Abstract
During a survey of the diversity of lignicolous fungi in Yunnan Province, China, we collected and identified five microfungi species from dead woody litters of Castanopsis trees in terrestrial habitats. Through both morphological comparisons and phylogenetic analyses of multi-gene sequences, we identified two taxa as new species and three collections as new host records within Pleosporales. Pseudolophiostomalincangensesp. nov. is introduced as a sexual morph in Lophiostomataceae, Pleopunctumbaoshanensesp. nov. is introduced as a hyphomycetous fungi in Phaeoseptaceae, and Paraphomaaquatica as a first report of sexual morph in Paraphoma. In addition, Occultibambusakunmingensis and Pleopunctummegalosporum were isolated for the first time from the dead twigs of Castanopsisdelavayi and C.calathiformis, respectively. Comprehensive morphological descriptions, illustrations, and phylogenetic analysis results are provided for the above-mentioned species.
Key words: Hyphomycetes, new species, phylogeny, sexual morph, taxonomy
Introduction
Castanopsis is an evergreen tree belonging to Fagaceae and represents one of the largest genera with approximately 134 species, predominantly distributed across tropical and subtropical regions of Asia (Tan et al. 2023). Notably, 58 species are native to China, with 30 being endemic. Castanopsis species are valued for their timber and edible nuts, contributing significantly to the economy (Huang et al. 1999; Tang et al. 2005). Fungal diversity associated with Castanopsis has been extensively documented across various countries, including China, Korea, India, Indonesia, Japan, Nepal, Papua New Guinea, Thailand, and the United States (Crawford et al. 1987; Tang et al. 2005; Duong et al. 2008; Osono et al. 2020; Jayawardena et al. 2020). The presence of fungi on Castanopsis trees has been widely introduced, with approximately 360 records worldwide representing 220 species across 35 different Castanopsis species (Duong et al. 2008). Tang et al. (2005) reported 38 fungal taxa during a study on decaying leaves of Castanopsisfissa in Hong Kong, China highlighting the rich fungal diversity within this genus. These fungi encompass endophytes and saprobes found on different parts of Castanopsis trees, such as ectomycorrhizal, woody branches, fallen trunks, bark, and leaves (Inácio et al. 2005; Gao et al. 2016; Hyde et al. 2016; Ren et al. 2022; Pang et al. 2023).
Yunnan Province, located in southwestern China, boasts significant biological diversity attributed to its complex topography, highly variable climate, and lush vegetation (Feng and Yang 2018). This region covers an extensive area of 394,000 square with approximately 94% comprising mountainous terrain (Asian Development Bank 2012). Over the past decade, there has been a surge in interest in studying microfungi in Yunnan Province, with numerous studies focusing on leaf litter fungi and lignicolous freshwater fungi (Cai et al. 2002; Luo et al. 2018; Hapuarachchi et al. 2019; Dong et al. 2020). Recent discoveries have unveiled several new taxa from Dothideomycetes and Sordariomycetes inhabiting woody litter in terrestrial habitats, such as Diatrypaceae, Didymosphaeriaceae, Hermatomycetaceae, Hysteriaceae, Monoblastiaceae and Phaeoseptaceae (Maharachchikumbura et al. 2021; Mortimer et al. 2021; Ren et al. 2022; Wanasinghe and Mortimer et al. 2022). However, many studies lack proper identification and phylogenetic data, underscoring the need to re-evaluate various species in this region (Mortimer et al. 2021).
Pleosporales was established by Barr (1987) and is recognized as the largest order within the class Dothideomycetes, constituting a quarter of all species (Wijayawardene et al. 2022). This order has a remarkable diversity, comprising 91 families and 614 genera (Wijayawardene et al. 2022). Taxonomically, pleosporalean taxa exhibit versatility in ecological niches, being found as epiphytes, endophytes, parasites, hyperparasites, lichenized organisms, or saprobes across a wide range of habitats worldwide (Wanasinghe et al. 2020; Yang et al. 2023). Studies have highlighted the discovery of numerous new pleosporalean species from freshwater, marine, and terrestrial environments (Brahmanage et al. 2020). Morphologically, the sexual morph of Pleosporales is characterized by perithecial ascomata, typically with a papillate apex, ostiolate, cellular pseudoparaphyses, and bitunicate asci. The asexual morphs encompass both coelomycetes and hyphomycetes (Hongsanan et al. 2020).
The present study aims to describe two novel fungal species and three new host records collected from dead woody litter in Baoshan and Lincang of Yunnan Province, China. This involves morphological illustrations and multi-gene phylogenetic analyses utilizing ML and BI methods to confirm the phylogenetic placement. The study aims to contribute to fungal diversity and ecology in Yunnan Province while providing valuable insights into the taxonomy and phylogenetics of woody litter fungi.
Materials and methods
Sample collection, observation, and isolation
Decayed woody samples were collected from mixed forest areas in China (Yunnan Province) during the rainy season (July) and brought to the laboratory in separate zip-lock plastic bags. Specimens were examined using a stereomicroscope (Olympus SZ61, Tokyo, Japan). Micro-morphological characteristics were photographed using a Canon EOS 600D (Tokyo, Japan) digital camera mounted on a Nikon ECLIPSE 80i (Tokyo, Japan) compound microscope. All microscopic measurements were taken using the Tarosoft (R) Image Frame Work v.09, and the measurements were reported as minimum–maximum values and average values. Images were processed with Adobe Photoshop CS6 software v.13 (Adobe Systems, San Jose, CA, USA). Single-spore isolation was used to obtain pure cultures, following the methods described by Ren et al. (2022). Herbarium materials were deposited at the Herbarium of Cryptogams Kunming Institute of Botany, Academia Sinica (HKAS), Kunming, China, and living cultures were deposited at the Kunming Institute of Botany Culture Collection (KUNCC), Kunming, China. Faces of fungi (Jayasiri et al. 2015) and Index Fungorum (2024) numbers were obtained for the new taxa.
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from the mycelium grown on PDA at 25 °C for four weeks using Biospin Fungus Genomic DNA Extraction Kit (BioFlux®) (Hangzhou, P.R. China). Five gene regions, including internal transcribed spacer region (ITS), large subunit nuclear ribosomal (LSU), small subunit ribosomal RNA (SSU), translation elongation factor 1-alpha gene (tef1-α), and RNA polymerase II second largest subunit (rpb2) were amplified with primers ITS5/ITS4 (White et al. 1990), LR0R/LR5 (Vilgalys and Hester 1990), NS1/NS4 (White et al. 1990), 983F/2218R (Rehner and Buckley 2005) and fRPB2-5F/fRPB2-7cR (Liu et al. 1999), respectively. The PCR thermal cycle programs for SSU, LSU, ITS, tef1-α, and rpb2 were set as described in Wanasinghe et al. (2021). The quality of PCR products was checked on 1% agarose gel electrophoresis stained with ethidium bromide. The PCR products were sent for sequencing at Qingke Company, Kunming City, Yunnan Province, China. The sequences were deposited in GenBank.
Phylogenetic analyses
Sequences exhibiting high similarities (>90%) were identified through BLASTn searches to determine the closest match to the taxa. Representative sequences were individually blasted and the initial results from BLASTn searches show our five taxa belong to Pseudolophiostoma in Lophiostomataceae, Occultibambusa in Occultibambusaceae, Pleopunctum in Phaeoseptaceae, and Paraphoma in Phaeosphaeriaceae. Thus, four different datasets were prepared and analysed in this study based on recent publications (Gomzhina et al. 2020; Phukhamsakda et al. 2020; Magaña-Dueñas et al. 2021; Guarnaccia et al. 2022; Yu et al. 2022; Xu et al. 2023). The sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/) and the accession numbers are listed in Table 1. The newly generated sequences were assembled by BioEdit 7.2.3 (Hall 1999). The individual gene regions were separately aligned in the MAFFT v.7 web server (http://mafft.cbrc.jp/alignment/server/) (Katoh et al. 2019). The alignments of each gene were improved by manually deleting the ambiguous regions plus gaps and combined using BioEdit 7.2.3. The final alignments were converted to NEXUS format (.nxs) using Clustal X version 1.81 (Thompson et al. 1997) and processed for Bayesian and maximum parsimony analysis. The FASTA format was changed into PHY format via the Alignment Transformation Environment (ALTER) online program (http://www.sing-group.org/ALTER/) and used for maximum likelihood (ML) analysis.
Table 1.
Names, strain numbers, and corresponding GenBank accession numbers of the taxa used in the phylogenetic analysis.
| Taxon name | Strain number | GenBank accession numbers | |||||
|---|---|---|---|---|---|---|---|
| SSU | ITS | LSU | tef1-α | rpb2 | tub2 | ||
| Alpestrisphaeriajonesii | GAAZ 54-1 | KX687755 | KX687757 | KX687753 | KX687759 | NA | NA |
| Alpestrisphaeriajonesii | GAAZ 54-2 | KX687756 | KX687758 | KX687754 | KX687760 | NA | NA |
| Alpestrisphaeriaterricola | SC-12T | JX985749 | JN662930 | JX985750 | NA | NA | NA |
| Angustimassarinaacerina | MFLUCC 14-0505T | NG_063573 | KP888637 | NR_138406 | KR075168 | NA | NA |
| Angustimassarinapopuli | MFLUCC 13-0034T | NG_061204 | KP888642 | KP899137 | KR075164 | NA | NA |
| Biappendiculisporajaponica | KT 573T | AB618686 | LC001728 | AB619005 | LC001744 | NA | NA |
| Biappendiculisporajaponica | KT 794 | AB618688 | LC001730 | AB619007 | LC001746 | NA | NA |
| Biappendiculisporajaponica | KT 686 | AB618687 | LC001729 | AB619006 | LC001745 | NA | NA |
| Brunneofusisporaclematidis | MFLUCC 17-2070 | MT226685 | MT310615 | MT214570 | MT394692 | MT394629 | NA |
| Brunneofusisporainclinatiostiola | CGMCC 3.20403 | MZ964884 | MZ964866 | MZ964875 | OK061075 | OK061069 | NA |
| Brunneofusisporasinensis | KUMCC 17-0030 | MH393556 | MH393558 | MH393557 | NA | MH395329 | NA |
| Capulatisporasagittiforme | KT 1934T | AB618693 | AB369268 | AB369267 | LC001756 | NA | NA |
| Coelodictyosporiummuriforme | MFLUCC 13-0351T | KP899127 | KP899136 | KP888641 | KR075163 | NA | NA |
