583 and 587 are essential for catalysis. FEBS Lett. 1988 Apr 11;231(1):54–58. doi: 10.1016/0014-5793(88)80701-4. [DOI] [PubMed] [Google Scholar]
- Dion N., Le Moual H., Crine P., Boileau G. Kinetic evidence that His-711 of neutral endopeptidase 24.11 is involved in stabilization of the transition state. FEBS Lett. 1993 Mar 8;318(3):301–304. doi: 10.1016/0014-5793(93)80533-z. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Knill-Jones J., Lowe D. M., Wilkinson A. J., Blow D. M., Brick P., Carter P., Waye M. M., Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985 Mar 21;314(6008):235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Fillenz M. Hypothesis for a neuronal mechanism involved in memory. Nature. 1972 Jul 7;238(5358):41–43. doi: 10.1038/238041a0. [DOI] [PubMed] [Google Scholar]
- Fournie-Zaluski M. C., Lucas E., Waksman G., Roques B. P. Differences in the structural requirements for selective interaction with neutral metalloendopeptidase (enkephalinase) or angiotensin-converting enzyme. Molecular investigation by use of new thiol inhibitors. Eur J Biochem. 1984 Mar 1;139(2):267–274. doi: 10.1111/j.1432-1033.1984.tb08003.x. [DOI] [PubMed] [Google Scholar]
- Fournie-Zaluski M. C., Perdrisot R., Gacel G., Swerts J. P., Roques B. P., Schwartz J. C. Inhibitory potency of various peptides on enkephalinase activity from mouse striatum. Biochem Biophys Res Commun. 1979 Nov 14;91(1):130–135. doi: 10.1016/0006-291x(79)90593-x. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Holland D. R., Tronrud D. E., Pley H. W., Flaherty K. M., Stark W., Jansonius J. N., McKay D. B., Matthews B. W. Structural comparison suggests that thermolysin and related neutral proteases undergo hinge-bending motion during catalysis. Biochemistry. 1992 Nov 24;31(46):11310–11316. doi: 10.1021/bi00161a008. [DOI] [PubMed] [Google Scholar]
- Hooper N. M. Families of zinc metalloproteases. FEBS Lett. 1994 Oct 31;354(1):1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Jongeneel C. V., Bouvier J., Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989 Jan 2;242(2):211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- Le Moual H., Devault A., Roques B. P., Crine P., Boileau G. Identification of glutamic acid 646 as a zinc-coordinating residue in endopeptidase-24.11. J Biol Chem. 1991 Aug 25;266(24):15670–15674. [PubMed] [Google Scholar]
- Le Moual H., Dion N., Roques B. P., Crine P., Boileau G. Asp650 is crucial for catalytic activity of neutral endopeptidase 24-11. Eur J Biochem. 1994 Apr 1;221(1):475–480. doi: 10.1111/j.1432-1033.1994.tb18760.x. [DOI] [PubMed] [Google Scholar]
- Lee S., Zambas E. D., Marsh W. L., Redman C. M. Molecular cloning and primary structure of Kell blood group protein. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6353–6357. doi: 10.1073/pnas.88.14.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay G., Waksman G., Roques B. P., Crine P., Boileau G. Fusion of a cleavable signal peptide to the ectodomain of neutral endopeptidase (EC 3.4.24.11) results in the secretion of an active enzyme in COS-1 cells. J Biol Chem. 1989 Sep 15;264(26):15620–15623. [PubMed] [Google Scholar]
- Llorens C., Gacel G., Swerts J. P., Perdrisot R., Fournie-Zaluski M. C., Schwartz J. C., Roques B. P. Rational design of enkephalinase inhibitors: substrate specificity of enkephalinase studied from inhibitory potency of various dipeptides. Biochem Biophys Res Commun. 1980 Oct 31;96(4):1710–1716. doi: 10.1016/0006-291x(80)91371-6. [DOI] [PubMed] [Google Scholar]
- Malfroy B., Kuang W. J., Seeburg P. H., Mason A. J., Schofield P. R. Molecular cloning and amino acid sequence of human enkephalinase (neutral endopeptidase). FEBS Lett. 1988 Feb 29;229(1):206–210. doi: 10.1016/0014-5793(88)80828-7. [DOI] [PubMed] [Google Scholar]
- Malfroy B., Schofield P. R., Kuang W. J., Seeburg P. H., Mason A. J., Henzel W. J. Molecular cloning and amino acid sequence of rat enkephalinase. Biochem Biophys Res Commun. 1987 Apr 14;144(1):59–66. doi: 10.1016/s0006-291x(87)80475-8. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Thayer M. M., Flaherty K. M., Pley H., Benvegnu D. Crystallographic structures of the elastase of Pseudomonas aeruginosa. Matrix Suppl. 1992;1:112–115. [PubMed] [Google Scholar]
