Abstract
The formation of new ribosomes is tightly coordinated with cell growth and proliferation. In eukaryotes, the correct assembly of all ribosomal proteins and RNAs follows an intricate scheme of maturation and rearrangement steps across three cellular compartments: the nucleolus, nucleoplasm, and cytoplasm. We demonstrate that usnic acid, a lichen secondary metabolite, inhibits the maturation of the large ribosomal subunit in yeast. We combine biochemical characterization of pre-ribosomal particles with a quantitative single-particle cryo-EM approach to monitor changes in nucleolar particle populations upon drug treatment. Usnic acid rapidly blocks the transition from nucleolar state B to C of Nsa1-associated pre-ribosomes, depleting key maturation factors such as Dbp10 and hindering pre-rRNA processing. This primary nucleolar block rapidly rebounds on earlier stages of the pathway which highlights the regulatory linkages between different steps. In summary, we provide an in-depth characterization of the effect of usnic acid on ribosome biogenesis, which may have implications for its reported anti-cancer activities.
Subject terms: Molecular biology, Biochemistry
Usnic acid is a lichen secondary metabolite and has a antitumor activity. Here the authors show that usnic acid rapidly blocks large ribosomal subunit formation. This hinders pre-rRNA processing and leads to depletion of key maturation factors such as Dbp10.
Introduction
Ribosomes are responsible for protein synthesis, which is crucial for all living organisms. They comprise two subunits, in eukaryotes a small 40S and a large 60S subunit which together contain a total of ~80 ribosomal proteins (r-proteins) and 4 ribosomal RNAs (rRNAs). The formation of ribosomes, called ribosome biogenesis, is one of the most complex and dynamic cellular pathways and requires the action of all three RNA polymerases (Pol I-III) as well as over 250 non-ribosomal assembly factors (reviewed in refs. 1–4). Maintaining a pool of translating ribosomes is an enormous resource and energy investment for a proliferating cell5. Ribosome biogenesis is consequently highly regulated in accordance with cell growth and proliferation. Therefore, defects or deregulation within this pathway can result in severe pathologies, including cancer and so-called ribosomopathies6–12.
The assembly of eukaryotic ribosomal subunits follows a stepwise and strictly hierarchical scheme and is coordinated between all three cellular compartments by the involved assembly factors. The assembly process is intensively studied in the yeast Saccharomyces cerevisiae, which is the best-established eukaryotic model organism in this research field. In the nucleolus, RNA Pol I transcribes a long 35S precursor (pre)-rRNA containing the sequences of the 18S rRNA of the small subunit and the 5.8S and 25S rRNAs of the large subunit along with internal (ITS) and external (ETS) transcribed spacers. After removal of the 35S 5’-ETS, the resulting 32S pre-rRNA is cleaved at the A2 site, enabling separate processing of the 40S and 60S rRNAs. Maturation of the 40S-specific 20S pre-rRNA is accomplished with a single endonucleolytic cleavage in the cytoplasm resulting in the mature 18S rRNA. In contrast, processing of the 60S-specific 27SA2 pre-rRNA to the mature 25S and 5.8S rRNAs requires several endo- and exonucleolytic processing steps in the nucleus and the cytoplasm (Supplementary Fig. 1).
The 60S assembly path is initiated by the construction of the solvent-exposed surface, followed by maturation of the polypeptide exit tunnel (PET), processing of the foot structure, and formation of the central protuberance (CP). The latter involves nucleolar incorporation of the 5S ribonucleoprotein (RNP). The 5S RNP contains the 5S rRNA, which is the third rRNA present in the large subunit and is independently transcribed by Pol III13. Construction of the peptidyl transferase center (PTC) on the subunit interface, which catalyzes peptide bond formation on the nascent protein, is only finished in the cytoplasm13–19.
Biochemical studies and high-resolution cryo-EM structures obtained in recent years have highlighted many highly conserved steps of ribosome biogenesis in yeast (most recently by ref. 20 and reviewed e.g., in ref. 1) and human cells18,21–25. Still, we have limited information on the regulatory networks that dynamically coordinate the single events along the pathway. Due to this interconnectivity, blocks in later stages of the pathway rebound on (are signaled back to) earlier steps. As described in more detail in ref. 26, such rebound effects are caused by hindered downstream release and recycling of assembly factors that are, therefore, missing at earlier stages.
We previously showed that the conversion of 27SA2 pre-rRNA to the mature forms takes about 15 mins27. Considering also the cytoplasmic steps, we estimate that the whole large subunit assembly might be finished within <20 mins. The fast progression of the process and the coordinating rebound effects complicate the differentiation of primary and secondary effects5,26,28. Particularly challenging is the investigation of the very rapidly conducted earliest steps in 60S assembly. Specific inhibitors bear the advantage of blocking the pathway within seconds and can, therefore, capture the primary effects of a block before rebound effects set in ref. 26. Well-characterized compounds can thus be key to unraveling the dynamic nature of ribosome biogenesis and the underlying regulatory linkages27,29–32, reviewed in refs. 26,33,34.
Here, we characterize the natural compound usnic acid as a novel ribosome biogenesis inhibitor. Usnic acid is one of the most prominent and most studied lichen secondary metabolites with various biological activities with medical potential (reviewed in refs. 35–40). Our mechanistic understanding of these effects is, however, in many cases incomplete. Here, we show that usnic acid inhibits early nucleolar steps in large ribosomal subunit assembly. Biochemical characterization of nucleolar pre-60S precursors combined with single-particle cryo-EM demonstrates a global shift of nucleolar pre-60S particle populations to earlier stages within minutes after drug treatment. More precisely, usnic acid blocks the progression of Nsa1-TAP-associated pre-60S particles from state B to C. A major player in this transition is the DEAD-box RNA helicase Dbp10, which is strongly depleted from nucleolar particles upon usnic acid treatment. Our data provide novel insights into nucleolar pre-60S maturation and uncover how the pathway responds to perturbation by usnic acid on a minute’s time scale. This effect of usnic acid on ribosome biogenesis can help to understand the biological activity of usnic acid in eukaryotic cells.
Results
Usnic acid blocks early pre-60S maturation within minutes
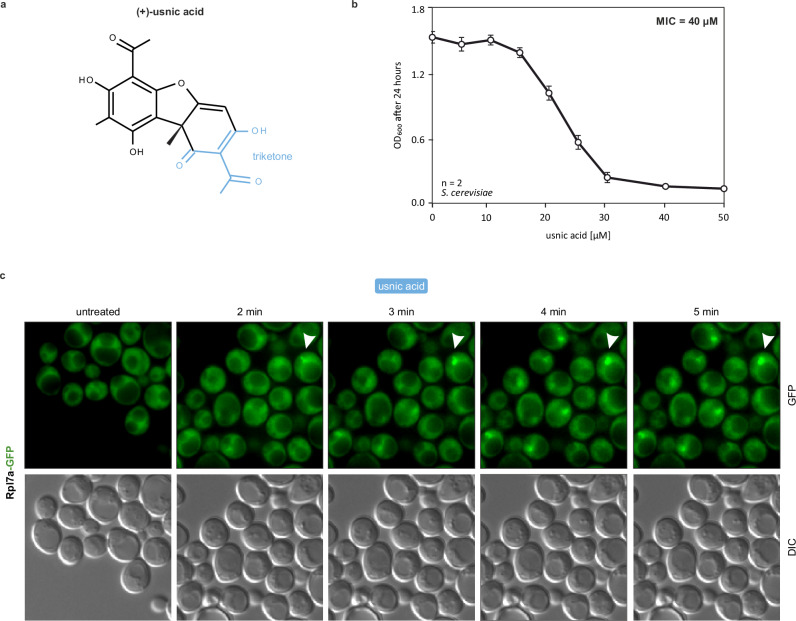
We recently identified usnic acid (Fig. 1a) as a novel candidate ribosome biogenesis inhibitor in yeast cells41. To characterize the effects of the drug on ribosome biogenesis in more detail, we determined the minimum inhibitory concentration (MIC) to be 40 µM (Fig. 1b) and then examined the nuclear accumulation kinetics of pre-60S particles upon usnic acid treatment. For this purpose, we treated a Rpl7a-GFP reporter strain for the localization of the large ribosomal subunit41 with usnic acid and monitored the localization of the fluorescence signal over a time range of 0 to 5 min (Fig. 1c). In untreated wildtype cells, Rpl7a-GFP was localized in the cytoplasm as expected for r-proteins mostly being part of mature ribosomes. Strikingly, after the addition of usnic acid, Rpl7a-GFP accumulated in the nucleus already after 2 min. In contrast, the 40S-reporter Rps9-GFP accumulated only after 30 min of usnic acid treatment in the nucleolus (Supplementary Fig. 2A). This suggests that usnic acid primarily acts on 60S maturation.
Fig. 1. Ribosome biogenesis inhibition kinetics of usnic acid.
a Chemical structure of (+)-usnic acid (triketone moiety highlighted in blue). b Minimum inhibitory concentration (MIC) of usnic acid for the yeast wildtype strain C303a grown in minimal media. Early logarithmic cells were diluted to OD600 0.01 and grown in microtiter plates in a Bioscreen device under continuous shaking and automated OD600 measurements every 30 min. The plotted values reflect the OD600 after 24 h growth in the presence of different concentrations of usnic acid. Mean and standard deviation of two biological replicates each measured in technical triplicates. c Kinetics of nuclear Rpl7a-GFP accumulation upon usnic acid treatment. The Rpl7a-GFP reporter strain41 was treated with 40 µM usnic acid and monitored by fluorescence microscopy in a time series (0–5 min). Source data are provided as a Source Data file for b and c.
Ribosome biogenesis is deeply embedded in the cell’s regulatory networks controlling growth and proliferation5. TORC1 kinase regulates the pathway at multiple stages42–46. To test if usnic acid treatment results in similar phenotypes as inhibition of the TOR-pathway, we treated a Nog1-GFP/Rpl7-mCherry reporter strain with the TORC1-specific inhibitor rapamycin and compared it to usnic acid (Supplementary Fig. 2B). As in case with the Rlp7-GFP reporter, effects of usnic acid were visible within the first minutes as indicated by nucleolar accumulation of both Rpl7-mCherry and Nog1-GFP. As reported previously46, nucleolar accumulation of Nog1-GFP was also observed upon rapamycin treatment, but in a much longer time frame than with usnic acid treatment. Strikingly, Rlp7-mCherry remained in the cytoplasm after rapamycin treatment. Taken together, these differences show that the effects of usnic acid on ribosome biogenesis are not caused by TORC1 inhibition.
To identify which functional groups of the compound are important for its inhibitory potential, we generated a set of usnic acid derivatives (Supplementary Fig. 3, Supplementary Table 1). Interestingly, most chemical modifications led to the loss of the inhibitory effect on 60S maturation. Only treatment with derivatives modified at the hydroxyl group at position 7 or 9, where either a methyl (Usnic_01, Usnic 02; Supplementary Fig. 3B) or a butyl group (Usnic_10, Usnic_11; Supplementary Fig. 3B) was attached, caused accumulation of Rpl7a-GFP in the nucleus and are hence active (Supplementary Fig. 3A). In contrast, attachment of a bulky benzyl group at these positions (Usnic_03-Usnic_05; Supplementary Fig. S3B) prohibited the shift of the Rlp7a-GFP signal (Supplementary Fig. 3A). The loss of inhibition after attaching amines to the carbonyl group within the triketone (Usnic_07-Usnic_09; Supplementary Fig. 3B) was consistent with the reported loss of toxicity by changing the triketone moiety in usnic acid37,38.