| Coelodictyosporiumpseudodictyosporium | MFLUCC 13-0451T | NA | KR025858 | KR025862 | NA | NA | NA |
| Crassiclypeusaquaticus | KT 970T | LC312472 | LC312501 | LC312530 | LC312559 | LC312588 | NA |
| Crassiclypeusaquaticus | KH 104 | LC312470 | LC312499 | LC312528 | LC312557 | LC312586 | NA |
| Decaisnellaformosa | BCC 25616 | GQ925833 | GQ925846 | NA | GU479851 | NA | NA |
| Decaisnellaformosa | BCC 25617 | GQ925834 | GQ925847 | NA | GU479850 | NA | NA |
| Desertiserpenticahydei | SQUCC 15092T | MW077163 | MW077147 | MW077156 | MW075773 | NA | NA |
| Dimorphiopsisbrachystegiae | CPC 22679T | NA | KF777160 | KF777213 | NA | NA | NA |
| Ernakulamiakrabiensis | MFLUCC 18-0237 | MK347880 | MK347773 | MK347990 | NA | NA | NA |
| Ernakulamiaxishuangbannaensis | KUMCC 17-0187 | MH260354 | MH275080 | MH260314 | NA | NA | NA |
| Flabellascomaaquaticum | KUMCC 15-0258 | MN304832 | MN304827 | MN274564 | MN328898 | MN328895 | NA |
| Flabellascomacycadicola | KT 2034T | LC312473 | LC312502 | LC312531 | LC312560 | LC312589 | NA |
| Flabellascomafusiforme | MFLUCC 18-1584 | NA | MN304830 | MN274567 | MN328902 | NA | NA |
| Guttulisporacrataegi | MFLUCC 14-0993 | KP899126 | KP899135 | KP888640 | KR075162 | NA | NA |
| Guttulisporacrataegi | MFLUCC 13-0442T | KP899125 | KP899134 | KP888639 | KR075161 | NA | NA |
| Kiskunsagiaubrizsyi | REF121T | MK589351 | JN859341 | MK589359 | MK599325 | NA | NA |
| Lentistomaaquaticum | MFLUCC 18-1275 | MT864320 | MT627697 | MN913723 | MT954370 | NA | NA |
| Lentistomabipolare | KT 3056 | LC312484 | LC312513 | LC312542 | LC312571 | LC312600 | NA |
| Lentistomabipolare | CBS 115375 | LC312477 | LC312506 | LC312535 | LC312564 | LC312593 | NA |
| Leptopariesmagnoliae | MFLU 18-1291 | ON870915 | ON878077 | ON870390 | NA | NA | NA |
| Leptopariespalmarum | KT 1653T | LC312485 | LC312514 | LC312543 | LC312572 | LC312601 | NA |
| Leptosphaeriaheterospora | AFTOL-ID 1036 | NA | GQ203795 | AY016369 | DQ497609 | DQ497615 | NA |
| Lignosphaeriafusispora | MFLUCC 11-0377T | NA | KP888646 | NR_164233 | NA | NA | NA |
| Lophiohelichrysumhelichrysi | IT-1296T | KT333437 | KT333435 | KT333436 | KT427535 | NA | NA |
| Lophiopoaceaparamacrostomum | MFLUCC 11-0463T | KP899122 | NA | KP888636 | NA | NA | NA |
| Lophiopoaceawinteri | KT 740 | AB618699 | JN942969 | AB619017 | LC001763 | JN993487 | NA |
| Lophiopoaceawinteri | KT 764 | AB618700 | JN942968 | AB619018 | LC001764 | JN993488 | NA |
| Lophiostomamultiseptatum | CBS 623.86 | GU296163 | NA | GU301833 | NA | GU371791 | NA |
| Lophiostomamultiseptatum | KT 604/JCM17668T | AB618684 | LC001726 | AB619003 | LC001742 | NA | NA |
| Lophiostomasemiliberum | KT 622 | AB618694 | JN942966 | AB619012 | LC001757 | JN993483 | NA |
| Lophiostomasemiliberum | KT 652 | AB618695 | JN942967 | AB619013 | LC001758 | JN993485 | NA |
| Neooccultibambusachiangraiensis | MFLUCC 12-0559 | KU712458 | KU712442 | KU764699 | NA | KU872761 | NA |
| Neooccultibambusakaiyangensis | CGMCC 3.20404 | MZ964886 | MZ964868 | MZ964877 | OK061077 | OK061071 | NA |
| Neooccultibambusatrachycarpi | CGMCC 3.20405 | MZ964888 | MZ964870 | MZ964879 | OK061079 | OK061073 | NA |
| Neopaucisporarosae-ecae | MFLUCC 17-0807T | MG829139 | MG828924 | MG829033 | MG829217 | NA | NA |
| Neotrematosphaeriabiappendiculatum | KTC 975 | GU205254 | NA | GU205228 | NA | NA | NA |
| Neotrematosphaeriabiappendiculatum | KTC 1124T | GU205256 | NA | GU205227 | NA | NA | NA |
| Neovaginatisporaclematidis | MFLUCC 17-2149 | MT226676 | MT310606 | MT214559 | MT394738 | NA | NA |
| Neovaginatisporafuckelii | MFLUCC 17-1334 | MN304833 | MN304828 | MN274565 | MN328899 | MN328896 | NA |
| Neovaginatisporafuckelii | KT 634 | AB618690 | LC001732 | AB619009 | LC001750 | NA | NA |
| Occultibambusaaquatica | MFLUCC 11-0006 | KX698112 | KX698114 | KX698110 | NA | NA | NA |
| Occultibambusabambusae | MFLUCC 13-0855 | KU872116 | KU940123 | KU863112 | KU940170 | KU940193 | NA |
| Occultibambusachiangraiensis | MFLUCC 16-0380 | KX655551 | NA | KX655546 | KX655566 | KX655561 | NA |
| Occultibambusafusispora | MFLUCC 11-0127 | NA | KU940125 | KU863114 | KU940172 | KU940195 | NA |
| Occultibambusahongheensis | KUMCC 21-0020 | MZ329029 | MZ329037 | MZ329033 | NA | MZ325467 | NA |
| Occultibambusajonesii | GZCC 16-0117 | KY628324 | NA | KY628322 | KY814758 | KY814756 | NA |
| Occultibambusakunmingensis | HKAS 102151 | MT864342 | MT627716 | MN913733 | MT878453 | MT954407 | NA |
| Occultibambusakunmingensis | KUNCC 21-0506 | PP779901 | PP779906 | PP779897 | PP778371 | NA | NA |
| Occultibambusamaolanensis | GZCC 16-0116 | KY628325 | NA | KY628323 | KY814759 | KY814757 | NA |
| Occultibambusapustula | MFLUCC 11-0502 | KU872118 | KU940126 | KU863115 | NA | NA | NA |
| Occultibambusasichuanensis | CGMCC 3.20938 | NA | ON332913 | ON332931 | ON383989 | ON381181 | NA |
| Occultibambusasichuanensis | UESTCC 22.0004 | NA | ON332914 | ON332932 | ON383990 | ON381182 | NA |
| Parapaucisporapseudoarmatispora | KT 2237 | LC100018 | LC100021 | LC100026 | LC100030 | NA | NA |
| Paraphomaaquatica | FMR 16956T | NA | OU612361 | OU612360 | OU612357 | OU612356 | OU612355 |
| Paraphomaaquatica | KUNCC 21-0523 | NA | PP779905 | PP779896 | PP778370 | NA | NA |
| Paraphomachlamydocopiosa | UMPc01 | NA | KU999072 | NA | NA | KU999080 | KU999084 |
| Paraphomachrysanthemicola | CBS 172.70 | NA | KF251165 | KF251669 | KF252173 | KF253123 | KF252660 |
| Paraphomachrysanthemicola | CBS 522.66T | NA | KF251166 | KF251670 | KF252174 | KF253124 | KF252661 |
| Paraphomaconvolvuli | MF-9.222 | NA | MG764055 | MG764069 | NA | NA | NA |
| Paraphomaconvolvuli | MF-9.265 | NA | MG764062 | MG764071 | MG779467 | NA | MG779457 |
| Paraphomaconvolvuli | MF-9.298.1 | NA | MG764057 | MG764074 | MG779468 | NA | MG779459 |
| Paraphomaconvolvuli | MF-9.300.1 | NA | MG764064 | MG764066 | MG779469 | NA | MG779460 |
| Paraphomaconvolvuli | MF-9.301.1 | NA | MG764060 | MG764075 | MG779470 | KF253126 | MG779461 |
| Paraphomadioscoreae | CPC 11361 | NA | KF251169 | KF251673 | KF252177 | KF253127 | KF252664 |
| Paraphomadioscoreae | CBS 135100 | NA | KF251167 | KF251671 | KF252175 | NA | KF252662 |
| Paraphomafimeti | CBS 170.70T | NA | KF251170 | KF251674 | KF252178 | KF253128 | KF252665 |
| Paraphomafimeti | CBS 368.91 | NA | KF251171 | KF251675 | KF252179 | KF253129 | KF252666 |
| Paraphomagaribaldii | CBS 148459 | NA | OL435708 | NA | NA | OL449256 | OL449254 |
| Paraphomagaribaldii | CBS 148460 | NA | OL435709 | NA | NA | OL449257 | OL449255 |
| Paraphomaledniceana | CBS 146533 | NA | MT371091 | MT371396 | MT372655 | MT372654 | MT372661 |
| Paraphomamelnikiae | MF-9.88 | NA | MG764063 | MG764065 | MG779466 | NA | MG779456 |
| Paraphomamelnikiae | MF-9.95 | NA | MG764054 | MG764067 | MG779462 | NA | NA |
| Paraphomamelnikiae | MF-9.182.1 | NA | MG764058 | MG764068 | MG779463 | NA | MG779454 |
| Paraphomamelnikiae | MF-9.240 | NA | MG764061 | MG764070 | MG779464 | NA | MG779453 |
| Paraphomamelnikiae | MF-9.296.1 | NA | MG764056 | NA | MG779465 | NA | MG779458 |
| Paraphomapye | UMPp04; BRIP 65171 | NA | KU999075 | NA | NA | NA | KU999087 |
| Paraphomapye | UMPp02 | NA | KU999073 | NA | NA | KU999081 | KU999085 |
| Paraphomaradicina | CBS 111.79 | NA | KF251172 | KF251676 | KF252180 | KF253130 | KF252667 |
| Paraphomaradicina | CBS 102875T | NA | KF251173 | KF251677 | KF252181 | KF253131 | KF252668 |
| Paraphomarhaphiolepidis | CBS 142524T | NA | KY979758 | KY979813 | KY979851 | KY979896 | KY979924 |
| Paraphomasalicis | CBS 146797 | NA | MW883437 | MW883829 | MW890069 | NA | MW890140 |
| Paraphomavariabilis | CBS 147695T | NA | LR993310 | LR993311 | LR993313 | NA | LR993314 |
| Paraphomavinacea | UMPV004 | NA | KU176887 | KU176891 | NA | NA | KU176895 |
| Paucisporakunmingense | MFLUCC 17-0932T | MF173430 | MF173432 | MF173428 | MF173434 | MF173436 | NA |
| Paucisporaquadrispora | KT 843T | AB618692 | LC001734 | AB619011 | LC001755 | NA | NA |
| Phaeoseptumaquaticum | CBS 123113T | NA | JN644072 | KY940803 | NA | NA | NA |
| Phaeoseptumcarolshearerianum | NFCCI-4221T | MK307816 | MK307813 | MK307810 | MK309874 | MK309877 | NA |
| Phaeoseptumcarolshearerianum | NFCCI-4384 | MK307818 | MK307815 | MK307812 | MK309876 | MK309879 | NA |
| Phaeoseptumhydei | MFLUCC 17-0801T | MT240624 | MT240623 | MT240622 | MT241506 | NA | NA |
| Phaeoseptummali | HKAS122916 | ON009082 | ON009098 | ON009114 | ON009257 | ON009282 | NA |
| Phaeoseptummali | HKAS122917 | ON009083 | ON009099 | ON009115 | ON009258 | ON009283 | NA |
| Phaeoseptummali | MFLUCC 17-2108T | NA | MK625197 | MK659580 | MK647990 | NA | NA |
| Phaeoseptummanglicola | NFCCI-4666 T | MK307817 | MK307814 | MK307811 | MK309875 | MK309878 | NA |
| Phaeoseptumterricola | MFLUCC 10-0102T | MH105780 | MH105779 | MH105778 | MH105781 | NA | NA |
| Platystomumcrataegi | MFLUCC 14-0925T | KT026113 | KT026117 | KT026109 | KT026121 | NA | NA |
| Platystomumrosae | MFLUCC 15-0633T | KT026115 | KT026119 | KT026111 | NA | NA | NA |
| Platystomumsalicicola | MFLUCC 15-0632T | KT026114 | KT026118 | KT026110 | NA | NA | NA |
| Pleopunctumbaoshanense | KUNCC 21-0494T | PP779898 | PP779902 | PP779893 | PP778367 | PP778372 | NA |
| Pleopunctumclematidis | MFLUCC 17-2091 | NA | MT214573 | MT310618 | MT394632 | MT394693 | NA |
| Pleopunctumellipsoideum | MFLUCC 19-0390T | MK804514 | MK804517 | MK804512 | MK828510 | NA | NA |
| Pleopunctumellipsoideum | MFLUCC 21-0064 | NA | OM258687 | OM250079 | NA | NA | NA |
| Pleopunctumguizhouense | GZCC 23-0595 | NA | OR091332 | OR098710 | NA | NA | NA |
| Pleopunctumheveae | MFLUCC 21-0146 | NA | OL782070 | OL780491 | NA | NA | NA |
| Pleopunctummegalosporum | KUNCC 10785T | NA | OQ146985 | ON261162 | OQ943186 | NA | NA |
| Pleopunctummegalosporum | KUNCC 10442 | NA | OQ146986 | OQ135180 | OQ943187 | NA | NA |
| Pleopunctummegalosporum | KUNCC 21-0622 | PP779899 | PP779903 | PP779894 | PP778368 | PP778373 | NA |
| Pleopunctummenglaense | KUMCC 21-0026T | ON009087 | ON009103 | ON009119 | ON009262 | ON009287 | NA |
| Pleopunctummenglaense | KUMCC 21-0025 | ON009086 | ON009102 | ON009118 | ON009261 | ON009286 | NA |
| Pleopunctummulticellularum | KUNCC 10789T | NA | OQ146989 | ON261166 | OQ943190 | NA | NA |
| Pleopunctummulticellularum | KUNCC 10781 | NA | OQ146981 | ON261158 | OQ943189 | NA | NA |