- Mierau I., Tan P. S., Haandrikman A. J., Mayo B., Kok J., Leenhouts K. J., Konings W. N., Venema G. Cloning and sequencing of the gene for a lactococcal endopeptidase, an enzyme with sequence similarity to mammalian enkephalinase. J Bacteriol. 1993 Apr;175(7):2087–2096. doi: 10.1128/jb.175.7.2087-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura T., Nakamura Y., Nishino J., Sawayama T., Komiya T., Deguchi T., Kita A., Nakamura H., Matsumoto J. A novel class of enkephalinase inhibitors containing a C-terminal sulfo group. J Med Chem. 1992 Feb 7;35(3):602–608. doi: 10.1021/jm00081a024. [DOI] [PubMed] [Google Scholar]
- Noël G., Zollinger L., Larivière N., Nault C., Crine P., Boileau G. Expression of porcine pro-opiomelanocortin cDNA in heterologous monkey kidney cells. Biosynthesis and secretion of the prohormone without processing. J Biol Chem. 1987 Feb 5;262(4):1876–1881. [PubMed] [Google Scholar]
- Pauptit R. A., Karlsson R., Picot D., Jenkins J. A., Niklaus-Reimer A. S., Jansonius J. N. Crystal structure of neutral protease from Bacillus cereus refined at 3.0 A resolution and comparison with the homologous but more thermostable enzyme thermolysin. J Mol Biol. 1988 Feb 5;199(3):525–537. doi: 10.1016/0022-2836(88)90623-7. [DOI] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J. Evolutionary families of peptidases. Biochem J. 1993 Feb 15;290(Pt 1):205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton J. M., Gee N. S., Matsas R., Turner A. J., Kenny A. J. Purification of endopeptidase-24.11 ('enkephalinase') from pig brain by immunoadsorbent chromatography. Biochem J. 1983 Dec 1;215(3):519–523. doi: 10.1042/bj2150519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick S. L., Fournie-Zaluski M. C., Roques B. P., Matthews B. W. Thiorphan and retro-thiorphan display equivalent interactions when bound to crystalline thermolysin. Biochemistry. 1989 Feb 21;28(4):1493–1497. doi: 10.1021/bi00430a011. [DOI] [PubMed] [Google Scholar]
- Roques B. P., Fournié-Zaluski M. C., Soroca E., Lecomte J. M., Malfroy B., Llorens C., Schwartz J. C. The enkephalinase inhibitor thiorphan shows antinociceptive activity in mice. Nature. 1980 Nov 20;288(5788):286–288. doi: 10.1038/288286a0. [DOI] [PubMed] [Google Scholar]
- Roques B. P., Lucas-Soroca E., Chaillet P., Costentin J., Fournié-Zaluski M. C. Complete differentiation between enkephalinase and angiotensin-converting enzyme inhibition by retro-thiorphan. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3178–3182. doi: 10.1073/pnas.80.11.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roques B. P., Noble F., Daugé V., Fournié-Zaluski M. C., Beaumont A. Neutral endopeptidase 24.11: structure, inhibition, and experimental and clinical pharmacology. Pharmacol Rev. 1993 Mar;45(1):87–146. [PubMed] [Google Scholar]
- Stark W., Pauptit R. A., Wilson K. S., Jansonius J. N. The structure of neutral protease from Bacillus cereus at 0.2-nm resolution. Eur J Biochem. 1992 Jul 15;207(2):781–791. doi: 10.1111/j.1432-1033.1992.tb17109.x. [DOI] [PubMed] [Google Scholar]
- Suda H., Aoyagi T., Takeuchi T., Umezawa H. Letter: A thermolysin inhibitor produced by Actinomycetes: phospholamidon. J Antibiot (Tokyo) 1973 Oct;26(10):621–623. doi: 10.7164/antibiotics.26.621. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer M. M., Flaherty K. M., McKay D. B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J Biol Chem. 1991 Feb 15;266(5):2864–2871. doi: 10.2210/pdb1ezm/pdb. [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan J., Kim Y. A., Jackson D., Orlowski M., Hersh L. B. Use of site-directed mutagenesis to identify valine-573 in the S'1 binding site of rat neutral endopeptidase 24.11 (enkephalinase). Biochemistry. 1990 Sep 4;29(35):8052–8056. doi: 10.1021/bi00487a009. [DOI] [PubMed] [Google Scholar]
- Wilkinson A. J., Fersht A. R., Blow D. M., Winter G. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry. 1983 Jul 19;22(15):3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- Xie J., Soleilhac J. M., Schmidt C., Peyroux J., Roques B. P., Fournié-Zaluski M. C. New kelatorphan-related inhibitors of enkephalin metabolism: improved antinociceptive properties. J Med Chem. 1989 Jul;32(7):1497–1503. doi: 10.1021/jm00127a017. [DOI] [PubMed] [Google Scholar]
- Xu D., Emoto N., Giaid A., Slaughter C., Kaw S., deWit D., Yanagisawa M. ECE-1: a membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell. 1994 Aug 12;78(3):473–485. doi: 10.1016/0092-8674(94)90425-1. [DOI] [PubMed] [Google Scholar]