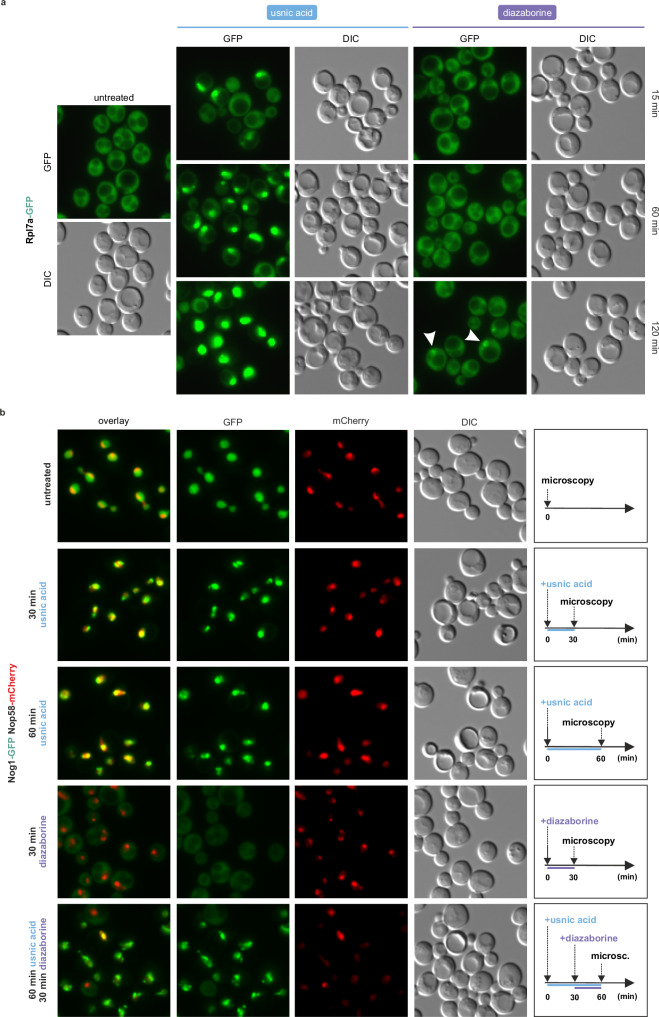
To relate the kinetics of 60S maturation inhibition by usnic acid to that of the well-characterized ribosome biogenesis inhibitor diazaborine27,47–49, we compared changes in the localization of r-proteins and shuttling factors (i.e., ribosome assembly factors that bind in the nucleus and are released in the cytoplasm) upon treatment (Fig. 2). In contrast to the fast onset of nucleolar Rlp7a-GFP retention upon usnic acid treatment, diazaborine treatment only resulted in nucleolar accumulation of the GFP signal after two hours and was less pronounced (Fig. 2a). This shows that the response of the reporter protein is dependent on the exact stage of perturbation. While diazaborine blocks the first 60S maturation step in the cytoplasm, usnic acid most likely acts earlier in maturation.
Fig. 2. Usnic acid shows different Rpl7a-GFP accumulation kinetics in the nucleus than diazaborine and leads to fragmented nucleolar Nog1-GFP signals.
a The Rpl7a-GFP reporter strain41 was treated with diazaborine (10 µg/ml) or usnic acid (40 µM) and the GFP signal was monitored via fluorescence microscopy. b Wildtype yeast cells expressing the chromosomally GFP-tagged shuttling factor Nog1 and the mCherry-tagged nucleolar factor Nop58 were treated with usnic acid (40 µM), diazaborine (10 µg/ml) or both inhibitors. Untreated cells served as control. Source data are provided as a Source Data file.
As shown previously, the GFP-tagged shuttling proteins Nog1, Bud20, and Tif6 are trapped in the cytoplasm upon diazaborine treatment (Fig. 2b and Supplementary Fig. 4)26,27,48. In contrast, usnic acid treatment did not shift Nog1-GFP to the cytoplasm even after 60 min but caused a dotted GFP signal in the nucleolus (Fig. 2b). Additionally, the Nop58-mCherry signal became more diffuse, indicating a disruption of nucleolar integrity likely as a sign of nucleolar stress. Fragmentation of nucleoli was observed previously upon inhibition of ribosome biogenesis in yeast and human cells10,33,47,50,51. This suggests that usnic acid causes a block in the nucleolus, and that the affected particles cannot progress further in the maturation cascade. Consequently, we hypothesized that the simultaneous addition of usnic acid and diazaborine should not lead to an accumulation of shuttling factors in the cytoplasm. Indeed, cells pretreated with usnic acid for 30 min prior to the addition of diazaborine retained Nog1-GFP in the nucleolus (Fig. 2b). We further analyzed two other shuttling factors, Tif6-GFP and Bud20-GFP, for their localization upon usnic acid treatment (Supplementary Fig. 4). Both proteins shifted to the cytoplasm with diazaborine treatment. However, when usnic acid was added prior to diazaborine, no shift was observed. While the Tif6-GFP strain treated with usnic acid showed similar granule structures as Nog1-GFP, Bud20-GFP showed the same overall nucleoplasmic localization as the untreated control. These results show that usnic acid blocks maturation after Nog1 and Tif6 incorporation but before Bud20 binds to the pre-ribosomal particle and the particle is exported. This confirms that usnic acid blocks maturation after Nog1 is incorporated and prior to the export of the particle to the cytoplasm.
These results show that, in contrast to diazaborine, usnic acid does not interfere with the recycling of these shuttling factors to the nucleolus but blocks the cascade before nucleolar pre-ribosomes are exported to the nucleoplasm. Usnic acid thus blocks the maturation cascade directly in the nucleolus.
Usnic acid treatment blocks nucleolar maturation prior to PET formation
Fluorescence microscopy hinted at an usnic acid-mediated block in nucleolar pre-60S maturation. To detect changes in particle composition, we isolated 60S precursors via the TAP-tagged assembly factor Nop7 (Fig. 3a). Nop7-TAP-containing particles mainly comprise 27SB pre-rRNA, but also low levels of 27SA2 and 7S pre-rRNA (Fig. 3b, Supplementary Fig. 5A). Thus, the Nop7-TAP particle population (summarizing all consecutive particle states that copurify with Nop7-TAP) spans from early nucleolar stages of 60S ribosome biogenesis until shortly after C2 cleavage in the nucleoplasm (Supplementary Fig. 1)52. Since usnic acid rapidly affected pre-60S maturation (Fig. 1), we purified and characterized Nop7-TAP particles after very short treatment periods (Fig. 3) to prevent secondary inhibition due to rebound effects26,27.
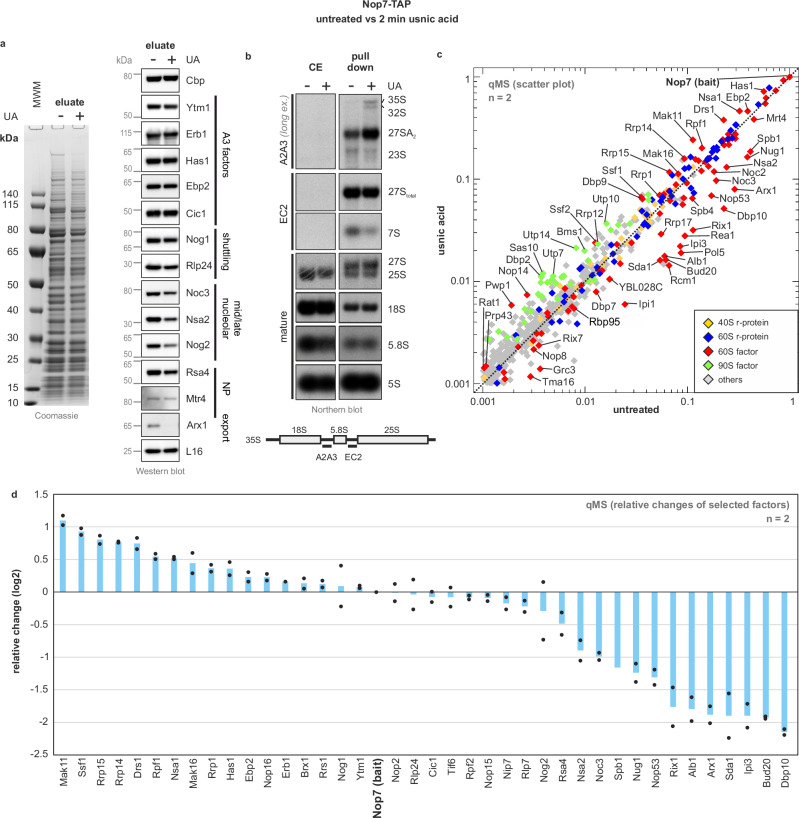
Fig. 3. Usnic acid causes accumulation of an early nucleolar population of Nop7-TAP pre-ribosomes and depletion of late maturation factors.
Early pre-ribosomal particles were isolated via Nop7-TAP as bait from untreated cells (−) or after 2 min of 60 µM usnic acid (UA) treatment (+). a Nop7-TAP eluates were analyzed using SDS-PAGE following Coomassie staining or Western blot (MWM = molecular weight marker). b Samples from a were investigated by Northern blotting, detected (pre)-rRNA species are marked on the right, the probes used are indicated on the left, with their binding sites depicted in the scheme. c Quantitative mass spectrometry (qMS) was used to determine the relative changes in protein levels due to usnic acid treatment. Normalized abundances from usnic acid-treated cells were plotted against the control sample from untreated cells in the form of a scatter plot. Selected maturation factors that increase (above the diagonal) or decrease (below the diagonal) upon inhibitor treatment are indicated. The protein levels were normalized to the bait Nop7, and all proteins with an abundance of >0.1% (>0.001) relative to the bait in the untreated sample are shown. The means of two biological replicates for each factor are shown (n = 2) (Supplementary Data 1). d The fold change (log2 transformed values) of the qMS data (from c) for selected maturation factors (>0.01 abundance to bait) is shown for two biological replicates (n = 2; mean plus individual points shown). Source data are provided as a Source Data file.
Purification of Nop7-TAP particles after 2 min of usnic acid treatment showed a marked decrease in late-joining nuclear maturation factors Noc3, Nsa2, Nog2 and Rsa4 (Fig. 3a). In addition, the shuttling protein Arx1 was depleted, which joins the pre-60S particle only after successful PET formation. In contrast, the levels of A3 factors (as tested for Ytm1, Erb1, Has1, Cic1 and Ebp2) mainly remained unaffected. Consistent with this result, Northern blotting showed a strong decrease in 7S and a slight decrease in the 27S(total) pre-rRNA, while earlier 27SA2, 32 A, and 35S pre-rRNAs accumulated on the particle (Fig. 3b and Supplementary Fig. 5A). The results of the Northern and Western blot suggest a blockage at the stage of 27SB pre-rRNA. The accumulation of the earlier 27SA2, 32S, and 35S pre-rRNAs likely already reflects rebound effects on earlier stages of ribosome biogenesis (see discussion26).
To obtain a more global picture of inhibition, we performed quantitative mass spectrometry (qMS) and monitored compositional changes in Nop7-TAP particles after short-term treatment (Fig. 3c, d, Supplementary Data 1). Consistent with a failed processing of 27SB pre-rRNA to 26S and 7S pre-rRNA and the Western blot data (Fig. 3a), we observed a strong decrease in late joining factors on the Nop7-TAP particle. This includes Arx1, the Rix1 complex, Sda1 but also Bud20 and Nop53, which already bind prior to the association of the Las1/Grc3 complex responsible for ITS2 cleavage14,17,53. Indeed, Nop53-TAP or Bud20-TAP purification showed that both proteins are mostly recovered in soluble form after usnic acid treatment instead of being associated with pre-ribosomal particles (Supplementary Fig. 6). This shows, that in the presence of usnic acid, precursor particles ready for Nop53 and Bud20 binding are lacking. Consequently, the drug-induced blockage must occur prior to their incorporation. In line with the decrease of Arx1, Rpl31, which is part of the PET rim, shows a severe reduction in Nop7-TAP particles (Supplementary Data 1). Consequently, the block occurs before this part of the 60S subunit is formed.