| Pleopunctummulticellularum | KUNCC 10778 | NA | OQ146978 | ON261155 | NA | NA | NA |
| Pleopunctumpseudoellipsoideum | MFLUCC 19-0391T | NA | MK804518 | MK804513 | MK828511 | NA | NA |
| Pleopunctumpseudoellipsoideum | HKAS122915 | ON009085 | ON009101 | ON009117 | ON009260 | ON009285 | NA |
| Pleopunctumrotundatum | KUNCC 10787T | NA | OQ146987 | ON261164 | OQ943194 | NA | NA |
| Pleopunctumrotundatum | KUNCC 10780 | NA | OQ146980 | ON261157 | OQ943193 | NA | NA |
| Pleopunctumthailandicum | MFLUCC 21-0039T | NA | MZ198896 | MZ198894 | MZ172461 | NA | NA |
| Pleopunctumpseudoellipsoideum | KUMCC 21-0820 | ON009084 | ON009100 | ON009116 | ON009259 | ON009284 | NA |
| Pseudocapulatisporaclematidis-subumbellatae | MFLUCC 17-2063 | MT226677 | MT310607 | MT214560 | MT394739 | MT394687 | NA |
| Pseudocapulatisporalongiappendiculatum | MFLUCC 17-1452T | MT214415 | MT214368 | MT214462 | MT235783 | NA | NA |
| Pseudocapulatisporalongiappendiculatum | MFLUCC 17-1457 | MT214416 | MT214369 | MT214463 | MT235784 | MT235821 | NA |
| Pseudolophiostomachiangraiense | MFLUCC 17–2076T | MT226678 | MT310608 | MT214561 | MT394740 | MT394688 | NA |
| Pseudolophiostomaclematidis | MFLUCC 17-2081 | MT226679 | MN393004 | MT214562 | MT394741 | MT394689 | NA |
| Pseudolophiostomacornisporum | KH 322T | LC312486 | LC312515 | LC312544 | LC312573 | LC312602 | NA |
| Pseudolophiostomalincangense | KUNCC 21-0606T | PP779900 | PP779904 | PP779895 | PP778369 | PP778374 | NA |
| Pseudolophiostomamangiferae | MFLUCC 17-2651T | MG931028 | MG931031 | MG931025 | NA | NA | NA |
| Pseudolophiostomamangiferae | MFLUCC 17-2653 | MG931029 | MG931032 | MG931026 | NA | NA | NA |
| Pseudolophiostomaobtusisporum | KT 3098 | LC312490 | LC312519 | LC312548 | LC312577 | LC312606 | NA |
| Pseudolophiostomaobtusisporum | KT 2838T | LC312489 | LC312518 | LC312547 | LC312576 | LC312605 | NA |
| Pseudolophiostomatropicum | KH 352 | LC312492 | LC312521 | LC312550 | LC312579 | LC312608 | NA |
| Pseudolophiostomatropicum | KT 3134T | LC312493 | LC312522 | LC312551 | LC312580 | LC312609 | NA |
| Pseudolophiostomavitigenum | HH 26930T | AB618697 | LC001735 | AB619015 | LC001761 | NA | NA |
| Pseudolophiostomavitigenum | HH 26931 | AB618698 | LC001736 | AB619016 | LC001762 | NA | NA |
| Pseudopaucisporabrunneospora | KH 227T | LC312494 | LC312523 | LC312552 | LC312581 | LC312610 | NA |
| Pseudoplatystomumscabridisporum | BCC 22835 | NA | NA | GQ925844 | GU479857 | GU479830 | NA |
| Pseudoplatystomumscabridisporum | BCC 22836 | NA | NA | GQ925845 | GU479856 | GU479829 | NA |
| Quintarialignatilis | CBS 117700 | GU296188 | NA | GU301865 | NA | GU371761 | NA |
| Quintarialignatilis | BCC 17444 | GU479764 | NA | GU479797 | GU479859 | GU479832 | NA |
| Seriascomabambusae | KUMCC 21-0021 | MZ329031 | MZ329039 | MZ329035 | MZ325470 | MZ325468 | NA |
| Seriascomadidymosporum | MFLUCC 11-0179 | KU872119 | KU940127 | KU863116 | KU940173 | KU940196 | NA |
| Seriascomayunnanense | MFLU 19-0690 | MN174694 | NA | MN174695 | MN210324 | MN381858 | NA |
| Setophomaterrestris | CBS 335.29 | NA | KF251246 | NA | NA | KF253196 | KF252729 |
| Sigarisporacoronillae | MFLUCC 14-0941T | KT026116 | KT026120 | KT026112 | NA | NA | NA |
| Sigarisporajunci | MFLUCC 14-0938T | MG829178 | MG828966 | MG829078 | NA | NA | NA |
| Sigarisporaravennicum | MFLUCC 14-0005T | KP698415 | KP698413 | KP698414 | NA | NA | NA |
| Sigarisporascrophulariicola | MFLUCC 17-0689T | NA | MG828969 | MG829081 | NA | NA | NA |
| Teichosporarubriostiolata | TR7 | NA | KU601590 | KU601590 | KU601609 | KU601599 | NA |
| Teichosporatrabicola | C134 | NA | KU601591 | KU601591 | KU601601 | KU601600 | NA |
| Thyridariamacrostomoides | GKM 224N | NA | GU385191 | NA | GU327777 | NA | NA |
| Thyridariamacrostomoides | GKM 1033 | NA | GU385190 | NA | GU327776 | NA | NA |
| Thyridariamacrostomoides | GKM 1159 | NA | GU385185 | NA | GU327778 | NA | NA |
| Vaginatisporaappendiculata | MFLUCC 13-0835T | KY264749 | NA | KY264745 | NA | NA | NA |
| Vaginatisporaaquatica | MFLUCC 11-0083T | KJ591575 | KJ591577 | KJ591576 | NA | NA | NA |
| Vaginatisporascabrispora | KT 2443T | LC312496 | LC312525 | LC312554 | LC312583 | LC312612 | NA |
| Versicolorisporiumtriseptatum | JCM 14775 | AB524501 | AB365596 | AB330081 | NA | NA | NA |
| Versicolorisporiumtriseptatum | UESTCC 21.0016 | OL741381 | OL741378 | OL741318 | NA | NA | NA |
The newly generated sequences are indicated in bold. T refers to ex-type strains, and NA refers to “no data in GenBank”.
The maximum likelihood (ML) analysis was performed on the CIPRES Science Gateway v.3.3 (http://www.phylo.org/portal2/; Miller et al. 2010) using RAxML-HPC2 on XSEDE v.8.2.12 (Stamatakis 2014) with parameters adjusted for 1000 bootstrap iterations and the GTRGAMMA substitution model. Bayesian inference was performed in MrBayes v.3.2.7a (Ronquist et al. 2012) using Markov chain Monte-Carlo sampling (BMCMC) to determine posterior probabilities (PPs) (Rannala and Yang 1996). The model of evolution for each gene was estimated using MrModeltest v.2.3 (Nylander et al. 2008) via PAUP v.4.0b10 (Ronquist and Huelsenbeck 2003). Six simultaneous Markov chains were run for 2,000,000 generations, with trees sampled at every 200 generations, until it was stopped when the standard deviation of split frequencies between the two simultaneous runs dropped below 0.01. Phylogenetic trees were visualized with FigTree v.1.4.0 (Rambaut 2012) and edited using Microsoft PowerPoint and Adobe Illustrator® CS6 v.26.0 (Adobe Systems, San Jose, CA, USA). The newly produced sequences were deposited in the GenBank nucleotide database (Table 1).
Results
Phylogenetic analysis
Analyses 1, Lophiostomataceae phylogeny, was based on combined SSU, LSU, ITS, tef1-α, and rpb2. The final alignment contained 4,230 characters used for the phylogenetic analyses, including alignment gaps. The alignment contained 74 strains, and the tree was rooted with Teichosporarubriostiolata (TR7) and T.trabicola (C134). The RAxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of -29697.884081. The matrix had 1,584 distinct alignment patterns with 26.07% undetermined characters or gaps. The estimated base frequencies were as follows; A = 0.249681, C = 0.245034, G = 0.268293, T = 0.236992; substitution rates AC = 1.509197, AG = 3.725453, AT = 1.272039, CG = 1.297966, CT = 7.905766, GT = 1.0; gamma distribution shape parameter α = 0.192719 and tree-length = 0.192719. The tree topologies of combined sequence data obtained from ML and BI analyses were not significantly different. Our isolate, Pseudolophiostomalincangense (KUNCC 21-0606), was closer and sister to P.vitigenum strains (HH 26930, HH 26931) but formed a separate lineage with 100% ML bootstrap and 1.00 BYPP support (Fig. 1).
Figure 1.
Phylogram generated from ML analysis based on SSU, LSU, ITS, tef1-α, and rpb2 sequence data representing the family Lophiostomataceae. Related sequences were obtained from Phukhamsakda et al. (2020). Bootstrap values for ML are equal to or greater than 75%, and posterior probability values are equal to or greater than 0.95 from the BYPP analysis labelled on the nodes. Strains of the newly described species are in red, while type strains are in bold. The tree is rooted with Teichosporarubriostiolata (TR7) and T.trabicola (C134).
Analyses 2, Occultibambusaceae phylogeny, was based on combined SSU, LSU, ITS, tef1-α, and rpb2. The final alignment contained 4,103 characters used for the phylogenetic analyses, including alignment gaps. The alignment contained 25 strains, and the tree was rooted with Ernakulamiakrabiensis (MFLUCC 18-0237) and E.xishuangbannaensis (KUMCC 17-0187). The RAxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of -16329.806421. The matrix had 1,047 distinct alignment patterns with 24.1% undetermined characters or gaps. The estimated base frequencies were as follows; A = 0.243761, C = 0.253253, G = 0.274093, T = 0.228893; substitution rates AC = 2.182200, AG = 2.182200, AT = 1.737406, CG = 1.445194, CT = 9.965966, GT = 1.0; gamma distribution shape parameter α = 0.155054 and tree-length = 0.951813. The tree topologies of combined sequence data obtained from ML and BI analyses were not significantly different. Our new isolate (KUNCC 21-0506) was nested with Occultibambusakunmingensis (HKAS 102151, type) and with 100% ML bootstrap and 1.00 BYPP support (Fig. 2).
Figure 2.
Phylogram generated from maximum likelihood analysis based on the combined SSU, LSU, ITS, tef1-α, and rpb2 dataset of Occultibambusaceae species. Related sequences were obtained from Yu et al. (2022). Bootstrap values for ML are equal to or greater than 75%, and posterior probability values are equal to or greater than 0.95 from the BYPP analysis labelled on the nodes. Strains of the newly described species are in red, while type strains are in bold. The tree is rooted with Ernakulamiakrabiensis (MFLUCC 18-0237) and E.xishuangbannaensis (KUMCC 17-0187).
Analyses 3, Phaeoseptaceae phylogeny, was based on combined SSU, LSU, ITS, tef1-α, and rpb2. The final alignment contained 3,195 characters used for the phylogenetic analyses, including alignment gaps. The alignment contained 37 strains, and the tree was rooted with Angustimassarinaacerina (MFLUCC 14-0505) and A.populi (MFLUCC 13-0034). The RaxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of -12325.441217. The matrix had 882 distinct alignment patterns with 30.9% undetermined characters or gaps. The estimated base frequencies were as follows: A = 0.249369, C = 0.247445, G = 0.266516, T = 0.236670; substitution rates AC = 1.406931, AG = 4.076513, AT = 1.212607, CG = 1.303216, CT = 8.242374, GT = 1.000000; gamma distribution shape parameter α = 0.201687 and tree-length = 3.128705. The tree topologies of combined sequence data obtained from ML and BI analyses were not significantly different. Our isolate, Pleopunctumbaoshanense (KUNCC 21-0494), constituted a strongly supported (100% ML and 1.00 BYPP) independent lineage basal to P.pseudoellipsoideum (MFLUCC 19-0391, KUMCC 21-0820, HKAS122915) and P.ellipsoideum (97% ML and 1.00 BYPP). While our other isolate (KUNCC 21-0622) was grouped together with P.menglaense strains (KUNCC 1442, KUNCC 10785) with 100% ML bootstrap and 1.00 BYPP support (Fig. 3).
Figure 3.
Phylogram generated from maximum likelihood analysis based on the combined SSU, LSU, ITS, tef1-α, and rpb2 dataset of Phaeoseptaceae species. Related sequences were obtained from Xu et al. (2023). Bootstrap values for ML are equal to or greater than 75%, and posterior probability values are equal to or greater than 0.95 from the BYPP analysis labelled on the nodes. Strains of the newly described species are in red, while type strains are in bold. The tree is rooted with Angustimassarinaacerina (MFLUCC 14-0505) and A.populi (MFLUCC 13-0034).
Analyses 4, Paraphoma phylogeny, was based on combined LSU, ITS, tef1-α, rpb2, and tub2. The final alignment contained 3,351 characters used for the phylogenetic analyses, including alignment gaps. The alignment contained 32 strains, and the tree was rooted with Setophomaterrestris (CBS 335.29). The RAxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of -13609.051798. The matrix had 929 distinct alignment patterns with 37.21% undetermined characters or gaps. The estimated base frequencies were as follows; A = 0.238385, C = 0.250275, G = 0.266033, T = 0.245306; substitution rates AC = 1.198498, AG = 2.769971, AT = 1.326456, CG = 0.861230, CT = 5.288549, GT = 1.0; gamma distribution shape parameter α = 0.274993 and tree-length = 1.180712. The tree topologies of combined sequence data obtained from ML and BI analyses were not significantly different. Our isolate, Paraphomabaoshanenses (KUNCC 21-0523), forms a distinct branch close to P.aquatica (FMR 16956) and with 100% ML bootstrap and 1.00 BYPP support (Fig. 4).
Figure 4.
Phylogram generated from maximum likelihood analysis based on the combined LSU, ITS, tef1-α, rpb2 and tub2 dataset of Paraphoma species. Related sequences were obtained from previous publications (Magaña-Dueñas et al. 2021, Guarnaccia et al. 2022, and Gomzhina et al. 2020). Bootstrap values for ML are equal to or greater than 75%, and posterior probability values are equal to or greater than 0.95 from the BYPP analysis labelled on the nodes. Strains of the newly described species are in red, while type strains are in bold. The tree is rooted with Setophomaterrestris (CBS 335.29).
Taxonomy
The present study introduces two new species and three new records. These taxa belong to the order Pleosporales and are described below.
. Pseudolophiostoma lincangense
G.C. Ren & Tibpromma sp. nov.
D9F25B40-C2D3-57C0-9DF5-D09FA7A3E16E
Index Fungorum: IF902122
Facesoffungi Number: FoF15842
Figure 5.
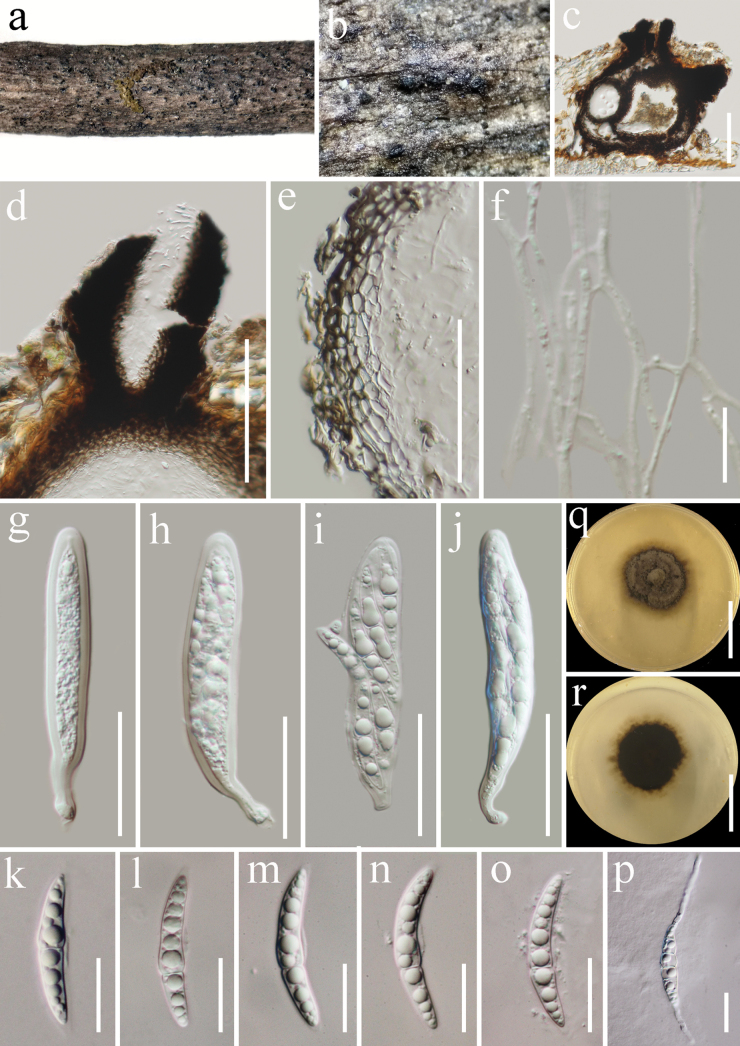
Pseudolophiostomalincangense (HKAS 122880, holotype) a, b appearance of ascomata on the host surface c section of ascoma d ostiole e peridium f pseudoparaphyses g–j asci k–o ascospores p germinated ascospore q, r culture characters on PDA (q = from above, r = from below). Scale bars: 100 μm (c, d); 50 μm (e); 10 μm (f); 30 μm (g–j); 15 μm (k–p); 30 mm (q, r).
Etymology.
The epithet refers to the location where the fungus was collected.
Holotype.
HKAS 122880.
Description.
Saprobic on dead twigs of Castanopsiscalathiformis (Fagaceae) in terrestrial habitat. Sexual morph: Ascomata 280–330 µm high, 230–290 μm diam. (x̄ = 310 × 260 μm, n = 5), solitary or gregarious, immersed, papilla erumpent through host surface, subglobose, single or two locular, coriaceous, brown to dark brown, ostiolate. Ostioles 110–160 µm long, 80–90 μm diam., carbonaceous, mostly central, with crest-like opening, filled with hyaline periphyses. Peridium 10–20 µm wide, comprising 4–9 layers, composed of dark brown outer layers, inner layers comprising hyaline, flattened, angular, thick-walled cells of textura angularis. Hamathecium composed of numerous, 1–2 µm wide, flamentous, septate, branched, cellular pseudoparaphyses. Asci 80–100 × 15–20 µm (x̄ = 94 × 16 µm, n = 20), 8-spored, bitunicate, fissitunicate, cylindrical, with pedicel, apically rounded, with a minute ocular chamber. Ascospores 32–40 × 5–5.8 µm (x̄ = 35 × 5.3 µm, n = 30), 1–2-seriate, fusiform, hyaline, straight or slightly curved, 1-septate, becoming 3-septate when germinated, constricted at the septa, narrower towards both end cells, upper cell longer than lower cell, guttulate, smooth-walled, with mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics.
Ascospores germinated on PDA within 24 h at room temperature (25 °C). Germ tubes produced from the apical or the second cell of ascospore. Colonies on PDA, reaching 25 mm diameter after two weeks at 20–25 °C, mycelia superficial, circular, flat, fimbriate, undulate edge, gray with white gray at the center; reverse, dark green, pale yellow at the center.
Material examined.
China, Yunnan Province, Lincang (24°5'30"N, 100°5'33"E, elevation: 1557.49 m) on dead woody twigs of Castanopsiscalathiformis (Fagaceae), 12 July 2020, G.C. Ren, LC25 (HKAS 122880, holotype), ex-type living culture KUNCC 21-0606.
Notes.
Multi-loci phylogenetic analyses based on a concatenated SSU, LSU, ITS, tef1-α, and rpb2 sequence dataset show that our new collection (KUNCC 21-0606) clusters sister to strains of Pseudolophiostomavitigenum (HH 26930 and HH 26931) with 100% ML and 1.00 BYPP support (Fig. 1). Sequence comparison for the ITS region between Pseudolophiostomalincangense (KUNCC 21-0606) and P.vitigenum (HH 26930, type) showed a 2.67% (14/524 bp) base pair difference, 0.24% (2/848 bp) base pair difference for LSU region, 1.97% (17/863 bp) base pair difference for tef1-α region, but we were unable to compare rpb2 gene of P.vitigenum as there was no sequence data. Comparatively, the morphological characterization of Pseudolophiostomalincangense is similar to P.vitigenum in having immersed ascomata with papilla; carbonaceous ostiole with crest-like opening, and filled with hyaline periphyses; cylindrical asci with a short truncate pedicel and a minute ocular chamber; fusiform, hyaline, constricted at the septa, 1-septate ascospores (Thambugala et al. 2015). However, our new collection differs from P.vitigenum in having single or two locular ascomata, peridium consists of two layer cells of textura angularis, smaller asci (94 × 16 μm vs. 129.8 × 20.1 μm) and ascospores (35 × 5.3 μm vs. 38.5 × 10.5 μm), and of the upper cell of the ascospores is longer than the lower cell (Thambugala et al. 2015). Therefore, we identify our collection as a new species from Yunnan Province, China.
. Occultibambusa kunmingensis
C.X. Liu, H. Zhang & K.D. Hyde, Fungal Diversity 105: 471.
4C3ABFCA-D6F9-5EC7-89FE-809A2AC9CCE2
Index Fungorum: IF557930
Facesoffungi Number: FoF09272
Figure 6.
Occultibambusakunmingensis (HKAS 122706) a host b ascomata on host surface c vertical section of ascoma d peridium e pseudoparaphyses f–j asci k–m ascospores n germinated ascospore o, p colonies on PDA (o = from above, p = from below). Scale bars: 100 µm (c); 20 µm (d–j); 10 µm (k–n); 20 mm (o, p).
Description.
Saprobic on dead twigs of Castanopsisdelavayi (Fagaceae) in terrestrial habitat. Sexual morph: Ascomata 160–180 μm high, 160–240 μm diam. (x̄ = 175 × 195 μm, n = 5), dark brown to black, solitary or gregarious, semi-immersed to superficial, coriaceous, subglobose, with a short papillate, ostiolate. Ostioles 50–65 µm long, 35–40 μm diam., black, short. Peridium 20–35 μm thick, thin at the base and becoming wider laterally, composed of several layers of brown to dark brown, thick-walled cells of textura angularis. Hamathecium 2.5–3.5 µm wide, hyphae-like, septate, cellular pseudoparaphyses, embedded in a gelatinous matrix. Asci (60)74–107(–116) × 12–14 μm (x̄ = 82 × 13 μm, n = 20), 8-spored, bitunicate, fissitunicate, clavate or cylindric-clavate, narrowly rounded at the apex, with a short truncate pedicel, apically rounded, with a minute ocular chamber. Ascospores 30–40 × 4–6 μm (x̄ = 35.6 × 5.3 μm, n = 20), overlapping 1–2-seriate, fusiform, straight or slightly curved, 1-septate, brown, constricted at the septa, guttulate, thin and smooth-walled, without mucilaginous sheaths and appendages. Asexual morph: Undetermined.
Culture characteristics.
Ascospores germinated on PDA within 24 h at room temperature (25 °C). Germ tubes produced from the basal and apical cells of ascospore. Colonies on PDA, reaching 25 mm diameter after 2 weeks at 20–25 °C, mycelia superficial, dense, circular, raised, entire edge, velvety, flossy; reverse black.
Material examined.
China, Yunnan Province, Baoshan (25°18'48"N, 99°09'50"E) on dead woody twigs of Castanopsisdelavayi (Fagaceae), 12 July 2020, G.C. Ren, BS17 (HKAS 122706), living culture KUNCC 21-0506.