Consistent with an early nucleolar block, qMS revealed a strong increase in early joining factors such as Mak11, Rrp14, Rrp15, and Drs1 (Fig. 3d). On the other hand, the increased levels of 90S factors such as Utp7, Utp10, and Utp14 indicate rebound effects on initial maturation steps, as already suggested by the accumulation of 32S and 35S pre-rRNA (Fig. 3b, c). Moreover, Nsa1, which is released by the AAA-ATPase Rix754, also increased together with the “Nsa1-module” proteins Mak16, Rpf1, and Rrp114. Factors that normally associate shortly before the Nsa1-module leaves (e.g., Noc3 and Spb1; transition state C to D14) decreased after usnic acid treatment, suggesting that the maturation path is blocked before their entry. Finally, a pronounced reduction was also observed for the GTPase Nug1 and its interaction partner Dbp10. Indeed, the DEAD-box RNA helicase Dbp10 was the protein showing the strongest decrease upon usnic acid treatment, suggesting that the drug-induced blockage occurs before Dbp10 binds.
Taken together, these results show that usnic acid inhibits an early nucleolar maturation step prior to PET formation. This effect is reminiscent of, but not fully identical to, the previously described depletion of the so-called B-factors55 (see discussion for more details). The stage of inhibition can be narrowed down to a step shortly after Nog1 incorporation but before Mak11 dissociates and Dbp10, Nug1 and Nsa2 bind at the transition from state B to C14.
The nucleolar usnic acid block rapidly rebounds on the earliest steps in pre-60S formation
The maturation factor Nsa1 was enriched on the Nop7-TAP particle after usnic acid treatment. To resolve compositional changes in the nucleolus more precisely, we purified particles via Nsa1-TAP (Fig. 4). As Nsa1 resides shorter on the pre-60S particle than Nop7, it co-purifies a narrower population of nucleolar intermediates. In addition, we also examined the effects on even earlier pre-60S particles using Noc2-TAP as bait56,57 to evaluate the effects on different nucleolar particle populations. To capture the primary effects of usnic acid treatment, we purified Noc2- and Nsa1-TAP particles after short-term treatment (2 min). Noc2-TAP thereby together with Noc1 co-purifies the earliest known, co-transcriptional pre-60S particles, but also associates in complex with Noc3 with late nuclear pre-ribosomes14,58. The prevalence of one of the two complexes in our purifications can be monitored by western blotting using specific antibodies to Noc1 and Noc3 (Fig. 4). To monitor time-displaced secondary effects, we also analyzed compositional changes occurring after 15 min of inhibitor treatment. This strategy allowed us to detect if and how the primary block set by usnic acid rebounds on earlier particles.
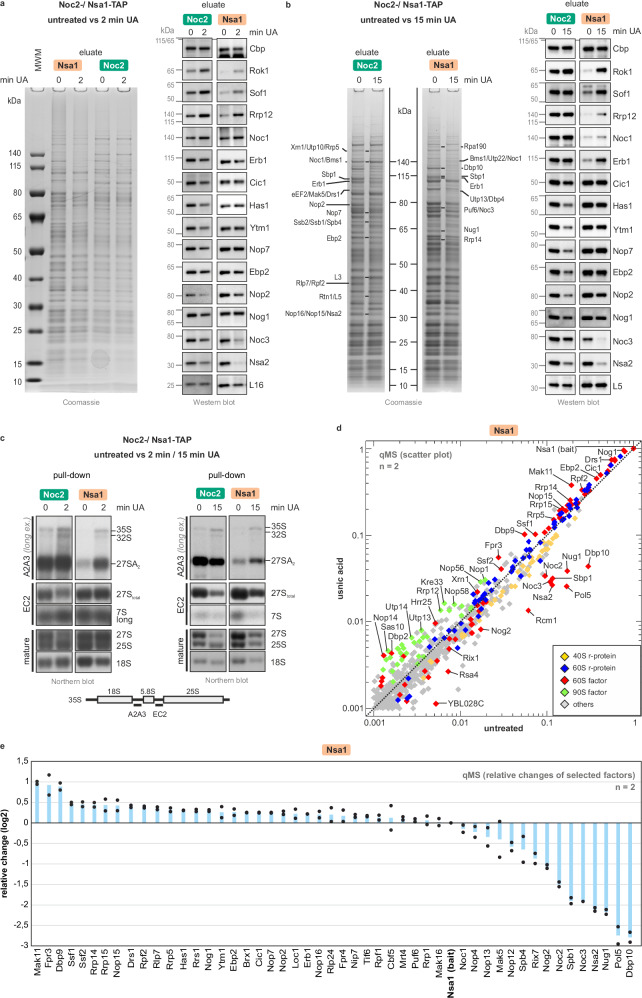
Fig. 4. The nucleolar usnic acid block rebounds on the earliest steps in pre-60S formation.
a, b Early nucleolar 60S particles were isolated either via Noc2-TAP or Nsa1-TAP from untreated cells (0, control) as well as from cells after short-term (a, 2 min) or long-term (b, 15 min) usnic acid (UA) treatment. Noc2-TAP and Nsa1-TAP eluates were analyzed by SDS-PAGE followed by Coomassie staining or Western Blot (MWM = molecular weight marker). For the 15 min treatment sample, selected bands were identified by mass spectrometry. c Noc2-TAP and Nsa1-TAP eluates (untreated, 2 or 15 min usnic acid) were analyzed by Northern blotting. Detected (pre)-rRNA species are marked on the right, and the probes used on the left with their binding sites depicted in the scheme. d Scatter plot analysis of Nsa1-TAP qMS data (untreated vs. 2 min usnic acid treated). Normalized abundances from treated cells were plotted against the untreated control sample. Protein levels were normalized to the bait protein Nsa1. Proteins with an abundance of > 0.1% (>0.001) relative to the bait protein in the untreated sample are shown. Selected maturation factors that increase (above the diagonal) or decrease (below the diagonal) upon inhibitor treatment are indicated. Means of two biological replicates for each factor are shown (n = 2) (Supplementary Data 2). e Fold change of selected maturation factors relative to the bait (>0.01 abundance to bait; log2 transformed values) of the qMS data shown for two biological replicates (n = 2; mean plus individual data points shown). Source data are provided as a Source Data file.
Consistent with increased 27SA2 pre-rRNA levels on Nop7-TAP particles (Fig. 3b), the 27SA2 pre-rRNA also accumulated on Nsa1-TAP particles within 2 min of usnic acid treatment. The 27S(total) pre-rRNA signal, in contrast, was unaffected (Fig. 4c, Supplementary Fig. 5B). In the Noc2-TAP particle, the 27SA2 pre-rRNA level was unaltered. Since the signal for 27S (total) pre-rRNA decreased, this indicates a shift to earlier 27SA2 pre-rRNA species (Fig. 4c). After 15 min of usnic acid treatment, the accumulation of 27SA2 pre-rRNA in Nsa1-TAP particles was even more pronounced. In the earlier Noc2-TAP particles, in contrast, both pre-rRNAs decreased.
In the Nsa1-TAP and Noc2-TAP particle populations, the 32S pre-rRNA increased after 2 min of usnic acid treatment, whereas after 15 min, only the progenitor, the 35S pre-rRNA, was detected. Thus, rebounding on earlier ribosome maturation steps occurs already after 2 min of usnic acid treatment and proceeds stepwise through 32S pre-rRNA to 35S pre-rRNA (Fig. 4c, Supplementary Fig. 1).
As obvious from the Western blots (Fig. 4a, b), the levels of the late joining factors Noc3 and Nsa2 decreased in Nsa1- and Noc2-TAP purifications. Furthermore, 90S factors Rok1, Rrp12, and Sof1 increased in both nucleolar particles after 2 and 15 min of usnic acid treatment, corroborating the increase in 35S and 32S pre-rRNA. The levels of Ytm1, Cic1, Has1, Nop7, Nog1, Ebp2, and Nop2 remained more or less unchanged in the Nsa1-TAP particle but decreased in the earlier Noc2-TAP particle after 2 and 15 min of usnic acid treatment. This indicates their entrapment on later Nsa1-TAP particles, which depletes them in earlier stages and might cause the observed rebound effect (Fig. 4b).
For a more global view, we analyzed purified Nsa1-TAP samples by qMS (untreated and 2 min usnic acid). Consistent with the Nop7-TAP purification, early binding pre-60S factors such as Mak11, Rrp14, Rrp15, Fpr3, and Dbp9 increased. In contrast, factors that already co-transcriptionally bind to 25S rRNA domains I and II (e.g., Erb1, Ytm1, Ebp2, Brx1) or to ITS2 (Cic1, Nop15, Rlp7 and Nop7) only slightly increased (Fig. 4d, e, Supplementary Data 2)14,15. Noc3, Spb1, Nsa2, Pol5, Nug1, and especially Dbp10 decreased, which is again consistent with the results from the Nop7-TAP purification (Fig. 3).
Impact of usnic acid on structure and composition of early nucleolar pre-60S particles
After characterizing the Nsa1-TAP population on a biochemical level, we were interested in structural changes upon usnic acid treatment. Therefore, we used single-particle cryo-EM to analyze Nsa1-TAP particles purified from untreated cells or cells treated with usnic acid for 2 min (Fig. 4). The micrographs of both datasets (Supplementary Table 2) were combined and (pre)-processed together (Supplementary Fig. 7). The final pre-60S particle stack was randomly split into three batches serving as technical replicates. We assessed compositional heterogeneity within our three replicates using the neural network-based algorithm cryoDRGN59 (Supplementary Fig. 8), representing the heterogeneity in the samples as a latent space. We used these latent space embeddings to cluster the particles using a Gaussian-Mixture Modeling (GMM) algorithm. We determined the number of clusters in our dataset in an unbiased manner by applying the elbow criterion method (Supplementary Fig. 9). This approach allows modeling of continuous particle distributions and a heuristic determination of the number of clusters/classes, in contrary to classical, highly discrete 3D classification algorithms. This workflow enabled us to systematically analyze the structural heterogeneity present in our datasets and assess the impact of usnic acid.