Known host, habitats, and distribution.
Bamboo and Castanopsisdelavayi from freshwater and terrestrial in China (Dong et al. 2020; Jiang et al. 2021; this study).
Notes.
According to the multi-gene phylogenetic analyses of combined SSU, LSU, ITS, tef1-α, and rpb2 sequence dataset, our new isolate (KUNCC 21-0506) nested with Occultibambusakunmingensis (HKAS 102151), which was isolated from decaying bamboo submerged in freshwater in China (Dong et al. 2020) with 100% ML and 1.00 BYPP bootstrap support (Fig. 2). Our new isolate fits well with the morphological characteristics of the holotype of O.kunmingensis in having semi-immersed to superficial ascomata with short papillate, clavate or cylindric-clavate asci, fusiform, 1-septate, brown ascospores without mucilaginous sheaths and appendages (Dong et al. 2020). Sequence comparison for the ITS and tef1-α region between our isolates (KUNCC 21-0506) and O.kunmingensis (HKAS 102151) showed no significant base pair differences. Therefore, we identified our taxon as a new host record of O.kunmingensis from Castanopsisdelavayi (Fagaceae) in China, and it is the first record from woody litter.
. Pleopunctum baoshanense
G.C. Ren & Tibpromma sp. nov.
9C3E4EA1-4C18-5462-9705-2C350BD58FB7
Index Fungorum: IF902123
Facesofungi Number: FoF15843
Figure 7.
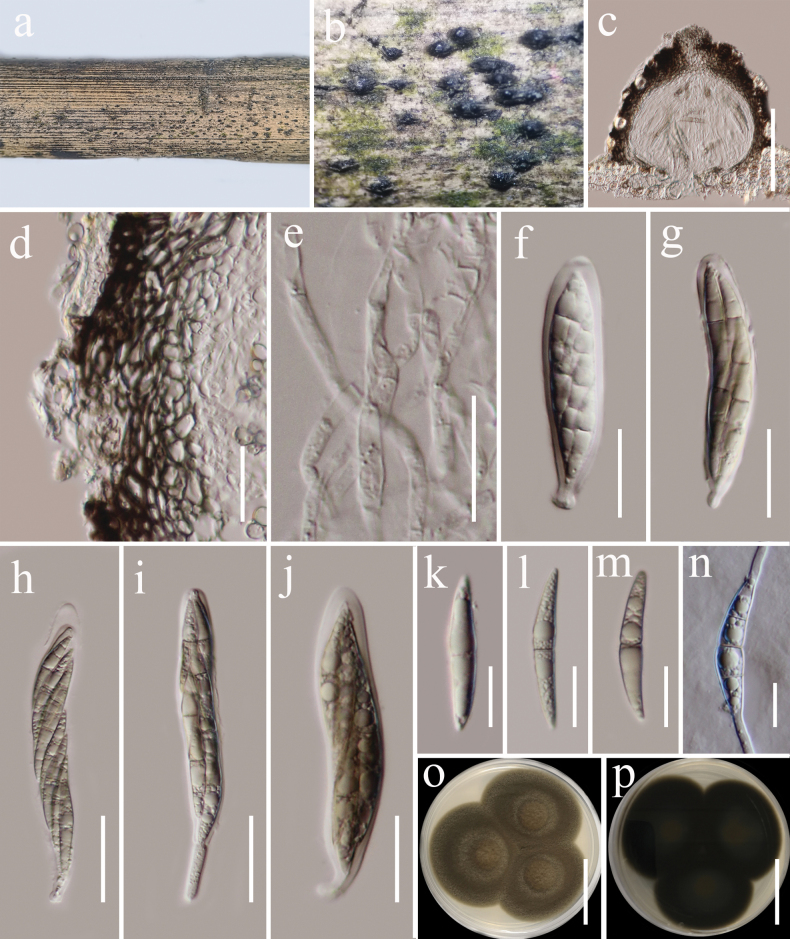
Pleopunctumbaoshanense (HKAS 134936, holotype) a host b colonies on the host surface c–f conidia with basal hyaline cells g germinated conidium h, i colony on PDA (h = from above, i = from below). Scale bars: 20 μm (c–g); 20 mm (h, i).
Etymology.
The specific epithet “baoshanense” reflects the location “Baoshan” where the holotype was collected.
Holotype.
HKAS 134936.
Description.
Saprobic on decaying wood of Castanopsiscalathiformis (Fagaceae) in terrestrial habitat. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies on natural substrate sporodochial, superficial, black, scattered, gregarious. Mycelium immersed in the substratum, composed of septate hyphae. Conidiophores 3–5 µm wide (x̄ = 3.8 µm, n = 15), macronematous, mononematous, cylindrical, unbranched, septate, hyaline and smooth-walled. Conidiogenous cells monoblastic, terminal, integrated, hyaline. Conidia 33–46 × 22.5–27.6 μm (x̄ = 39 × 25 μm, n = 40), acrogenous, solitary, muriform, oval to ellipsoidal, smooth-walled, broadly obtuse at apex and dark brown, truncate at base and paler brown, often with a hyaline, elliptical to globose basal cell, 10–14 × 13–15 μm (x̄ = 12 × 13 μm, n = 30).
Culture characteristics.
Conidia germinated on PDA within 24 h at room temperature (25 °C). Germ tubes produced from the basal cells of conidia. Colonies on PDA, reaching 30 mm diameter after 2 weeks at 20–25 °C, mycelia superficial, irregular, umbonate at the center, fimbriate, undulate entire, grey, radially furrowed, with hyaline, glistening, rough, wrinkled, granular droplets of oil; reverse, grey, radially furrowed, grey white at the margin.
Material examined.
China, Yunnan Province, Baoshan (25°18'48"N, 99°09'50"E), on dead woody twigs of Castanopsiscalathiformis (Fagaceae), 12 July 2020, G.C. Ren, B194 (HKAS 134936, holotype), ex-type living culture KUNCC 21-0494.
Notes.
Multi-loci phylogenetic analyses based on a concatenated SSU, LSU, ITS, tef1-α, and rpb2 sequence dataset show that our new collection (KUNCC 21-0494) clusters sister to strains of Pleopunctumpseudoellipsoideum (MFLUCC 19-0391, KUMCC 21-0820, HKAS 122915) and P.ellipsoideum (MFLUCC 19-0390, MFLUCC 21-0064) with solid support (100% ML and 1.00 BYPP, Fig. 3). Sequence comparison for the ITS region between Pleopunctumbaoshanense (KUMCC 21-0494) and P.pseudoellipsoideum (MFLUCC 19-0391) showed a 1.54% (8/520 bp, without the gaps) base pair difference, 2.26% (21/930 bp, without the gaps) base pair difference for the tef1-α region. Sequence comparison for the ITS region between Pleopunctumbaoshanense (KUNCC 21-0494) and P.ellipsoideum (MFLUCC 19-0390) showed a 1.65% (8/486 bp, without the gaps) base pair difference, 1.81% (15/827 bp, without the gaps) base pair difference for tef1-α region. Pleopunctumbaoshanense, P.pseudoellipticum, and P.ellipsoideum are morphologically similar, they have sporodochial conidiomata; septate hyphae mycelium; mononematous, cylindrical conidiophores; monoblastic, terminal, hyaline conidiogenous cells and muriform, oval to ellipsoidal conidia often with a hyaline, elliptical to globose basal cell, but can be distinguished from P.pseudoellipsoideum and P.ellipsoideum in having hyaline conidiophores and conidiogenous cells, different sizes of conidia (39 × 25 μm vs. 50 × 24 μm vs. 45 × 20 μm), while both of them have medium brown conidiophores and conidiogenous cells (Liu et al. 2019; Xu et al. 2023). Therefore, based on morphology and phylogenetic distinctiveness, we introduce Pleopunctumbaoshanense as a new species.
. Pleopunctum megalosporum
R.J. Xu, Q. Zhao & Boonmee Journal of Fungi 9 (5, no. 560): 9 (2023)
98D6AF85-D36C-514A-8C17-BDC3149E1A0F
Index Fungorum: IF847826
Facesofungi Number: FoF14063
Figure 8.
Pleopunctummegalosporum (HKAS 134935) a host b colonies on the host surface c conidia on substrate d–h, k–p β conidia with basal cells and conidiophores i, j α conidia showing remnant of conidiogenous cells at base q germinated conidium r, s colony on PDA (r = from above, s = from below). Scale bars: 100 μm (c); 30 μm (d–q); 20 mm (s, r).
Description.
Saprobic on dead twigs of Castanopsiscalathiformis (Fagaceae) in terrestrial habitat. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies on host, sporodochial, superficial, light brown, scattered, and punctiform. Mycelium immersed in the substratum, composed of septate, branched, sub hyaline to light brown hyphae. Conidiophores 13–66 × 2.6–4.2 µm (x̄ = 37.7 × 3.5, n = 20), macronematous, mononematous, hyaline to light brown, cylindrical, unbranched, septate, smooth-walled. Conidiogenous cells 3.9–5.5 × 3.6–4.7 µm (x̄ = 4.6 × 4 µm, n = 20), monoblastic, terminal, light brown. The conidia are dimorphic, acrogenous, and solitary. α conidia 30–40 × 10–15 µm (x̄ = 33 × 12.5 µm, n = 30), hyaline to light brown, muriform, oblong to obovate, constricted at septa, slightly obtuse to rounded at apex. β conidia 48–60 × 24–31 µm (x̄ = 54.7 × 27.3 µm, n = 30), brown, muriform, oval to long elliptical, slightly constricted at septa, often with a hyaline, elliptical to globose, 0–multiple-basal cells, 12.5–17.6 × 7.7–11 µm (x̄ = 14.7 × 9.2 µm, n = 20).
Culture characteristics.
Conidia germinated on PDA within 24 h at room temperature (25 °C). Germ tubes produced from the basal cells of conidia. Colonies on PDA, reaching 30 mm diameter after 2 weeks at 20–25 °C, mycelia superficial, irregular, slightly umbonate at the center, fimbriate, undulate edge, grey at the margin, grey white at the center, with hyaline, glistening, granular droplets of oil; reverse, atrovirens, notably radially furrowed, golden brown at the margin.
Material examined.
China, Yunnan Province, Lincang (24°5'30"N, 100°5'33"E, elevation: 1557.49 m), on dead woody twigs of Castanopsiscalathiformis (Fagaceae), 12 July 2020, G.C. Ren, LC62 (HKAS 134935), living culture KUNCC 21-0622.
Known host, habitats, and distribution.
Cryptocaryaacutifolia, freshwater and terrestrial, China (Xu et al. 2023; this study).
Notes.
Pleopunctummegalosporum was introduced by Xu et al. (2023) from submerged decaying wood in a freshwater stream in China. Our collection (KUMCC 21-0622) resembles P.megalosporum (KUMCC22-10799) in having sporodochial conidiomata; septate, subhyaline to light brown mycelium; mononematous, cylindrical, light brown conidiophores; monoblastic, terminal, light brown conidiogenous cells and muriform, oval to long elliptical conidia often with a hyaline, elliptical to globose, 0–multiple-basal cell (Xu et al. 2023). Multi-loci phylogenetic analyses based on a concatenated SSU, LSU, ITS, tef1-α, and rpb2 sequence dataset show that our new collection (KUNCC 21-0622) clusters with Pleopunctummegalosporum (KUNCC 10785, KUNCC 10442) with strong support (100% ML and 1.00 BYPP, Fig. 3). Sequence comparison for the ITS and tef1-α region between our isolate (KUNCC 21-0622) and Pleopunctummegalosporum (KUNCC 10785) showed no significant base pair differences. Therefore, we introduce our collection as the first record of P.megalosporum from Castanopsiscalathiformis (Fagaceae) in China.
. Paraphoma aquatica
Magaña-Dueñas, Stchigel & Cano-Lira Journal of Fungi 7(12, no. 1102): 9 (2021)
CEB03285-7CA4-5B1C-B15D-01062F4A87E4
Index Fungorum number: IF841364
FacesofFungi number: FoF15844
Figure 9.
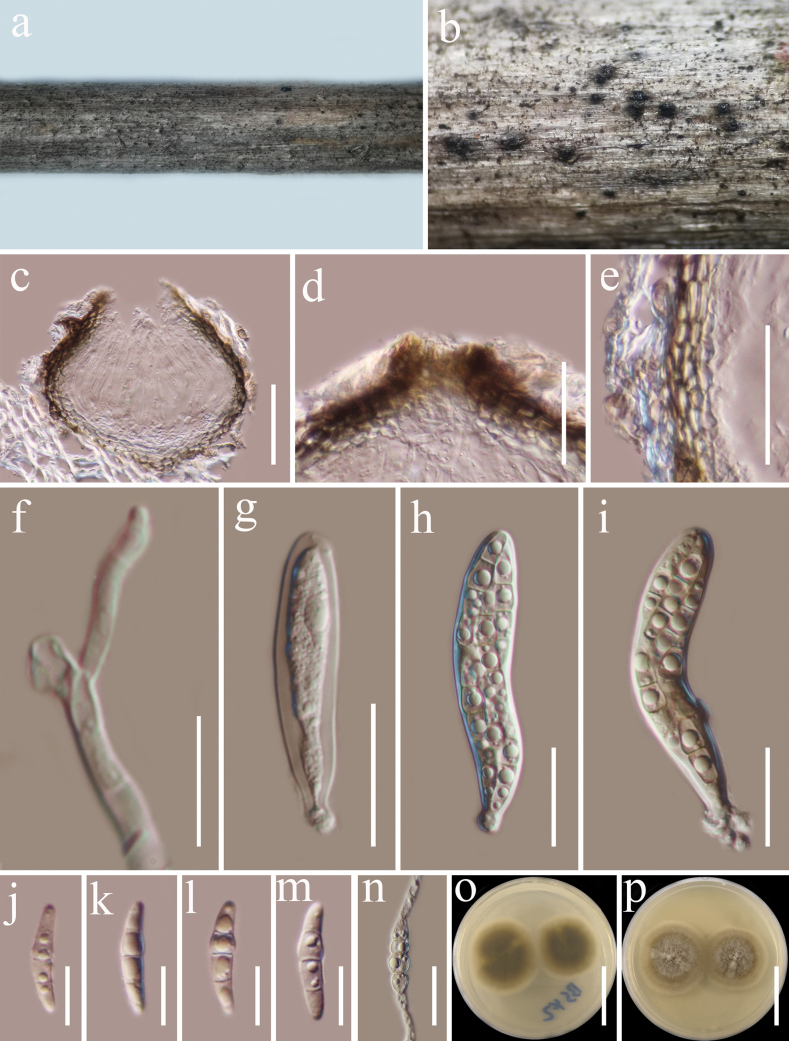
Paraphomaaquatica (HKAS 122713) a host b ascomata on the host surface c vertical section of ascoma d ostiole e peridium f pseudoparaphyses g–i asci j–m ascospores n germinated ascospore o, p culture characters on PDA (o = from above, p = from below). Scale bars: 50 µm (c); 30 µm (d, e); 10 µm (f, j–n); 20 µm (g–i); 20 mm (o, p).
Description.
Saprobic on dead woody twigs of Castanopsisdelavayi (Fagaceae). Sexual morph: Ascomata 110–130 × 120–130 μm (x̄ = 122 × 126 μm, n = 5), solitary, scattered, erumpent to immersed, uni-loculate, globose to subglobose, black. Ostioles central. Peridium 7–12 μm wide, thin, comprising 3–4 layers of light brown to brown cells of textura prismatica. Hamathecium 1.5–2.5 μm wide, cylindrical, filiform, hyaline, septate, branched, cellular pseudoparaphyses, embedded in a gelatinous matrix. Asci 45–63 × 9–11 μm (x̄ = 55 × 9.5 μm, n = 20), 8-spored, bitunicate, fissitunicate, clavate, slightly broad at center, apically rounded, with short and rounded pedicel, minute ocular chamber. Ascospores 19–22 × 4–4.6 μm (x̄ = 20.7 × 4.2 μm, n = 30), overlapping, biseriate, hyaline, narrowly fusiform, straight or slightly curved, with 3 transverse septa, enlarged at the second cell, constricted at the septa, smooth-walled, guttulate, without a mucilaginous sheath. Asexual morph: see Magaña-Dueñas et al. (2021).
Culture characteristics.
Ascospores germinated on PDA within 24 h at room temperature (25 °C). Germ tubes produced from the basal and apical cells of ascospore. Colonies on PDA, reaching 25 mm diameter after 2 weeks at 20–25 °C, mycelia superficial, medium density mycelia, entire margin, umbonate at center, band, rough surface, velvety, raised, grayish yellow, white mycelium attached to the central surface; reverse atrovirens.
Material examined.
China, Yunnan Province, Baoshan (25°18'48"N, 99°09'50"E), on dead woody twigs of Castanopsisdelavayi (Fagaceae), 12 July 2020, G.C. Ren, BS42 (HKAS 122713), living culture KUNCC 21-0523.
Known host, habitats, and distribution.
Capafonts and Castanopsisdelavayi, freshwater submerged plant debris and terrestrial, China and Spain (Magaña-Dueñas et al. 2021; this study).
Notes.
Multi-loci phylogenetic analyses based on a concatenated LSU, ITS, tef1-α, rpb2, and tub2 sequence dataset show that our new collection (KUNCC 21-0523) formed a sister lineage to the ex-type strain of Paraphomaaquatica (FMR 16956) with solid support (100% ML and 1.00 BYPP, Fig. 4). Sequence comparison for the ITS region between Paraphomabaoshanenses (KUNCC 21-0523) and P.aquatica (FMR 16956) showed a 6.17% (30/486 bp) base pair difference, 0.12% (2/848 bp) base pair difference for LSU region, 2.06% (9/437 bp) base pair difference for the tef1-α region. Unfortunately, the morphology could not be compared as Paraphomaaquatica was reported in its asexual morph with no information on its sexual morph (Magaña-Dueñas et al. 2021). The species of Paraphoma were introduced from its asexual morph (Morgan-Jones and White 1983; de Gruyter et al. 2010; Quaedvlieg et al. 2013; Moslemi et al. 2016, 2018; Crous et al. 2017, 2021a, 2021b; Gomzhina et al. 2020; Magaña-Dueñas et al. 2021; Guarnaccia et al. 2022), while we introduced our new collection from its sexual morph. Therefore, we could not compare our new collection with other Paraphoma species. However, based on the phylogenetic distinctiveness, we introduce our collection as the first record of P.aquatica from Castanopsisdelavayi (Fagaceae) in China, and our species is the first sexual morph recorded in this genus.
Discussion
This study introduces two new species of woody litter fungi: Pseudolophiostomalincangense, and Pleopunctumbaoshanense from Yunnan Province, China. We also report new host records of Occultibambusakunmingensis on Castanopsisdelavayi, Pleopunctummegalosporum on Castanopsiscalathiformis for the first time in China, and Paraphomaaquatica is the first sexual morph of Paraphoma on Castanopsisdelavayi.
Pseudolophiostoma was introduced by Thambugala et al. (2015), with P.vitigenum designated as the type species. Currently, this genus comprises seven accepted species (Species Fungorum 2024). Pseudolophiostoma species are typically saprobes found on various herbaceous, woody, or vine plants such as Bidenspilosavar.radiata, Clematisfulvicoma, Livistonaboninensis, Stachytarphetajamaicensis, and Vitiscoignetiae, and are distributed across China (Taiwan), Japan, and Thailand (Thambugala et al. 2015; Hashimoto et al. 2018; Tennakoon et al 2018; Phukhamsakda et al. 2020). Pseudolophiostoma species are known only by their sexual morph, and their asexual morph has not been discovered yet (Thambugala et al. 2015). Therefore, the asexual morph of Pseudolophiostoma remains uncertain and thus, further studies with additional collections are needed to understand the asexual morph.
Occultibambusa, introduced by Dai et al. (2017) with O.bambusae as the type species, is commonly found on bamboo culms. These species have a wide distribution, particularly in China and Thailand, where they inhabit both freshwater and terrestrial habitats (Hyde et al. 2016; Dai et al. 2017; Zhang et al. 2017; Dong et al. 2020; Jiang et al. 2021; Yu et al. 2022). Currently, Species Fungorum (2024) recognizes 10 Occultibambusa species. Most of these species have been reported based on their sexual morphs, with only O.fusispora known from its holomorph, and the coelomycetous asexual morph discovered in culture (Dai et al. 2017; Jiang et al. 2021). In this study, our collection also reports sexual morph and thus, further studies are needed to understand the asexual morph with additional fresh collections.
Pleopunctum was introduced as the first hyphomycetous genus in Phaeoseptaceae by Liu et al. (2019) to include two species: P.ellipsoideum and P.pseudoellipticum. Subsequently, nine Pleopunctum species viz. P.bauhiniae, P.clematidis, P.guizhouense, P.heveae, P.megalosporum, P.menglaense, P.multicellularum, P.rotundatum and P.thailandicum were accepted (Phukhamsakda et al. 2020; Boonmee et al. 2021; Koukol and Delgado 2021; Senwanna et al. 2021; Wanasinghe et al. 2021; Xu et al. 2023; Zhang et al. 2023). Currently, eleven Pleopunctum species are accepted in Species Fungorum (2024) and all have been reported in their asexual morph (Liu et al. 2019). Pleopunctum species distributed in China and Thailand and saprobic on dead wood of Bauhiniavariegata (Fabaceae), Clematissikkimensis (Ranunculaceae), Heveabrasiliensis (Euphorbiaceae) and some unknown woody litter in freshwater and terrestrial habitats (Hyde et al. 2019; Phukhamsakda et al. 2020; Senwanna et al. 2021; Xu et al. 2023). Pleopunctumbauhiniae, P.clematidis, P.heveae, P.megalosporum, and P.menglaense have dimorphic conidia on the natural substrate (Hyde et al. 2019; Phukhamsakda et al. 2020; Senwanna et al. 2021; Wanasinghe et al. 2021; Xu et al. 2023). However, P.ellipsoideum, P.guizhouense, P.multicellularum, P.pseudoellipticum, P.rotundatum and P.thailandicum are characterized by one conidium type and share very similar morphological characteristics (Liu et al. 2019; Boonmee et al. 2021; Koukol and Delgado 2021; Xu et al. 2023). The phenotypic variation among strains, influenced by environmental factors, can make morphological differentiation challenging (Hyde et al. 2020). However, our research has shown that molecular sequence data are a reliable tool for identifying Pleopunctum species. This confidence-inspiring finding, supported by previous publications and our study (Boonmee et al. 2021; Koukol and Delgado 2021; Xu et al. 2023).
Paraphoma was established in 1983 with P.radicina as the type species (Morgan-Jones and White 1983). Subsequently, fourteen species are accepted in Paraphoma. Currently, fifteen Paraphoma species are accepted in Species Fungorum (2024). Paraphoma species are widely distributed worldwide, for instance in Australia, China, Czech Republic, Italy, the Netherlands, New Zealand, Russia, South Korea, Spain, Switzerland, Ukraine, the United Kingdom and the United States. These species include endophytes, pathogens, and saprobes on the plant of Atractylodeslancea, Buxussempervirens, Chrysanthemummorifolium, Campanularapunculoides, Convolvulaceae sp., Dioscoreatokoro, Juniperuscommunis, Paraphomavinacea, Rhaphiolepsisindica, Strawberry, Salixcf.alba, Tanacetumcinerariifolium, soil and dung (de Gruyter et al. 2010; Bensch et al. 2012; Quaedvlieg et al. 2013; Crous et al. 2017, 2021b; Moslemi et al. 2018; Gomzhina et al. 2020; Guarnaccia et al. 2022). Currently, only asexual morph has been reported from Paraphoma species (Quaedvlieg et al. 2013; Gomzhina et al. 2020). However, we have discovered the sexual morph of Paraphomaaquatica, which is saprobic on dead wood of Castanopsiscalathiformis (Fagaceae), and this is the first sexual morph recorded in Paraphoma.