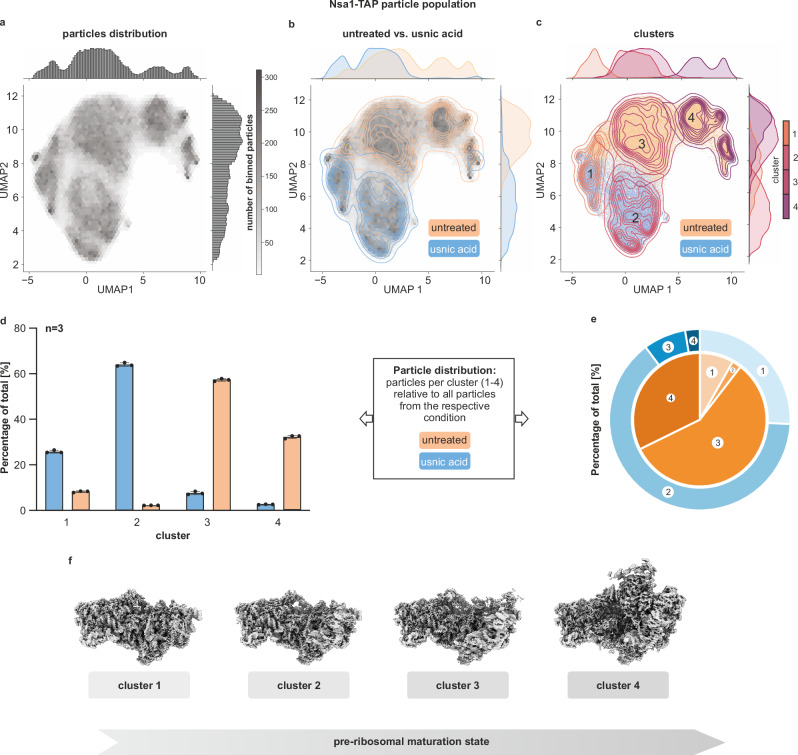
In our cryoDRGN analysis we identified and clustered 4 main maturation states present in each triplicate (Fig. 5 and Supplementary Fig. 8). The numbering from 1 to 4 reflects the degree of maturation, which is assessed by the compaction of the rRNA domains and the presence of maturation factors. To understand at which point usnic acid interferes with nucleolar pre-60S maturation, we wanted to display how the block globally affects the Nsa1-associated pre-ribosome population. To that end, we calculated how many particles of each dataset (treated/untreated) contributed to the reconstruction of each cluster (Fig. 5). In the untreated sample, the early clusters 1 and 2 represented only a minor part of all particles from this condition. The majority was found in clusters 3 and 4, with ~57% and ~32% of total particles, respectively. After usnic acid treatment, however, clusters 3 and 4 were strongly reduced and instead cluster 1 and 2 were predominant, with ~25% and ~64% of total particles, respectively. We thus detected a strong shift from cluster 3 to cluster 2. The accumulation of cluster 2 and depletion of subsequent states indicates that this subpopulation cannot be matured further. In agreement with our biochemical data, this demonstrates a massive shift in the steady-state distribution of the Nsa1-TAP particle population due to stalled further processing of cluster 2.
Fig. 5. Usnic acid blocks the maturation of nucleolar pre-60S ribosomes.
Early nucleolar 60S particles were isolated via Nsa1-TAP from untreated and 2 min usnic acid-treated cells. For each sample, one single-particle cryo-EM dataset was collected, and these datasets were merged for processing and quantitative analysis (a detailed description of the data processing workflow is given in the method section and Supplementary Figs. 7, 8). Structural heterogeneity was analyzed using cryoDRGN in three equally sized particle batches. Clustering using Gaussian Mixture Modeling (GMM) was performed to identify different pre-60S maturation states present in our datasets. a–c Representative cryoDRGN analysis of triplicate batch 1 (2D UMAP of batches 2 and 3 are shown in Supplementary Fig. 10). a Representative 2D UMAP of the particles’ hexagonal binned 8-dimensional latent space embeddings. b Kernel Density Estimate (kde) plot indicates particle embeddings from treated and untreated datasets. c Scatter plot, with each datapoint representing a single particles’ embedding and kde-plot representing clusters identified using GMM. Number of binned particles is indicated in gray scale, 2 min usnic acid treatment in blue, untreated in orange. The 4 clusters are represented in different shades of purple. d, e Relative particle contributions to each cluster from the usnic acid or untreated dataset. Source data are provided as a Source Data file. d Depicted as bar chart is the mean of the three batches, data points represent the three batches and error bars indicate standard deviation to the mean (n = 3). e Particles distribution as pie chart with mean percentage of total. f CryoEM reconstructions of the 4 clusters found in the cryoDRGN analysis, ordered by progressing ribosome maturation.
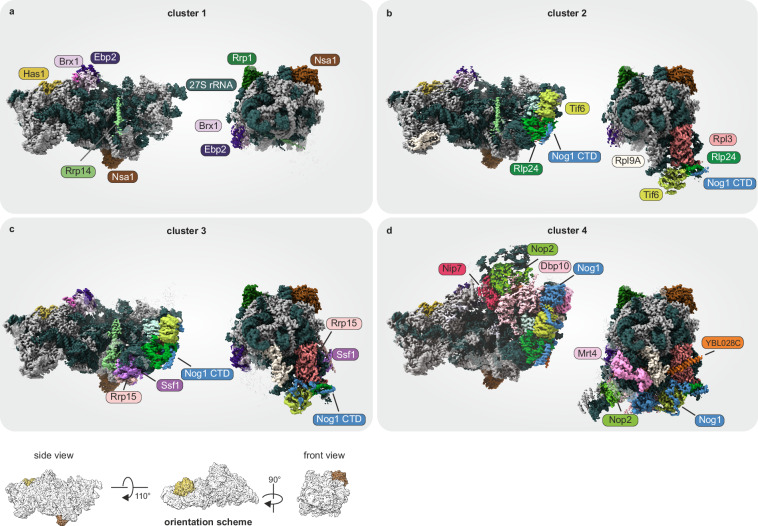
These four observed clusters mainly differ in the degree of 25S domain VI maturation and the presence of associated factors (Fig. 6). They largely resemble previously published high-resolution structures14,15. These models were fitted into our cryo-EM maps by flexible fitting and used for coloring of our maps. A more detailed description can be found in the material and methods section.
Fig. 6. GMM-clusters recapitulate nucleolar pre-60S biogenesis states.
Cryo-EM reconstructions of cluster 1 to 4 (a–d) ordered by biogenesis progression. Clusters are fitted with and colored based on PDB models 6EM3, 6EM4, 6C0F, and 8V87, respectively. Key factors changing between the clusters are highlighted. More detailed information about structural changes can be found in the main text and Supplementary Table 3. Density that has not been fitted with a model is shown transparently. As source data, the maps are uploaded to EMDB.
The least matured cluster 1, is defined by a still very flexible domain VI, which is not yet stably incorporated into the core particle. Cluster 1 closely resembles the published nucleolar state A (Fig. 6a; pdb 6em314). The foot structure is already fully assembled and Has1 is clearly visible from this stage on. Class 2 strongly resembles state B (Fig. 6b; pdb 6em414), characterized by beginning stabilization of domain VI with resolved densities corresponding to Rpl3, Tif6, the C-terminal Domain of Nog1 and Rlp24 (Fig. 6b). Cluster 3 is characterized by further stabilized domain VI. The distinct feature of cluster 3 is the further stabilization of the middle segment of Nog1 (approx. residue 347 to 448), the N-terminal part of Rrp14 together with Ssf1 and Rrp15, recapitulating state 2 (Fig. 6c; pdb 6C0F15) except that we did not resolve Mak11 in our structure. According to our biochemical data, these factors already interact at earlier stages, but cannot be visualized in cryo-EM due to flexibility.
In cluster 4 we observe the incorporation of Rpl19, Nsa2 and N-terminal parts of Nog1 (Fig. 6d14) and identified densities corresponding to Mrt4, the uncharacterized protein YBL028C (UniProt ID YBC8), and the core domain of Nog1. Accompanied by a more stable incorporation of Ebp2 and Nop2, the first steps of the L1 stalk assembly are clearly visible in cluster 4. In addition, we detect the helicase domains of Dbp10 at the location between Nop2 and the Nog1 GTPase domain where it was also found in human, S. cerevisiae, and Chaetomium thermophilum pre-ribosomes20,60,61. Cluster 4 thus closely resembles these recently published Dbp10-bound pre-60S particles.
We additionally refined the particles originating from the treated and untreated dataset for each cluster separately (Supplementary Fig. 11A). Comparing the combined, treated, and untreated reconstructions, we observed no major differences for clusters 1 to 3. Cluster 4, however, showed pronounced differences between the combined reconstruction and the reconstruction of the particles from the treated strain. The cryo-EM map of the particles from the treated strain resembles the published state D (Supplementary Fig. 11B14), characterized by a folded domain III and densities for Erb1 and Ytm1. Therefore, the minor fraction of treated particles in cluster 4 (~2.6%) is slightly further matured than their untreated fraction counterparts of cluster 4. In the particles from the untreated strain, this minor particle population might be shaded by averaging with particles of the more abundant Dpb10-containing state.
Discussion
Chemical probing with usnic acid dissects early nucleolar 60S assembly steps
Treatment of yeast cells with usnic acid rapidly sets a block in the nucleolar maturation cascade of the large ribosomal subunit. Our biochemical characterization and cryo-EM data both showed that after 2 min, the nucleolar Nsa1-TAP particle population shifted to earlier maturation stages. After this short treatment period, pre-60S particles are stalled in the nucleolus (Fig. 1c) and rRNA processing defects can be detected as well as pronounced compositional changes of purified nucleolar Nsa1-particles (Fig. 4–6).
When we compared usnic acid treatment with previously published effects of diazaborine27, we found that these two inhibitors block adjacent nucleolar stages of 60S maturation. However, diazaborine only acts indirectly on nucleolar maturation by prohibiting shuttling factor recycling from the cytoplasm. Through the absence of these factors in the nucleolus, the effect of diazaborine thus only rebounds on the nucleolar stage. Usnic acid, in contrast, seems to directly interfere with the compaction of pre-60S particles in the nucleolus after Nog1, Rlp24 and Tif6 associate and prior to Dbp10 association. This is corroborated by the very fast onset of usnic acid inhibition in the nucleolus contrasted by a delayed effect by diazaborine.
Besides the fact that the shuttling factors Nog1, Tif6 and Rlp24 were present on nucleolar particles from usnic acid treated cells, we also observed a pronounced increase in Mak11 (Figs. 3, 4). This contrasts with the decrease in Mak11 observed after diazaborine treatment and upon depletion of Nsa227,55. The accumulation of Mak11 indicates that the maturation path is blocked shortly before Mak11 leaves the particle. This step clearly precedes the incorporation of Nug1 and Dbp10 which are crucial to drive the transition from state C to state D and which both strongly decreased in our analysis (Figs. 3, 4, 7).
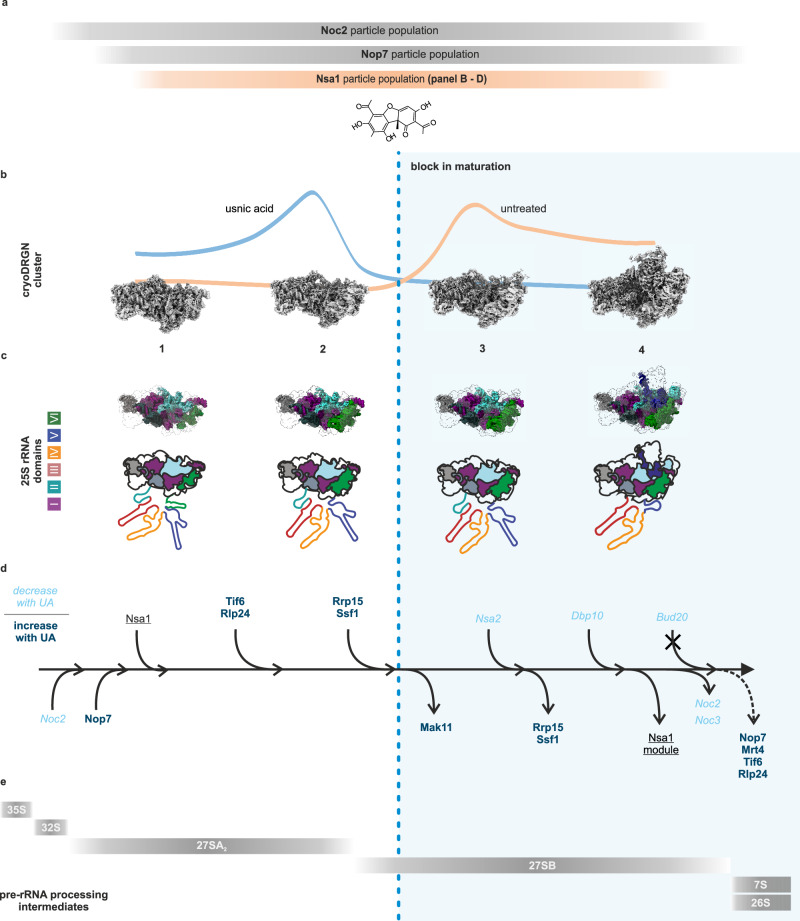
Fig. 7. Summarized effects of usnic acid on the Nsa1-TAP particle population.
a Comparison of the different particle populations isolated by different bait proteins used in this study. b–d Effects of usnic acid on the Nsa1-TAP particle population. b The (shifting) particle populations and the cryo-EM map of the respective clusters (1–4) (see Figs. 5, 6), c their 25S rRNA domain folding state, d maturation factor association/dissociation (see Fig. 4a, b, d, e) as well as e pre-rRNA processing states. The dashed blue line indicates the block in ribosome formation due to usnic acid treatment. For simplicity, rebound effects on earlier stages are not shown.
Usnic acid blocks the pre-60S particle transition from state B to C
With our quantitative cryo-EM approach using cryoDRGN, we could visualize how the global “landscape” of the Nsa1-TAP particle population changes with usnic acid. Predominant was the shift from cluster 3 containing the most particles, to the earlier cluster 2, indicating that the maturation cascade is blocked at this stage and cannot proceed (Figs. 5, 7). This shift is consistent with our qMS of the Nsa1-TAP particles that demonstrated a decrease in later-acting maturation factors (e.g., Spb1, Nug1, Nsa2, Noc3 or Dbp10), while earlier-acting factors (e.g., Mak11, Drs1 or Rrp5) increased.
The usnic acid-mediated blockage in cluster 2 (resembles State B14) to cluster 3 (resembles State C14) transition also results in a strong decrease of downstream particles in the treated strain as represented in cluster 4. This explains the strong reduction of Dbp10 in the qMS analysis (Fig. 4), which is found in cluster 4. Only very recently, structures from Chaetomium thermophilum20, S. cerevisiae14 and human cells25 revealed the exact binding site of Dbp10 on nucleolar pre-60S particles between the L1 stalk module and the other B-factors (e.g., Rlp24, Nsa2, Nog1, Nog2, Spb4 and Mak11). Since the Nog1 and Rlp24 levels did not change significantly after usnic acid treatment, these factors are present already prior to State B to C transition. In contrast, Mak11 shows a strong enrichment in the qMS, demonstrating that it already associated with the pre-60S particle prior state C. For Nsa2, we find a strong reduction in the qMS which is consistent with its presence in cluster 4. The overall changes in nucleolar particles isolated via Nop7-TAP after usnic acid treatment were highly similar to effects described for depletion of B-factors. The similar patterns of changes indicate that in both cases the maturation cascade is blocked at a similar stage. However, there were also significant differences between B-factor depletion and usnic acid treatment. While Nsa2 depletion resulted in decreased Mak11 levels on Nop7-TAP particles55, we observed a strong accumulation of Mak11 upon usnic acid treatment. This shows that the usnic acid-mediated block is upstream of Nsa2 binding as well as the release of Mak11. Mak11 accumulation was also observed after depletion of Nog162. However, as we still find Nog1 bound to Nsa1-TAP particles from usnic acid treated cells, the absence of Nog1 cannot explain the accumulation of Mak11 in our experiments. Thus, while the effects observed after usnic acid treatment are not fully congruent with depletion of B-factors like Nsa2 or Nog1, these similarities corroborate our hypothesis that usnic acid blocks transition from Nsa1-TAP state C to D.
Notably, our cryo-EM analysis show that the N-terminal parts of Nog1 are sequentially integrated from cluster 2 to 4. The complete incorporation of these parts of Nog1 might precede the release of Mak11 and represent a prerequisite for the binding of factors that are reduced upon usnic acid treatment (e.g. Dbp10 and Nsa2). We hypothesize that usnic acid may directly interfere with the incorporation of the N-terminal domain of Nog1, potentially explaining both Mak11 accumulation and depletion of later-binding factors. Further experimental validation is required to confirm this hypothesis.
Inhibition of state B to C transition by usnic acid rapidly rebounds on earlier maturation steps
After short-term treatment with usnic acid (2 min), we observed a block at the 27SB level, which rapidly caused an accumulation of 27SA2 pre-rRNA. This fast onset of inhibition allowed us to reconstruct how this rebounding effect is transmitted from the 27SB pre-rRNA-containing particles to earlier forms. The rapid reduction in A3 factors on early Noc2-TAP particles after 2 min demonstrates how fast recycling defects propagate when these factors are entrapped on downstream particles (Fig. 4). Consequently, all factors required for A2 and A3 processing are sequestered from emerging particles explaining the defect in 27SA2 pre-rRNA processing. The small steady-state levels of 27SA2 and 27SA3 pre-rRNAs (demonstrated by primer extension experiments of wildtype strains (e.g., shown in refs. 63–66)), highlight the fast processing of 27SA2 to 27SB pre-rRNA. In contrast, the 27SB pre-rRNA represents the prevailing form of the 27S(total) pre-rRNA (Supplementary Fig. 5). These 27SB pre-rRNA containing particles thus engage a large fraction of available early pre-60S maturation factors. A blockage at this stage will entrap these factors and therefore be transmitted rapidly to earlier particle intermediates (e.g., 27SA2 containing pre-ribosomes) that would require these factors for further maturation. Since these intermediates are present only in minor amounts in the cell, these secondary blockages will rapidly lead to further rebound effects on the very first stages of ribosome biogenesis and result in accumulation of the 35S pre-rRNA after 15 min of treatment. Interestingly, the central hub protein Erb1 showed a strong accumulation and a strong depletion on the Nsa1-TAP particles and the earlier Noc2-TAP particles, respectively (Fig. 4). This could indicate that Erb1 plays a major role in rebounding. We speculate that this arrangement evolved to coordinate the maturation steps at different stages of pre-ribosome assembly to avoid executing resource-demanding maturation steps when the pathway is blocked downstream. Besides the above discussed shifts in pre-rRNA species to earlier forms, we did not detect elevated levels of aberrant pre-rRNA species (e. g., 24S, 23S, 22S, 21S pre-rRNA) or degraded pre-rRNAs (Figs. 3, 4, Supplementary Fig. 5), therefore usnic acid likely does not cause disassembly of a pre-ribosomal intermediate, which usually involves (aberrant) pre-rRNA degradation by the exosome67,68.
Quantitative analysis of the structural heterogeneity using cryoDRGN
The cryoDRGN analysis of our single-particle analysis data proved to be a valuable tool to visualize and quantify global shifts of pre-ribosomal particle populations. This can help to foster our understanding of the consequences of blocks in ribosome maturation downstream of transcription, caused by substances or mutations. It can strengthen the understanding that pre-ribosomal particle purifications do not contain one single, sharply separated maturation state, but a mixture of all states associated with the used bait protein. Our quantitative analysis highlighted that all Nsa1-TAP-associated states in our sample were affected, as the maturation process works like a (mostly) linear assembly line with regulatory linkages between up- and downstream processes.
This analysis however bears certain limitations and possible pitfalls that need to be considered. First, the cryoDRGN encoder is susceptible to outlier particles such as ice contaminations, empty picks and broken particles. To prevent these outliers from impacting downstream clustering, they must be carefully excluded during pre-processing. This can lead to differences in the final particles of separately processed datasets, distorting their quantitative analysis. Therefore, we ensured comparability of our datasets by combining both conditions (usnic acid treated and untreated control) during pre-processing.
Further, we focused on modeling the structural heterogeneity in our dataset in an unbiased and reproducible manner. We avoided overfitting of our data by determining the number of clusters in our dataset in a heuristic manner. This, however, might lead to remaining structural heterogeneity within the clusters presumably reflected in the differences between treated and untreated particles of cluster 4.
One general limitation in single-particle cryo-EM analyses of pre-ribosomes is that averaging is only possible for ribosomal proteins, rRNA domains or assembly factors when and if they are stably incorporated. Consequently, such analyses are missing proteins that are associated loosely with the particles or within flexible rRNA domains. Additionally, proteins can dissociate during cryo-EM sample preparation and, thus, escape their structural investigation. Indeed, we observe enrichment of some factors (e.g., Nog1, the Ssf1-Rrp15-Rrp14 module and Mak11) in our qMS analysis following usnic acid treatment, while they are not, or only fuzzily depicted in the cryo-EM maps. Discrepancies in detecting individual factors with different approaches (e.g., cryo-EM and LC-MS/MS), reflect technical limitations of these methodologies. Deviations to previous published structures are most likely caused by differences in the sample preparation, in cell growth (e.g. starvation) or due to different purification conditions (e.g. composition of the tag or variances in the buffer)14,15. Thus, complementary biochemical and cell biological methods are needed for an in-depth assessment of pre-ribosomal particles.
Ribosome biogenesis inhibitors and their potential for cancer therapy
Inhibitors blocking ribosome biogenesis downstream of transcription are promising for cancer therapy (reviewed in refs. 7,69–73)74. Most tumors upregulate ribosome biogenesis to meet demands of rapid proliferation which can be used as prognostic parameter74–79. Non-malignant cells can sustain temporal inhibition of this pathway due to ribosome stability. This selectivity against tumor cells results in a favorable therapeutic window, as already demonstrated for the Pol I inhibitor CX-546180.
However, compounds that block the pathway downstream of transcription bear additional therapeutic potential. Such inhibitors could burden tumor cells more than Pol I inhibitors by stabilizing p53 through unincorporated ribosomal proteins (reviewed in refs. 81,82). They may avoid DNA-damage response, reducing side effects and relapses. Indeed, usnic acid and various of its derivatives showed pronounced inhibition of tumor cells38,83–87. The inhibitory effect of usnic acid on eucaryotic ribosome biogenesis shown here, is a step further in the mechanistic understanding of this potential cancer therapy.
Methods
Yeast strains
All yeast strains used in this study are listed in Supplementary Table 4. Chromosomal gene fusions were generated by homologous recombination using linear PCR-amplified DNA fragments to transform the respective yeast strain as described88,89.
Media and growth conditions
Yeast strains were grown in synthetic dextrose (SD, 20 g/l dextrose, 5 g/l (NH4)2SO4, 1.4 g/l yeast nitrogen base) medium supplemented with the appropriate supplement mix (CSM, in complete or drop-out formulation, MP Biomedicals) at 30 °C, adjusted to pH5.5. For microscopy, additional 0.4 mM adenine was added to prevent formation of the red pigment characteristic of ade2 strains.
Inhibitor preparation
Diazaborine (6-methyl-2(propane-1-sulfonyl)-2H-thieno[3,2-D][1,2,3]diazaborinin-1-OL, Novartis) was prepared as a 100 mg/ml stock solution in ddH2O. (+)-Usnic acid (2,6-diacetyl-7,9-dihydroxy-8,9b-dimethyldibenzofuran-1,3-(2H,9bH)-dion, Sigma Aldrich) was dissolved at 5 mg/ml in ddH2O upon the addition of 0.5 % NaOH (10 M) to improve solubility. Usnic acid derivatives were modified at distinct residues with different modifications, including methylation, benzylation and acylation (Supplementary Table 1) and dissolved in DMSO. Rapamycin (LC-Laboratories) was prepared as 5 mg/ml stock in DMSO and used for microscopy at 200 nM concentration.
Determination of the MIC
Yeast strains were grown to the exponential growth phase (OD600 of 0.5–0.6). Right after dilution with fresh medium to an OD600 of 0.01 (1.5 × 105 cfu), cells were treated with the indicated concentrations of usnic acid. Optical density (OD600) was periodically measured over 48 h, while shaking the cultures at 30 °C in a Bioscreen C Pro Automated Microbiology Growth Curve Analysis system (Oy Growth Curves Ab Ltd.) in technical triplicates. For determining the minimal inhibitory concentration, the mean values of the OD600 after 24 h (Kovalska-Krochmal and Dudek-Wicher, 2021) were plotted over the concentration of the inhibitor.