Supplementary Material
Citation
Ren G-C, Tibpromma S, Dong K-X, Gao C-X, Zhang C-S, Karunarathna SC, Elgorban AM, Gui H (2024) Unveiling fungi associated with Castanopsis woody litter in Yunnan Province, China: Insights into Pleosporales (Dothideomycetes) species. MycoKeys 108: 15–45. https://doi.org/10.3897/mycokeys.108.127560
Funding Statement
The Yunnan Revitalization Talents Support Plan (High-End Foreign Experts Program) and the Key Laboratory of Yunnan Provincial Department of Education of the Deep-Time Evolution on Biodiversity from the Origin of the Pearl River for their support. The authors extend their appreciation to the Researchers supporting Project Number (RSP2024R56) King Saud University, Riyadh, Saudi Arabia.
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
Saowaluck Tibpromma and Samantha C. Karunarathna thank the Yunnan Revitalization Talents Support Plan (High-End Foreign Experts Program) and the Key Laboratory of Yunnan Provincial Department of Education of the Deep-Time Evolution on Biodiversity from the Origin of the Pearl River for their support. The authors extend their appreciation to the Deputyship for Research and Innovation, "Ministry of Education" in Saudi Arabia for funding this research (IFKSUOR3-299-17).
Author contributions
Conceptualization: GCR, Data curation: GCR, Investigation: GCR, ST, SCK, Project administration: GCR, ST, SCK, Software: GCR, Supervision: ST, SCK, Writing – review and editing: GCR, ST, KXD, CXG, CSZ, SCK, AME, HG.
Author ORCIDs
Guang-Cong Ren https://orcid.org/0000-0001-9923-2626
Saowaluck Tibpromma https://orcid.org/0000-0002-4706-6547
Samantha C. Karunarathna https://orcid.org/0000-0001-7080-0781
Abdallah M. Elgorban https://orcid.org/0000-0003-3664-7853
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Asian Development Bank (2012) Greater Mekong Subregion Atlas of the Environment (2nd edn). Asian Development Bank. http://hdl.handle.net/11540/92
- Barr ME. (1987) Prodromus to class Loculoascomycetes. Amherst. University of Massachusetts, Massachusetts.
- Bensch K, Braun U, Groenewald JZ, Crous PW. (2012) The genus Cladosporium. Studies in Mycology 72: 1–401. 10.3114/sim0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonmee S, Wanasinghe DN, Calabon MS, Huanraluek N, Chandrasiri SKU, Jones GEB, Rossi W, Leonardi M, Singh SK, Rana S, Singh PN, Maurya DK, Lagashetti AC, Choudhary D, Dai Y-C, Zhao C-L, Mu Y-H, Yuan H-S, He S-H, Phookamsak R, Jiang H-B, Martín MP, Dueñas M, Telleria MT, Kałucka IL, Jagodziński AM, Liimatainen K, Pereira DS, Phillips AJL, Suwannarach N, Kumla J, Khuna S, Lumyong S, Potter TB, Shivas RG, Sparks AH, Vaghefi N, Abdel-Wahab MA, Abdel-Aziz FA, Li G-J, Lin W-F, Singh U, Bhatt RP, Lee HB, Nguyen TTT, Kirk PM, Dutta AK, Acharya K, Sarma VV, Niranjan M, Rajeshkumar KC, Ashtekar N, Lad S, Wijayawardene NN, Bhat DJ, Xu R-J, Wijesinghe SN, Shen H-W, Luo Z-L, Zhang J-Y, Sysouphanthong P, Thongklang N, Bao D-F, Aluthmuhandiram JVS, Abdollahzadeh J, Javadi A, Dovana F, Usman M, Khalid AN, Dissanayake AJ, Telagathoti A, Probst M, Peintner U, Garrido-Benavent I, Bóna L, Merényi Z, Boros L, Zoltán B, Stielow JB, Jiang N, Tian C-M, Shams E, Dehghanizadeh F, Pordel A, Javan-Nikkhah M, Denchev TT, Denchev CM, Kemler M, Begerow D, Deng C-Y, Harrower E, Bozorov T, Kholmuradova T, Gafforov Y, Abdurazakov A, Xu J-C, Mortimer PE, Ren G-C, Jeewon R, Maharachchikumbura SSN, Phukhamsakda C, Mapook A, Hyde KD. (2021) Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 111(1): 1–335. 10.1007/s13225-021-00489-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmanage RS, Dayarathne MC, Wanasinghe DN, Thambugala KM, Jeewon R, Chethana KWT, Samarakoon MC, Tennakoon DS, De Silva NI, Camporesi E, Raza M, Yan JY, Hyde KD. (2020) Taxonomic novelties of saprobic Pleosporales from selected dicotyledons and grasses. Mycosphere: Journal of Fungal Biology 11(1): 2481–2541. 10.5943/mycosphere/11/1/15 [DOI] [Google Scholar]
- Cai L, Tsui CKM, Zhang KQ, Hyde KD. (2002) Aquatic fungi from Lake Fuxian, Yunnan, China. Fungal Diversity 9: 57–70. https://fungaldiversity.org/fdp/sfdp/9-4.pdf [Google Scholar]
- Crawford RH, Carpenter SE, Mayfield J, Martin RE. (1987) Fungi from foliage of Arctostaphylospatula, Castanopsischrysophylla, and Ceanothusvelutinus. US Department of Agriculture, Forest Service, Pacific Northwest Research Station. 10.5962/bhl.title.70795 [DOI]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GEStJ, Barber PA, Alvarado P, Barnes CW, Buchanan PK, Heykoop M, Moreno G, et al. (2017) Fungal Planet description sheets: 558–624. Persoonia 38: 240–384. 10.3767/003158517X698941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Osieck ER, Jurjević Ž, Boers J, van Iperen AL, Starink-Willemse M, Dima B, Balashov S, Bulgakov TS, Johnston PR, Morozova OV, Pinruan U, Sommai S, Alvarado P, Decock CA, Lebel T, McMullan-Fisher S, Moreno G, Shivas RG, Zhao L, Abdollahzadeh J, Abrinbana M, Ageev DV, Akhmetova G, Alexandrova AV, Altés A, Amaral AGG, Angelini C, Antonín V, Arenas F, Asselman P, Badali F, Baghela A, Bañares A, Barreto RW, Baseia IG, Bellanger JM, Berraf-Tebbal A, Biketova AYu, Bukharova NV, Burgess TI, Cabero J, Câmara MPS, Cano-Lira JF, Ceryngier P, Chávez R, Cowan DA, de Lima AF, Oliveira RL, Denman S, Dang QN, Dovana F, Duarte IG, Eichmeier A, Erhard A, Esteve-Raventós F, Fellin A, Ferisin G, Ferreira RJ, Ferrer A, Finy P, Gaya E, Geering ADW, Gil-Durán C, Glässnerová K, Glushakova AM, Gramaje D, Guard FE, Guarnizo AL, Haelewaters D, Halling RE, Hill R, Hirooka Y, Hubka V, Iliushin VA, Ivanova DD, Ivanushkina NE, Jangsantear P, Justo A, Kachalkin AV, Kato S, Khamsuntorn P, Kirtsideli IY, Knapp DG, Kochkina GA, Koukol O, Kovács GM, Kruse J, Kumar TKA, Kušan I, Læssøe T, Larsson E, Lebeuf R, Levicán G, Loizides M, Marinho P, Luangsa-ard JJ, Lukina EG, Magaña-Dueñas V, Maggs-Kölling G, Malysheva EF, Malysheva VF, Martín B, Martín MP, Matočec N, McTaggart AR, Mehrabi-Koushki M, Mešić A, Miller AN, Mironova P, Moreau P-A, Morte A, Müller K, Nagy LG, Nanu S, et al. (2021a) Fungal Planet description sheets: 1284–1382. Persoonia 47: 178–374. 10.3767/persoonia.2021.47.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK, Cowan DA, Maggs-Kölling G, Marais E, Wingfield MJ, Yilmaz N, Adan OCG, Akulov A, Duarte E, Berraf-Tebbal A, Bulgakov TS, Carnegie AJ, de Beer ZW, Decock C, Dijksterhuis J, Duong TA, Eichmeier A, Hien LT, Houbraken JAMP, Khanh TN, Liem NV, Lombard L, Lutzoni FM, Miadlikowska JM, Nel WJ, Pascoe IG, Roets F, Roux J, Samson RA, Shen M, Spetik M, Thangavel R, Thanh HM, Thao LD, van Nieuwenhuijzen EJ, Zhang JQ, Zhang Y, Zhao LL, Groenewald JZ. (2021b) New and Interesting Fungi. 4. Fungal Systematics and Evolution 7(1): 255–343. 10.3114/fuse.2021.07.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82: 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- de Gruyter J, Woudenberg JHC, Aveskamp MM, Verkley GJM, Groenewald JZ, Crous PW. (2010) Systematic reappraisal of species in PhomasectionParaphoma, Pyrenochaeta and Pleurophoma. Mycologia 102(5): 1066–1081. 10.3852/09-240 [DOI] [PubMed] [Google Scholar]
- Dong W, Wang B, Hyde KD, McKenzie EHC, Raja HA, Tanaka K, Abdel-Wahab MA, Abdel-Aziz FA, Doilom M, Phookamsak R, Hongsanan S, Wanasinghe DN, Yu XD, Wang GN, Yang H, Yang J, Thambugala KM, Tian Q, Luo ZL, Yang JB, Miller AN, Fournier J, Boonmee S, Hu DM, Nalumpang S, Zhang H, Fournier J, Boonmee S, Hu DM, Nalumpang S, Zhang H. (2020) Freshwater Dothideomycetes. Fungal Diversity 105(1): 319–575. 10.1007/s13225-020-00463-5 [DOI] [Google Scholar]
- Duong LM, McKenzie EHC, Lumyong S, Hyde KD. (2008) Fungal succession on senescent leaves of Castanopsisdiversifolia in Doi Suthep-Pui National Park, Thailand. Fungal Diversity 30: 23–36. [Google Scholar]
- Feng B, Yang Z. (2018) Studies on diversity of higher fungi in Yunnan, southwestern China: A review. Plant Diversity 40(4): 165–171. 10.1016/j.pld.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Liu F, Cai L. (2016) Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14(1): 102–117. 10.1080/14772000.2015.1101027 [DOI] [Google Scholar]
- Gomzhina MM, Gasich EL, Khlopunova LB, Gannibal PB. (2020) Paraphoma species associated with Convolvulaceae. Mycological Progress 19(3): 185–194. 10.1007/s11557-020-01558-8 [DOI] [Google Scholar]
- Guarnaccia V, Martino I, Tabone G, Crous PW, Gullino ML. (2022) Paraphomagaribaldii sp. nov. causing leaf spot disease of Campanularapunculoides in Italy. Fungal Systematics and Evolution 9(1): 19–26. 10.3114/fuse.2022.09.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hapuarachchi KK, Karunarathna SC, Phengsintham P, Yang HD, Kakumyan P, Hyde KD, Wen TC. (2019) Ganodermataceae (Polyporales): Diversity in Greater Mekong Subregion countries (China, Laos, Myanmar, Thailand, and Vietnam). Mycosphere 10(1): 221–309. 10.5943/mycosphere/10/1/6 [DOI] [Google Scholar]
- Hashimoto A, Hirayama K, Takahashi H, Matsumura M, Okada G, Chen CY, Huang JW, Kakishima M, Ono T, Tanaka T. (2018) Resolving the Lophiostomabipolare complex: Generic delimitations within Lophiostomataceae. Studies in Mycology 90(1): 161–189. 10.1016/j.simyco.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, McKenzie EHC, Sarma VV, Boonmee S, Lücking R, Bhat DJ, Liu NG. (2020) Refned families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11(1): 1553–2107. 10.5943/mycosphere/11/1/13 [DOI] [Google Scholar]
- Huang CC, Zhang YT, Bartholomew B. (1999) Fagaceae. In: Wu ZY, Raven PH. (Eds) Flora of China, vol.4. Science Press, Beijing & Missouri Botanical Garden Press, St. Louis, 314–400.
- Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui BK, Daranagama DA, Das K, Dayarathne MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Go’es-Neto A, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li WJ, Lin CG, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, Santiago ALCMA, Drechsler-Santos ER, Senanayake IC, Tanaka K. (2016) Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Wanasinghe DN, Lücking R, Aptroot A, Cáceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, de Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo Z-L, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Bart B, Randrianjohany E, Hofstetter V, Gibertoni TB, Soares AMS, Plautz Jr HL, Sotão HMP, Xavier WKS, Bezerra JDP, de Oliveira TGL, de Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang S-N, Bao D-F, Samarakoon MC, Pem D, Karunarathna A, Lin C-G, Yang J, Perera RH, Kumar V, Huang S-K, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao Y, Konta S, Niskanen T, Liimatainen K, Dai Y-C, Ji X-H, Tian X-M, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu Y-Z, Jiang H-B, Zhang J-F, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei D, Réblová M, Fournier J, Nekvindová J, do Nascimento Barbosa R, dos Santos JEF, de Oliveira NT, Li G-J, Ertz D, Shang Q-J, Phillips AJL, Kuo C-H, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng X-Y, Fryar S, Tkalčec Z, Liang J, Li G, Wen T-C, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao R-L, Zhao Q, Kirk PM, Liu J-K, Yan JY, Mortimer PE, Xu J, Doilom M. (2019) Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96(1): 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Hyde KD, Jeewon R, Chen YJ, Bhunjun CS, Calabon MS, Jiang HB, Lin CG, Norphanphoun C, Sysouphanthong P, Pem D, Tibpromma S, Zhang Q, Doilom M, Jayawardena RS, Liu JK, Maharachchikumbura SSN, Phukhamsakda C, Phookamsak R, Al-Sadi AM, Naritsada Thongklang N, Wang Y, Gafforov Y, Jones EBG, Lumyong S. (2020) The numbers of fungi: Is the descriptive curve ffattening? Fungal Diversity 103(1): 219–271. 10.1007/s13225-020-00458-2 [DOI]
- Inácio CA, Cannon PF, Ferry BF. (2005) Revision of the genus Placostromella and inclusion of Palawaniellacastanopsis as a third species. Mycological Progress 4(2): 133–137. 10.1007/s11557-006-0116-6 [DOI] [Google Scholar]
- Index Fungorum (2024) Index Fungorum. http://www.indexfungorum.org/names/Names.asp
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu J-K, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo J-M, Ghobad-Nejhad M, Nilsson H, Pang K-L, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen T-C, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li W-J, Perera RH, Phookamsak R, de Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao R-L, Zhao Q, Kang J-C, Promputtha I. (2015) The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74(1): 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Jayawardena RS, Hyde KD, Chen YJ, Papp V, Palla B, Papp D, Bhunjun CS, Hurdeal VG, Senwanna C, Manawasinghe IS, Harischandra DL, Gautam AK, Avasthi S, Chuankid B, Goonasekara ID, Hongsanan S, Zeng XY, Liyanage KK, Liu NG, Karunarathna A, Hapuarachchi KK, Luangharn T, Raspé O, Brahmanage R, Doilom M, Lee HB, Mei L, Jeewon R, Huanraluek N, Chaiwan N, Stadler M, Wang Y. (2020) One stop shop IV: taxonomic update with molecular phylogeny for important phytopathogenic genera: 76–100 (2020). Fungal Diversity 103: 87–218. 10.1007/s13225-020-00460-8 [DOI] [Google Scholar]
- Jiang HB, Phookamsak R, Hyde KD, Mortimer PE, Xu JC, Kakumyan P, Karunarathna SC, Kumla J. (2021) A taxonomic appraisal of bambusicolous fungi in Occultibambusaceae (Pleosporales, Dothideomycetes) with new collections from Yunnan Province, China. Life (Basel, Switzerland) 11(9): 932. 10.3390/life11090932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukol O, Delgado G. (2021) Why morphology matters: The negative consequences of hasty descriptions of putative novelties in asexual ascomycetes. IMA Fungus 12(1): 1–8. 10.1186/s43008-021-00073-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16(12): 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Liu NG, Hyde KD, Bhat DJ, Jumpathong J, Liu JK. (2019) Morphological and phylogenetic studies of Pleopunctum gen. nov. (Phaeoseptaceae, Pleosporales) from China. Mycosphere 10(1): 757–775. 10.5943/mycosphere/10/1/17 [DOI] [Google Scholar]
- Luo Z, Hyde KD, Bhat DJ, Jeewon R, Maharachchikumbura SSN, Bao DF, Li WL, Su XJ, Yang XY, Su HY. (2018) Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycological Progress 17(5): 511–530. 10.1007/s11557-018-1377-6 [DOI] [Google Scholar]
- Magaña-Dueñas V, Cano-Lira JF, Stchigel AM. (2021) New Dothideomycetes from freshwater habitats in Spain. Journal of Fungi (Basel, Switzerland) 7(12): 1102. 10.3390/jof7121102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharachchikumbura SSN, Wanasinghe DN, Cheewangkoon R, Al-Sadi AM. (2021) Uncovering the hidden taxonomic diversity of fungi in Oman. Fungal Diversity 106(1): 229–268. 10.1007/s13225-020-00467-1 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Morgan-Jones G, White JF. (1983) Studies in the genus Phoma. iii: Paraphoma, a new genus to accomodate Phomaradicina 1983. Mycotaxon 18: 57–65. [Google Scholar]
- Mortimer PE, Jeewon R, Xu JC, Lumyong S, Wanasinghe DN. (2021) Morpho-phylo taxonomy of novel dothideomycetous fungi associated with dead woody twigs in Yunnan Province, China. Frontiers in Microbiology 12: 654683. 10.3389/fmicb.2021.654683 [DOI] [PMC free article] [PubMed]
- Moslemi A, Ades PK, Groom T, Crous PW, Nicolas ME, Taylor PWJ. (2016) Paraphoma crown rot of pyrethrum (Tanacetumcinerariifolium). Plant Disease 100(12): 2363–2369. 10.1094/PDIS-05-16-0628-RE [DOI] [PubMed] [Google Scholar]
- Moslemi A, Ades PK, Crous PW, Groom T, Scott JB, Nicolas ME, Taylor PWJ. (2018) Paraphomachlamydocopiosa sp. nov. and Paraphomapye sp. nov., two new species associated with leaf and crown infection of pyrethrum. Plant Pathology 67(1): 124–135. 10.1111/ppa.12719 [DOI] [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. (2008) AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics (Oxford, England) 24(4): 581–583. 10.1093/bioinformatics/btm388 [DOI] [PubMed] [Google Scholar]
- Osono T, Matsuoka S, Hirose D. (2020) Diversity and geographic distribution of ligninolytic fungi associated with Castanopsissieboldii leaf litter in Japan. Frontiers in Microbiology 11: 595427. 10.3389/fmicb.2020.595427 [DOI] [PMC free article] [PubMed]
- Pang W, Zhang P, Zhang Y, Zhang X, Huang Y, Zhang T, Liu B. (2023) The ectomycorrhizal fungi and soil bacterial communities of the five typical tree species in the Junzifeng national nature reserve, Southeast China. Plants 12(22): 3853. 10.3390/plants12223853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukhamsakda C, McKenzie EHC, Phillips AJL, Jones EBG, Bhat DJ, Marc S, Bhunjun CS, Wanasinghe DN, Thongbai B, Camporesi E, Ertz D, Jayawardena RS, Perera RH, Ekanayake AH, Tibpromma S, Doilom M, Xu J, Hyde KD. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102(1): 1–203. 10.1007/s13225-020-00448-4 [DOI] [Google Scholar]
- Quaedvlieg W, Verkley GJM, Shin HD, Barreto RW, Alfenas AC, Swart WJ, Groenewald JZ, Crous PW. (2013) Sizing up septoria. Studies in Mycology 75: 307–390. 10.3114/sim0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2012) FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. Journal of Molecular Evolution 43(3): 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97(1): 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- Ren G, Wanasinghe DN, de Farias ARG, Hyde KD, Yasanthika E, Xu J, Balasuriya A, Chethana KWT, Gui H. (2022) Taxonomic novelties of woody litter fungi (Didymosphaeriaceae, Pleosporales) from the Greater Mekong Subregion. Biology (Basel) 11(11): 1660. 10.3390/biology11111660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senwanna C, Mapook A, Samarakoon MC, Karunarathna A, Wang Y, Tang AMC, Haituk S, Suwannarach N, Hyde KD, Cheewangkoon R. (2021) Ascomycetes on Para rubber (Heveabrasiliensis). Mycosphere: Journal of Fungal Biology 12(1): 1334–1512. 10.5943/mycosphere/12/1/18 [DOI] [Google Scholar]
- Species Fungorum (2024) Species Fungorum. https://www.speciesfungorum.org/Names/Names.asp [Accessed on May 15, 2024]
- Stamatakis A. (2014) RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan WH, Ong L, Strijk JS. (2023) Castanopsiscorallocarpus (Fagaceae), a new species from Royal Belum (Perak) in Peninsular Malaysia. PhytoKeys 219: 1–10. 10.3897/phytokeys.219.95991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AMC, Jeewon R, Hyde KD. (2005) Succession of microfungal communities on decaying leaves of Castanopsisfissa. Canadian Journal of Microbiology 51(11): 967–974. 10.1139/w05-086 [DOI] [PubMed] [Google Scholar]
- Tennakoon DS, Kuo CH, Jeewon R, Thambugala KM, Hyde KD. (2018) Saprobic Lophiostomataceae (Dothideomycetes): Pseudolophiostomamangiferae sp. nov. and Neovaginatisporafuckelii, a new record from Mangiferaindica. Phytotaxa 364(2): 157–171. 10.11646/phytotaxa.364.2.3 [DOI] [Google Scholar]
- Thambugala KM, Hyde KD, Tanaka K, Tian Q, Wanasinghe DN, Ariyawansa HA, Jayasiri SC, Boonmee S, Camporesi E, Hashimoto A, Hirayama K, Schumacher RK, Promputtha I, Liu ZY. (2015) Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Diversity 74(1): 199–266. 10.1007/s13225-015-0348-3 [DOI] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24(24): 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Mortimer PE. (2022) Taxonomic and phylogenetic insights into novel Ascomycota from forest woody litter. Biology (Basel) 11(6): 889. 10.3390/biology11060889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Mortimer PE, Senwanna C, Cheewangkoon R. (2020) Saprobic Dothideomycetes in Thailand: Phaeoseptumhydei sp. nov., a new terrestrial ascomycete in Phaeoseptaceae. Phytotaxa 449(2): 149–163. 10.11646/phytotaxa.449.2.3 [DOI] [Google Scholar]
- Wanasinghe DN, Mortimer PE, Xu JC. (2021) Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaeaviscosa in Honghe (China). Journal of Fungi (Basel, Switzerland) 7(3): 180. 10.3390/jof7030180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: A Guide to Methods and Applications. Academic Press, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdoğdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M. (2022) Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13(1): 53–453. 10.5943/mycosphere/13/1/2 [DOI] [Google Scholar]
- Xu RJ, Zhu YA, Liu NG, Boonmee S, Zhou DQ, Zhao Q. (2023) Taxonomy and phylogeny of hyphomycetous muriform conidial taxa from the Tibetan Plateau, China. Journal of Fungi (Basel, Switzerland) 9(5): 560. 10.3390/jof9050560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu LL, Jones EBG, Hyde KD, Liu ZY, Bao DF, Liu NG, Li WL, Shen HW, Yu XD, Liu JK. (2023) Freshwater fungi from karst landscapes in China and Thailand. Fungal Diversity 119(1): 1–212. 10.1007/s13225-023-00514-7 [DOI] [Google Scholar]
- Yu XD, Zhang SN, Liu JK. (2022) Morpho-phylogenetic evidence reveals novel Pleosporalean taxa from Sichuan Province, China. Journal of Fungi (Basel, Switzerland) 8(7): 720. 10.3390/jof8070720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JF, Liu JK, Hyde KD, Yang W, Liu ZY. (2017) Fungi from Asian Karst formations II. Two new species of Occultibambusa (Occultibambusaceae, Dothideomycetes) from karst landforms of China. Mycosphere: Journal of Fungal Biology 8(4): 550–559. 10.5943/mycosphere/8/4/4 [DOI] [Google Scholar]
- Zhang YZ, Chen QL, Ma J, Lu YZ, Chen HB, Liu NG. (2023) Morphological and multigene phylogenetic analyses reveal five new hyphomycetes from freshwater habitats. Frontiers in Microbiology 14: 1253239. 10.3389/fmicb.2023.1253239 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.