Fluorescence microscopy
Yeast strains were grown to the exponential growth phase (OD600 0.4–0.6). Subsequently, the respective inhibitors (diazaborine (10 µg/ml) and/or usnic acid (40 µM or 60 µM) or usnic acid derivates (60 µM) were added and incubation continued for the indicated periods of time. Fluorescence microscopy experiments were performed using a Leica DM6 B Microscope equipped with a ×100/1.4 Plan APO objective and a high-resolution DFC9000GT camera. For imaging, LASX premium software (Leica) was used.
Tandem affinity purification
Cultivation, treatment and harvesting
To purify pre-60S particles of distinct maturation stages, yeast cells expressing C-terminal TAP-tag fusions of the assembly factors Noc2, Nsa1, Nop7, Nop53, and Bud20 were used. Cells were grown to late logarithmic growth phase (OD600 of 0.9–1.1) and treated with 60 µM usnic acid for the indicated time periods. 60 µM usnic acid were used to ensure complete inhibition in baffled flasks with very high aeration. This is based on our experience with the inhibitor diazaborine, that showed higher drug tolerance of yeast cells under such growth conditions. After inhibitor treatment, cells were harvested by centrifugation at 6000 × g for 3 min at 4 °C. Cells were washed with 20 ml cold water and centrifuged at 1250 × g for 10 min at 4 °C, before the pellets were stored at −80 °C.
Purification
For particle isolation, cell pellets corresponding to 2 L of culture were resuspended in one volume of lysis buffer (20 mM HEPES/KOH pH 7.5, 10 mM KCl, 2.5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.5 mM PMSF and 1x FY-Protease inhibitor (Serva)). Cells were disrupted in a bead mill (Merkenschlager) with 1.5× pellet volume of 0.6 mm glass beads (Sartorius AG) for 4 min with periodic CO2 cooling. After centrifugation (41,000 × g, 30 min, 4 °C), the supernatant was incubated for 60–90 min with magnetic IgG beads, which were prepared by crosslinking rabbit IgG to epoxy-activated magnetic beads as described27,90,91. After extensive washing, 20 % of the beads were removed and stored for subsequent RNA isolation and analysis. Pre-ribosomal particles were eluted from the residual beads with tobacco etch virus (TEV) protease containing cleavage buffer (lysis buffer with 100 mM NaCl). TEV-eluates were analyzed by SDS-PAGE, Western blotting and qMS.
SDS-PAGE and western blotting
Proteins were separated using precast 4–12% NuPAGE gels (Novex Life Technologies) and visualized using the NOVEX® Colloidal Blue Staining Kit (Invitrogen). Selected bands were identified by MS analysis.
For Western blotting, the proteins were blotted on a polyvinylidene fluoride membrane (Carl Roth GmbH) using the tank-blot-system (Trans-BlotTMCell, Bio-Rad) with CAPS transfer buffer (10 mM CAPS, pH 11.0, 10% Methanol). All antibodies were diluted in TST buffer with 1% milk powder (50 mM Tris/HCl, pH 7.4, 0.1% Tween20, 0.15% NaCl) (Supplementary Table 5). Detection via chemiluminescence was performed with the Clarity™ Western Blotting Detection Reagent (Bio-Rad) and the ChemiDoc™ Touch Imaging System (Bio-Rad). The PageRuler™ Prestained Protein Ladder from Thermo Fisher was used as molecular weight marker.
Northern blot analysis of RNAs present in pre-ribosomal particles
For analysis of the RNAs present in the TAP-purified pre-60S particles, 20% of washed magnetic IgG beads with bound ribosomal precursor particles were used for RNA isolation by phenol chloroform and isoamylalcohol (P:C:I, 25:24:1) extraction and ethanol precipitation. The resulting RNA was dissolved in ddH2O and diluted with RNA loading buffer for subsequent Northern blotting.
RNA samples were denatured at 65 °C for 10 min and separated on a 1.5% agarose gel (containing 20 mM 3-(N-morpholino) propanesulfonic acid, 5 mM sodium acetate, 1 mM EDTA, 0.75% formaldehyde, pH 7.0) at 60 V for 7 to 8 h. The RNA was transferred overnight onto Hybond-N+ nylon membranes (GE Healthcare) by capillary transfer in 20× SSC (3 M NaCl, 0.3 M Na3 - citrate). Northern blot probes (Supplementary Table 6) were radiolabeled with 32Pγ-ATP (30 µCi, Hartmann Analytic) via the T4-Polynucleotid Kinase reaction at 37 °C for 2–3 h in reaction buffer A for T4 PNK (Thermo Scientific). Hybridization with labeled probes was performed overnight at 42 °C (EC2 probe 37 °C) in buffer containing 0.5 M Na2HPO4, pH 7.2, 7% SDS, and 1 mM EDTA. After three subsequent washing steps with buffer containing 40 mM Na2HPO4, pH 7.2, and 1% SDS, signals were detected by exposing X-ray films. Membranes were recovered using 1% SDS prior to the next hybridization.
Statistics, reproducibility, and integrity
All microscopy data as well as pre-ribosomal particle purifications, including subsequent SDS-PAGE, western blot and Northern blot analysis (main and Supplementary Figs.), were independently repeated at least twice with the same output.
Uncropped gels, blots and microscopy images for all Figures and Supplementary Figs. are shown in the source data file. For each western or Northern blot one single membrane was used to consecutively detect all antibodies or probes.
Mass spectrometric analysis
Two datasets (two samples each) were analyzed by MS and both datasets contained two independent experiments each (n = 2), resulting in a total of eight analyzed samples. The samples were eluates from tandem affinity purifications as described in detail above, therefore the control samples always were pre-ribosomal particles from untreated cells.
Proteins were proteolysed with LysC and trypsin with a filter-aided sample preparation procedure (FASP) as described92,93. Acidified eluted peptides were analyzed on a QExactive HF or HF-X mass spectrometer (Thermo Fisher Scientific) coupled online to a UItimate 3000 RSLC nano-HPLC (Dionex). Samples were automatically injected and loaded onto the C18 trap cartridge and after 5 min eluted and separated on the C18 analytical column (nanoEase MZ HSS T3, 100 Å, 1.8 µm, 75 µm × 250 mm; Waters) by a 95 min non-linear acetonitrile gradient at a flow rate of 250 nl/min. MS spectra were recorded at a resolution of 60,000 with an automatic gain control (AGC) target of 3e6 and a maximum injection time of 30 or 50 ms from 300 to 1500 m/z. From the MS scan, the 10 or 15 most abundant peptide ions were selected for fragmentation via HCD with a normalized collision energy of 27 or 28, an isolation window of 1.6 m/z, and a dynamic exclusion of 30 s. MS/MS spectra were recorded at a resolution of 15,000 with an AGC target of 1e5 and a maximum injection time of 50 ms. Unassigned charges and charges of +1 and > +8 were excluded from precursor selection.
Protein identification and label-free quantification
Raw spectra were imported into Progenesis QI software (version 4.1). After feature alignment and normalization, spectra were exported as Mascot Generic files and searched against the yeast UniProt database (6721 sequences) with Mascot (Matrix Science, version 2.6.2) with the following search parameters: 10 ppm peptide mass tolerance and 20 mmu fragment mass tolerance, one missed cleavage allowed, carbamidomethylation was set as fixed modification, methionine oxidation, and asparagine or glutamine deamidation were allowed as variable modifications. A Mascot-integrated decoy database search calculated an average false discovery rate <0.5% for PSMs when searches were performed applying the Mascot percolator score and a significance threshold p < 0.05.
Peptide assignments were reimported into Progenesis QI software, and the abundances of all unique peptides allocated to each protein were summed. The resulting normalized abundances of the individual proteins were used for calculation of fold-changes of protein ratios between untreated and usnic acid treated samples. Therefore, normalized abundances of each detected protein were then referenced to the bait protein in each sample and the values in Log scale were shown in a scatter plot using Statgraphics 18 software. Only the proteins with an abundance above 0.001 are shown. For a rough overview, the detected proteins were manually assigned to following groups: 40S r-protein, 60S r-protein, 60S factor, 90S factor and others.
Cryo-EM
Grid preparation
For single-particle cryo-EM analysis, pre-ribosomal particles were purified from untreated cells as well as after 2 min of treatment with 60 µM usnic acid, as described above. A 4 µl sample of both treated or untreated Nsa1-particles was added onto Pre-floated Quantifoil R3.5/1 grids (2 nm carbon film) and incubated for 30 s at 4 °C and 75% humidity. Frontside blotting was performed for using the automatic plunge freezer EM GP1 (Leica) for 2 s before plunging the grids into liquid ethane. Grids were stored in liquid nitrogen until data collection.
Data collection
Cryo-EM data were collected on an FEI Titan Krios G3i operating at 300 kV in nanoprobe energy-filtered transmission electron microscopy at a nominal magnification of ×81,000 with 1.07 Å/pixel using a Gatan K3 BioQuantum direct electron detector with a slit width of 20 eV. The detector was operated in counting mode using hardware binning and dose fractionation, recording movies with an electron dose of 60 e-/Å2 and 54 frames, resulting in 3608 (Usnic) and 3987 (untreated) movies, respectively. Both datasets were collected on two consecutive days.
Image processing
Initial image processing was performed in cryoSPARC v3.294. Motion correction and CTF estimation were performed using Patch Motion Correction and CTFFIND495, respectively. The datasets were merged and curated for cryolo picking (v1.696). Here ~600 particles from 6 micrographs of each dataset were used to fine tune cryolo’s general model. Resulting particle coordinates were used to extract the particles with a box size of 448 pixels binned to 166 pixels (2.89 Å/px). Particle order was randomized and two rounds of 2D classification were performed. Ab-initio models were used to further clean the particle stack by heterogeneous refinement. Pre-60S particles were reextracted without binning and subjected to another round of heterogeneous refinement followed by local CTF refinement and homogeneous refinement.
Analysis of structural heterogeneity with CryoDRGN
Structural heterogeneity was analyzed using the neural network algorithm cryoDRGN (v0.3.2 & v3.159). Poses generated by homogeneous refinement and respective particles (downsampled to a box size of 128 (3.75 Å/pixel)) were used for training of the network with a latent variable of 10 dimensions for 50 epochs. Ice-contaminated and outlier particles were filtered by the absolute of the latent variable ||z|| > 2 stdev(||z||) + mean(||z||). Particles were reimported into cryoSPARC (v4.4.1) and refined. We excluded less well aligning particles which interfered with the clustering via their cryoSPARC-cross-correlation values. This value has an arbitrary unit and is found in the particle.cs file after refinement. Using the cryoSPARC python library cryoSPARC tools (v4.4.1, https://github.com/cryoem-uoft/), we excluded particles with a value below 250 and refined remaining particles. This was repeated twice, before local CTF refinement followed by homogeneous refinement. Next, particles were split into three equally sized batches serving as technical replicates. Each batch was homogeneously refined in cryoSPARC and used for another round of cryoDRGN analysis with a latent variable of 8 dimensions and particles downsampled to a box size of 128 pixel (3.75 Å/pixel). Dimensionality reduction was performed by UMAP for latent space visualization.
Gaussian Mixture Modeling in cryoDRGN latent space
Clustering was performed using Gaussian-Mixture Modeling on the latent space embeddings created by cryoDRGN. The number of clusters was determined using the elbow method after assessing cluster variation by calculating sum of squared errors (SSE) (Supplementary Fig. 9). We clustered with 1 to 20 clusters thrice with a randomly generated seed for each iteration. The SSE was calculated via Eq. (1).
| 1 |
Where is the value of each particle and is the mean within the respective cluster. As suggested by the elbow method, we used 4 clusters to cluster each batch with a unique randomly generated seed. The resulting particle stacks were refined in cryoSPARC, and clusters of identical maturation were combined for further refinements. The particle stacks were separated into treated and untreated stacks and refined. Resulting half maps were used to estimate the local resolution (Supplementary Figs. 12–14). Maps were post-processed using DeepEMhancer using the ‘highRes’ model provided by the developers97.
Quantitative analysis of structural heterogeneity
To visualize the global shift of the Nsa1-TAP particle population, we assessed the number of particles originating from the treated and untreated datasets contributing to each cluster and replicate. These values are listed in Supplementary Table 7. For each cluster, the percentage of the total was calculated for each replicate. These values were used to calculate the mean and standard deviation for each cluster.
Model fitting
The EM-density maps of clusters 1–4 were fitted with published PDB models 6EM3, 6EM4, 6C0F, and 8V87, respectively, using rigid body fitting in ChimeraX (v1.7)98,99. Cluster 1 and 2 were complemented with the atomic model of Rrp14, Has1 and Brx1 originating from PDB-6C0F, which were modeled either incompletely or not at all in the original models. Cluster 2 was complemented with the C-terminal Nog-1 helix from PDB-6C0F. Cluster 3 was complemented with the model of the 60S ribosomal protein L9A from PDB-6EM1. Cluster 4 was complemented with Nsa1, Rpf1 and Rrp1 from PDB-6EM1. Proteins and rRNA that had no apparent density in our maps were deleted from the fitted models. In the case of cluster 4, this includes the Ytm1/Erb1 module as well as proteins and rRNA of domain IV. A complete list of proteins fitted in our maps can be found in Supplementary Table 3.
The resulting models were further fitted into our experimental maps by flexible fitting using the namdinator pipeline100 and refined using phenix real-space refinement executed on the Namdinator web server (https://namdinator.au.dk/) with 2000 minimization steps for classes 3 and 4 and 3000 minimization steps for classes 1 and 2. The resulting models were then used to color our experimental maps using the color zone function in ChimeraX.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Michael A. McAlear, Micheline Fromont-Racin, Philipp Milkereit, Arlen W. Johnson, Sabine Rospert, Ed Hurt, C. Yam, Günter Daum, Wolfgang Zachariae, Katrin Karbstein, Juan P. G. Ballesta, Mercedes Dosil, Miguel Remacha und Jesus de la Cruz for sharing strains or providing antibodies. We thank the members of the Bergler lab and the Haselbach lab for their helpful discussion. We thank Ellen Zhong for helpful discussions about the quantitative cryoDRGN analysis. This research was supported by the Scientific Service Units of IST Austria through resources provided by the Electron Microscopy Facility. This research was funded in whole, or in part, by the Austrian Science Foundation grants [10.55776/P32977], [10.55776/P29451] and [10.55776/P32536] (to H.B.). Research at the IMP is generously supported by Boehringer Ingelheim and the Austrian Research Promotion Agency (Headquarter grant FFG-852936). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author contributions
L.K. and H.B. designed the study. L.K., M.G., G.Z. purified pre-ribosomal particles as well as L.K., M.G. isolated and analyzed RNA for all biochemical characterization. L.K., M.G. performed the fluorescence microscopy. J.M.-P. and S.M.H. performed the qMS analysis. M.V. and R.B. synthesized usnic acid derivatives. I.G. and V.-V.H. collected single-particle cryo-EM data. L.G., M.P., H.B., and M.G. processed the cryo-EM data. L.G. and D.H. designed the cryo DRGN-based quantitative analysis. L.G. performed clustering and particle refinements. L.K., M.P., M.G., L.G., and H.B. wrote the manuscript and prepared the figures. All authors have read and approved the final version of the manuscript.
Peer review
Peer review information
Nature Communications thanks David Taylor, John Woolford and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Data availability
Strains generated in this study are available from the corresponding authors upon request. For cryo-EM model building, published pre-60S models PDB-6EM3, PDB-6EM4, PDB-6C0F, PDB-6EM1 and PDB-8V87 were used. Motion-corrected micrographs with annotated particle locations were deposited in EMPIAR with the accession number EMPIAR-12237. The cryo-EM maps of clusters 1 to 4 with the combined datasets were deposited in the EMDB with the accession numbers EMD-51122, EMD-51126, EMD-51128, and EMD-51129. The reconstructions for the treated dataset were deposited with the accession numbers EMD-51175, EMD-51176, EMD-51177, and EMD-51178, while the untreated reconstructions of reported clusters can be found via EMD-51164, EMD-51165, EMD-51166, EMD-51167. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE101 partner repository with the dataset identifier PXD046691. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lisa Kofler, Lorenz Grundmann.
Contributor Information
David Haselbach, Email: david.haselbach@imp.ac.at.
Helmut Bergler, Email: helmut.bergler@uni-graz.at.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51754-3.
References
- 1.Klinge, S. & Woolford, J. L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol.20, 116–131 (2019). 10.1038/s41580-018-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baßler, J. & Hurt, E. Eukaryotic ribosome assembly. Annu. Rev. Biochem.88:8.1–8.26, 26 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack, K. E. & Bohnsack, M. T. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J.38, e100278 (2019). 10.15252/embj.2018100278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dörner, K., Ruggeri, C., Zemp, I. & Kutay, U. Ribosome biogenesis factors—from names to functions. EMBO J.n/a, e112699 (2023). 10.15252/embj.2022112699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warner, J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci.24, 437–440 (1999). 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- 6.Kang, J. et al. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Sig. Transduct. Target Ther.6, 1–22 (2021). 10.1038/s41392-021-00728-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penzo, M., Montanaro, L., Treré, D. & Derenzini, M. The ribosome biogenesis—cancer connection. Cells8, 55 (2019). 10.3390/cells8010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turi, Z., Lacey, M., Mistrik, M. & Moudry, P. Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging (Albany NY)11, 2512–2540 (2019). 10.18632/aging.101922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elhamamsy, A. R., Metge, B. J., Alsheikh, H. A., Shevde, L. A. & Samant, R. S. Ribosome biogenesis: a central player in cancer metastasis and therapeutic resistance. Cancer Res.82, 2344–2353 (2022). 10.1158/0008-5472.CAN-21-4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lafita-Navarro, M. C. & Conacci-Sorrell, M. Nucleolar stress: from development to cancer. Semin. Cell Dev. Biol.136, 64–74 (2023). 10.1016/j.semcdb.2022.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt, K. E., Macintosh, J., Bernard, G. & Trainor, P. A. RNA polymerases I and III in development and disease. Semin. Cell Dev. Biol.136, 49–63 (2023). 10.1016/j.semcdb.2022.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jerome, M. S., Nanjappa, D. P., Chakraborty, A. & Chakrabarty, S. Molecular etiology of defective nuclear and mitochondrial ribosome biogenesis: Clinical phenotypes and therapy. Biochimie10.1016/j.biochi.2022.11.001 (2022). [DOI] [PubMed]
- 13.Gamalinda, M. et al. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev.28, 198–210 (2014). 10.1101/gad.228825.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kater, L. et al. Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell171, 1599–1610.e14 (2017). 10.1016/j.cell.2017.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanghai, Z. A. et al. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature556, 126–129 (2018). 10.1038/nature26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, Y., Musalgaonkar, S., Johnson, A. W. & Taylor, D. W. Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome. Nat. Commun.10, 958 (2019). 10.1038/s41467-019-08880-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kater, L. et al. Construction of the central protuberance and l1 stalk during 60S subunit biogenesis. Mol. Cell10.1016/j.molcel.2020.06.032 (2020). [DOI] [PubMed]
- 18.Liang, X. et al. Structural snapshots of human pre-60S ribosomal particles before and after nuclear export. Nat. Commun.11, 3542 (2020). 10.1038/s41467-020-17237-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kargas, V. et al. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. eLife8, e44904 (2019). 10.7554/eLife.44904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau, B. et al. Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis. EMBO Rep.n/a, e56910 (2023). 10.15252/embr.202356910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ameismeier, M., Cheng, J., Berninghausen, O. & Beckmann, R. Visualizing late states of human 40S ribosomal subunit maturation. Nature558, 249–253 (2018). 10.1038/s41586-018-0193-0 [DOI] [PubMed] [Google Scholar]
- 22.Cheng, J. et al. The nucleoplasmic phase of pre-40S formation prior to nuclear export. Nucleic Acids Res.50, 11924–11937 (2022). 10.1093/nar/gkac961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper, N. J., Burnside, C. & Klinge, S. Principles of mitoribosomal small subunit assembly in eukaryotes. Nature614, 175–181 (2023). 10.1038/s41586-022-05621-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh, S., Broeck, A. V., Miller, L., Chaker-Margot, M. & Klinge, S. Nucleolar maturation of the human small subunit processome. Science373, eabj5338 (2021). 10.1126/science.abj5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broeck, A. V. & Klinge, S. Principles of human pre-60S biogenesis. Science381, eadh3892 (2023). [DOI] [PubMed]
- 26.Kofler, L., Prattes, M. & Bergler, H. From snapshots to flipbook—resolving the dynamics of ribosome biogenesis with chemical probes. Int. J. Mol. Sci.21, 2998 (2020). 10.3390/ijms21082998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zisser, G. et al. Viewing pre-60S maturation at a minute’s timescale. Nucleic Acids Res.46, 3140–3151 (2018). 10.1093/nar/gkx1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kos, M. & Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell37, 809–820 (2010). 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lempiäinen, H. & Shore, D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol.21, 855–863 (2009). 10.1016/j.ceb.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 30.Kawashima, S. A. et al. Potent, reversible, and specific chemical inhibitors of eukaryotic ribosome biogenesis. Cell167, 512–524.e14 (2016). 10.1016/j.cell.2016.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shore, D. & Albert, B. Ribosome biogenesis and the cellular energy economy. Curr. Biol.32, R611–R617 (2022). 10.1016/j.cub.2022.04.083 [DOI] [PubMed] [Google Scholar]
- 32.Shore, D., Zencir, S. & Albert, B. Transcriptional control of ribosome biogenesis in yeast: links to growth and stress signals. Biochem. Soc. Trans.49, 1589–1599 (2021). 10.1042/BST20201136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albert, B. et al. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. eLife8, e45002 (2019). 10.7554/eLife.45002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tye, B. W. et al. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. eLife8, e43002 (2019). 10.7554/eLife.43002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araújo, A. A. S. et al. Review of the biological properties and toxicity of usnic acid. Nat. Prod. Res.29, 2167–2180 (2015). 10.1080/14786419.2015.1007455 [DOI] [PubMed] [Google Scholar]
- 36.Alahmadi, A. A. Usnic acid biological activity: history, evaluation and usage. Int. J. Basic Clin. Pharmacol.6, 2752–2759 (2017). 10.18203/2319-2003.ijbcp20175072 [DOI] [Google Scholar]
- 37.Luzina, O. A. & Salakhutdinov, N. F. Usnic acid and its derivatives for pharmaceutical use: a patent review (2000–2017). Expert Opin. Ther. Pat.28, 477–491 (2018). 10.1080/13543776.2018.1472239 [DOI] [PubMed] [Google Scholar]
- 38.Guzow-Krzemińska, B., Guzow, K. & Herman-Antosiewicz, A. Usnic acid derivatives as cytotoxic agents against cancer cells and the mechanisms of their activity. Curr. Pharm. Rep.5, 429–439 (2019). 10.1007/s40495-019-00202-8 [DOI] [Google Scholar]
- 39.Solárová, Z. et al. Anticancer potential of lichens’ secondary metabolites. Biomolecules10, 87 (2020). 10.3390/biom10010087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, H., Xuan, M., Huang, C. & Wang, C. Advances in research on bioactivity, toxicity, metabolism, and pharmacokinetics of usnic acid in vitro and in vivo. Molecules27, 7469 (2022). 10.3390/molecules27217469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Awad, D. et al. Inhibiting eukaryotic ribosome biogenesis. BMC Biol.17, 46 (2019). 10.1186/s12915-019-0664-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kos-Braun, I. C., Jung, I. & Koš, M. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLOS Biol.15, e2000245 (2017). 10.1371/journal.pbio.2000245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talkish, J., Zhang, J., Jakovljevic, J., Horsey, E. W. & Woolford, J. L. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucl. Acids Res.40, 8646–8661 (2012). 10.1093/nar/gks609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laribee, R. N., Hosni-Ahmed, A., Workman, J. J. & Chen, H. Ccr4-not regulates RNA polymerase I transcription and couples nutrient signaling to the control of ribosomal RNA biogenesis. PLOS Genet.11, e1005113 (2015). 10.1371/journal.pgen.1005113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiter, A. et al. Reduction in ribosomal protein synthesis is sufficient to explain major effects on ribosome production after short-term TOR inactivation in Saccharomyces cerevisiae. Mol. Cell Biol.31, 803–817 (2011). 10.1128/MCB.01227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honma, Y. et al. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J.25, 3832–3842 (2006). 10.1038/sj.emboj.7601262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pertschy, B. et al. Diazaborine treatment of yeast cells inhibits maturation of the 60S ribosomal subunit. Mol. Cell. Biol.24, 6476–6487 (2004). 10.1128/MCB.24.14.6476-6487.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loibl, M. et al. The drug diazaborine blocks ribosome biogenesis by inhibiting the AAA-ATPase Drg1. J. Biol. Chem.289, 3913–3922 (2014). 10.1074/jbc.M113.536110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prattes, M. et al. Structural basis for inhibition of the AAA-ATPase Drg1 by diazaborine. Nat. Commun.12, 3483 (2021). 10.1038/s41467-021-23854-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol.22, 165–182 (2021). 10.1038/s41580-020-0272-6 [DOI] [PubMed] [Google Scholar]
- 51.Yang, K., Yang, J. & Yi, J. Nucleolar Stress: hallmarks, sensing mechanism and diseases. Cell Stress2, 125–140 (2018). 10.15698/cst2018.06.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harnpicharnchai, P. et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell8, 505–515 (2001). 10.1016/S1097-2765(01)00344-6 [DOI] [PubMed] [Google Scholar]
- 53.Bassler, J. et al. The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol. Cell. Biol.32, 4898–4912 (2012). 10.1128/MCB.00910-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kressler, D., Roser, D., Pertschy, B. & Hurt, E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol.181, 935–944 (2008). 10.1083/jcb.200801181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biedka, S. et al. Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J. Cell Biol.217, 2503–2518 (2018). 10.1083/jcb.201711037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milkereit, P. et al. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell105, 499–509 (2001). 10.1016/S0092-8674(01)00358-0 [DOI] [PubMed] [Google Scholar]
- 57.Chaker-Margot, M., Hunziker, M., Barandun, J., Dill, B. D. & Klinge, S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol.22, 920–923 (2015). 10.1038/nsmb.3111 [DOI] [PubMed] [Google Scholar]
- 58.Sanghai, Z. A., Piwowarczyk, R., Broeck, A. V. & Klinge, S. A co-transcriptional ribosome assembly checkpoint controls nascent large ribosomal subunit maturation. Nat. Struct. Mol. Biol.30, 594–599 (2023). 10.1038/s41594-023-00947-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong, E. D., Bepler, T., Berger, B. & Davis, J. H. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods18, 176–185 (2021). 10.1038/s41592-020-01049-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanden Broeck, A. & Klinge, S. Principles of human pre-60S biogenesis. Science381, eadh3892 (2023). 10.1126/science.adh3892 [DOI] [PubMed] [Google Scholar]
- 61.Cruz, V. E., Weirich, C. S., Peddada, N. & Erzberger, J. P. The DEAD-Box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60s ribosome biogenesis. 10.1101/2023.11.01.565222 (2023). [DOI] [PMC free article] [PubMed]
- 62.Saveanu, C. et al. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol.23, 4449–4460 (2003). 10.1128/MCB.23.13.4449-4460.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kressler, D., de la Cruz, J., Rojo, M. & Linder, P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol.18, 1855–1865 (1998). 10.1128/MCB.18.4.1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torchet, C. & Denmat, S. H.-L. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA6, 1498–1508 (2000). 10.1017/S1355838200000558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunbar, D. A., Dragon, F., Lee, S. J. & Baserga, S. J. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl Acad. Sci.97, 13027–13032 (2000). 10.1073/pnas.97.24.13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pertschy, B. et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol.27, 6581–6592 (2007). 10.1128/MCB.00668-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kilchert, C., Wittmann, S. & Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol.17, 227–239 (2016). 10.1038/nrm.2015.15 [DOI] [PubMed] [Google Scholar]
- 68.Zinder, J. C. & Lima, C. D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev.31, 88–100 (2017). 10.1101/gad.294769.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Derenzini, M., Montanaro, L. & Trerè, D. Ribosome biogenesis and cancer. Acta Histochem.119, 190–197 (2017). 10.1016/j.acthis.2017.01.009 [DOI] [PubMed] [Google Scholar]
- 70.Gaviraghi, M., Vivori, C. & Tonon, G. How cancer exploits ribosomal RNA biogenesis: a journey beyond the boundaries of rRNA transcription. Cells8, 1098 (2019). 10.3390/cells8091098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gilles, A. et al. Targeting the human 80S ribosome in cancer: from structure to function and drug design for innovative adjuvant therapeutic strategies. Cells9, 629 (2020). [DOI] [PMC free article] [PubMed]
- 72.Bursać, S., Prodan, Y., Pullen, N., Bartek, J. & Volarević, S. Dysregulated ribosome biogenesis reveals therapeutic liabilities in cancer. Trends Cancer7, 57–76 (2021). 10.1016/j.trecan.2020.08.003 [DOI] [PubMed] [Google Scholar]
- 73.Ni, C. & Buszczak, M. Ribosome biogenesis and function in development and disease. Development150, dev201187 (2023). 10.1242/dev.201187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Derenzini, M., Montanaro, L. & Treré, D. What the nucleolus says to a tumour pathologist. Histopathology54, 753–762 (2009). 10.1111/j.1365-2559.2008.03168.x [DOI] [PubMed] [Google Scholar]
- 75.Song, J. et al. Functional role of RRS1 in breast cancer cell proliferation. J. Cell Mol. Med.22, 6304–6313 (2018). 10.1111/jcmm.13922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao, P. et al. Genomic gain of RRS1 promotes hepatocellular carcinoma through reducing the RPL11-MDM2-p53 signaling. Sci. Adv.7, eabf4304 (2021). 10.1126/sciadv.abf4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan, X. et al. NOP14-mediated ribosome biogenesis is required for mTORC2 activation and predicts rapamycin sensitivity. J. Biol. Chem.300, 105681–105696 (2024). [DOI] [PMC free article] [PubMed]
- 78.Lu, C., Liao, W., Huang, Y., Huang, Y. & Luo, Y. Increased expression of NOP14 is associated with improved prognosis due to immune regulation in colorectal cancer. BMC Gastroenterol.22, 207 (2022). 10.1186/s12876-022-02286-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tian, Q.-S., Cheng, J. & Bao, Z.-J. NOP14 as a potential predictor of adult-type diffuse glioma prognosis and immunotherapy, is related to cell migration, proliferation, and CD8+T cell infiltration. Front. Biosci.29, 104 (2024). 10.31083/j.fbl2903104 [DOI] [PubMed] [Google Scholar]
- 80.Drygin, D. et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res.71, 1418–1430 (2011). 10.1158/0008-5472.CAN-10-1728 [DOI] [PubMed] [Google Scholar]
- 81.Golomb, L., Volarevic, S. & Oren, M. p53 and ribosome biogenesis stress: the essentials. FEBS Lett.588, 2571–2579 (2014). 10.1016/j.febslet.2014.04.014 [DOI] [PubMed] [Google Scholar]
- 82.Liu, Y., Deisenroth, C. & Zhang, Y. RP–MDM2–p53 pathway: linking ribosomal biogenesis and tumor surveillance. Trends Cancer2, 191–204 (2016). 10.1016/j.trecan.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geng, X. et al. Usnic acid induces cycle arrest, apoptosis, and autophagy in gastric cancer cells in vitro and in vivo. Med. Sci. Monit.24, 556–566 (2018). 10.12659/MSM.908568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun, T. X. et al. Usnic acid suppresses cervical cancer cell proliferation by inhibiting PD-L1 expression and enhancing T-lymphocyte tumor-killing activity. Phytother. Res.35, 3916–3935 (2021). 10.1002/ptr.7103 [DOI] [PubMed] [Google Scholar]
- 85.Yang, Y. et al. Inhibitory activity of (+)-usnic acid against non-small cell lung cancer cell motility. PLoS One11, e0146575 (2016). 10.1371/journal.pone.0146575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pyrczak-Felczykowska, A. et al. Synthesis of usnic acid derivatives and evaluation of their antiproliferative activity against cancer cells. J. Nat. Prod.82, 1768–1778 (2019). 10.1021/acs.jnatprod.8b00980 [DOI] [PubMed] [Google Scholar]
- 87.Kumar, K., Mishra, J. P. N. & Singh, R. P. Usnic acid induces apoptosis in human gastric cancer cells through ROS generation and DNA damage and causes up-regulation of DNA-PKcs and γ-H2A.X phosphorylation. Chem. Biol. Interact.315, 108898 (2020). 10.1016/j.cbi.2019.108898 [DOI] [PubMed] [Google Scholar]
- 88.Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast14, 953–961 (1998). [DOI] [PubMed] [Google Scholar]
- 89.Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast21, 947–962 (2004). 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- 90.Oeffinger, M. et al. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods4, 951–956 (2007). 10.1038/nmeth1101 [DOI] [PubMed] [Google Scholar]
- 91.Ohmayer, U. et al. Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One8, e68412 (2013). 10.1371/journal.pone.0068412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wiśniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods6, 359–362 (2009). 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- 93.Grosche, A. et al. The proteome of native adult Muller glial cells from murine retina. Mol. Cell Proteomics15, 462–480 (2015). [DOI] [PMC free article] [PubMed]
- 94.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 95.Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol.192, 216–221 (2015). 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol.2, 1–13 (2019). 10.1038/s42003-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol.4, 1–8 (2021). 10.1038/s42003-021-02399-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci.30, 70–82 (2021). 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kidmose, R. T. et al. Namdinator—automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ6, 526–531 (2019). 10.1107/S2052252519007619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2022). 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Strains generated in this study are available from the corresponding authors upon request. For cryo-EM model building, published pre-60S models PDB-6EM3, PDB-6EM4, PDB-6C0F, PDB-6EM1 and PDB-8V87 were used. Motion-corrected micrographs with annotated particle locations were deposited in EMPIAR with the accession number EMPIAR-12237. The cryo-EM maps of clusters 1 to 4 with the combined datasets were deposited in the EMDB with the accession numbers EMD-51122, EMD-51126, EMD-51128, and EMD-51129. The reconstructions for the treated dataset were deposited with the accession numbers EMD-51175, EMD-51176, EMD-51177, and EMD-51178, while the untreated reconstructions of reported clusters can be found via EMD-51164, EMD-51165, EMD-51166, EMD-51167. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE101 partner repository with the dataset identifier PXD046691. Source data are provided with this paper.