Abstract
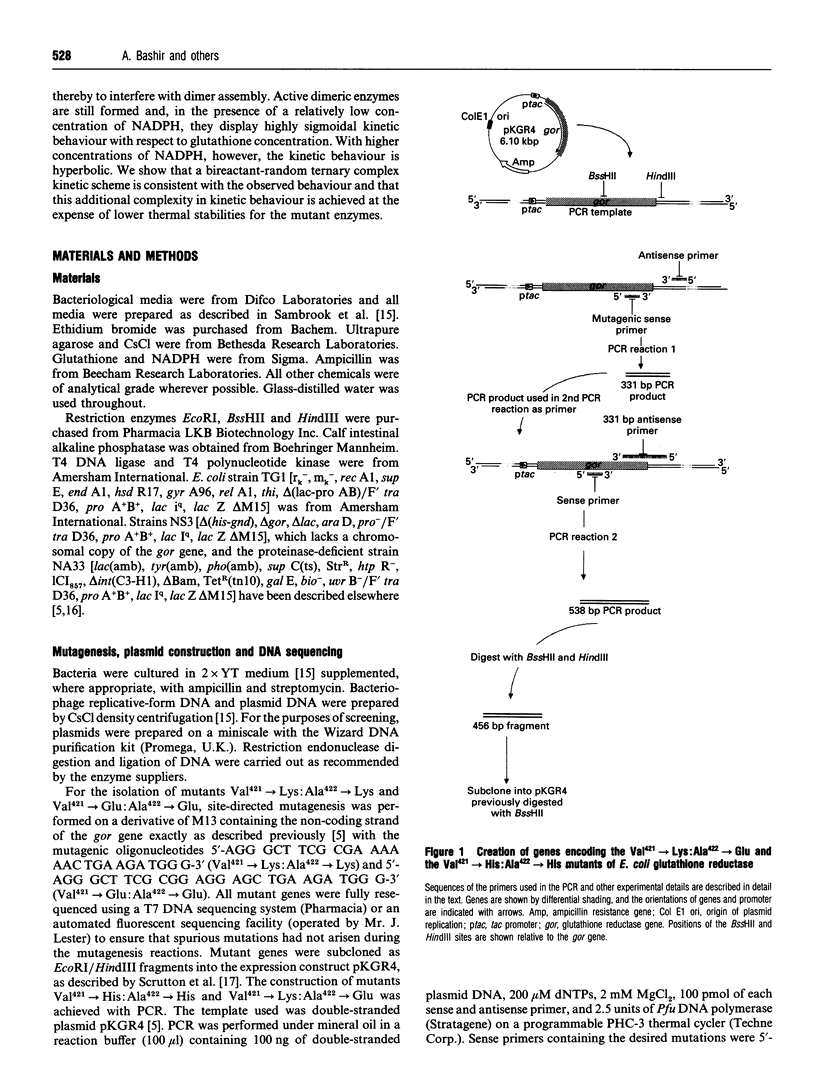
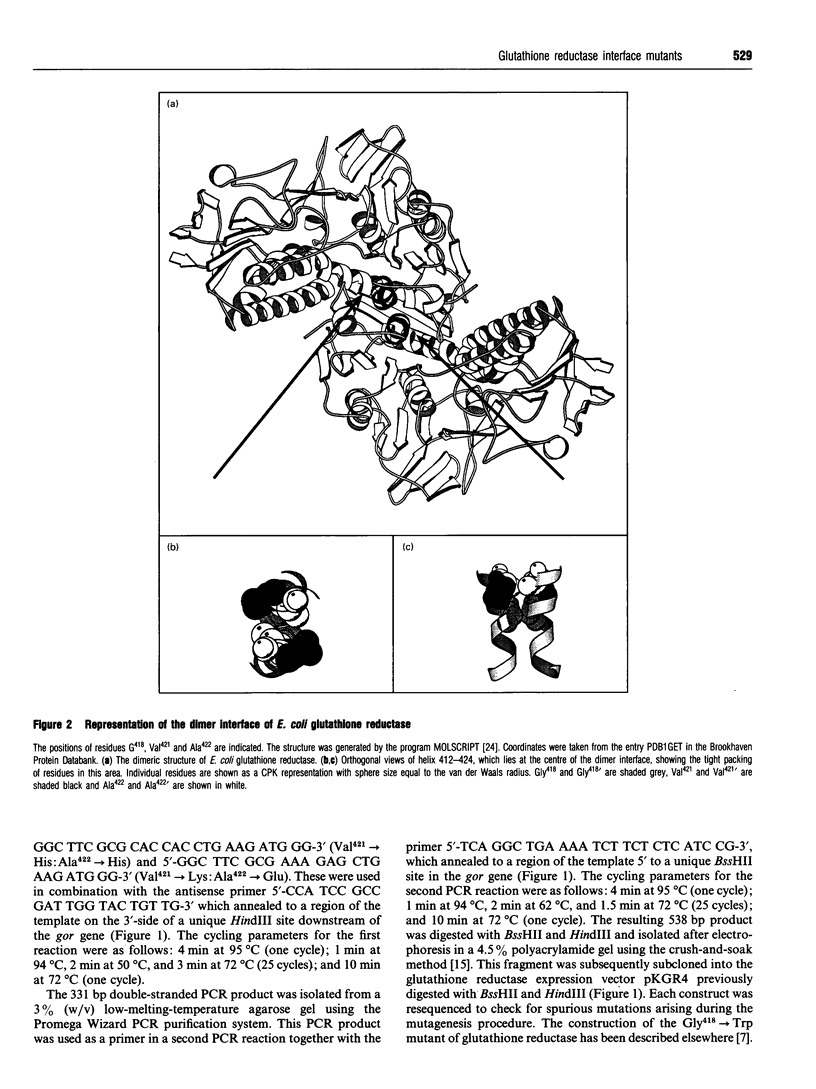
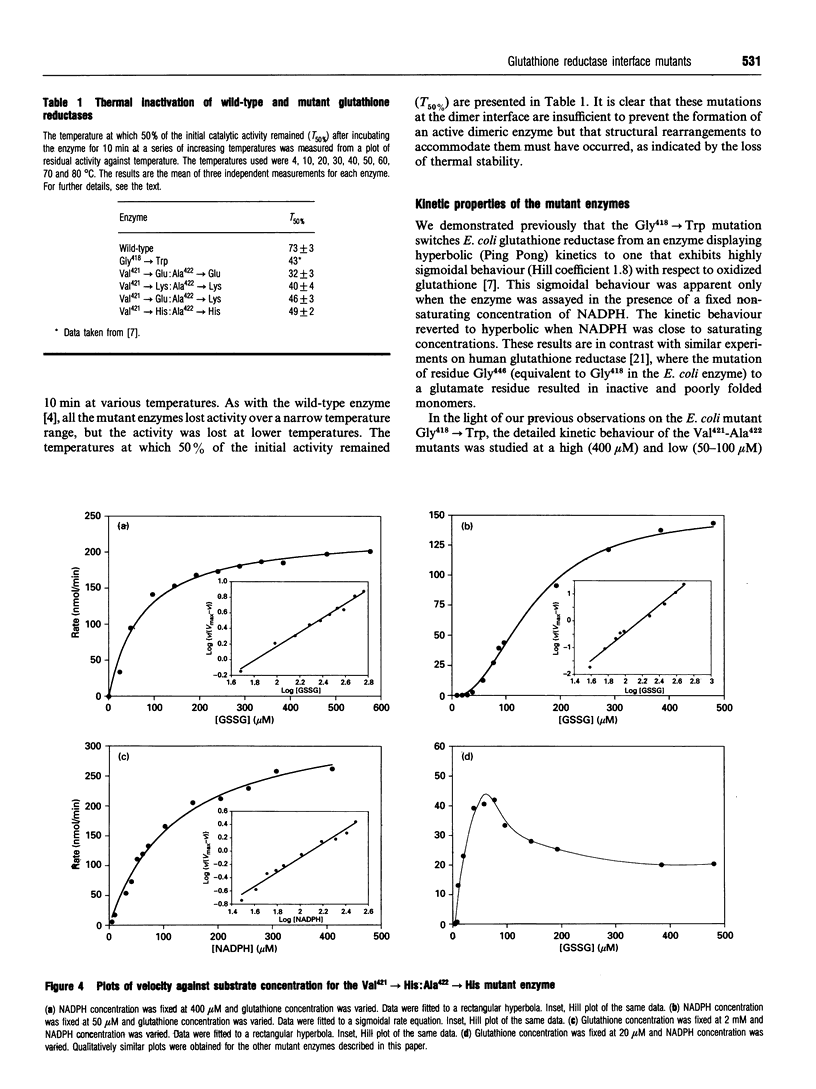
In wild-type glutathione reductase from Escherichia coli residues Val421 and Ala422 are located in an alpha-helix in a densely packed and hydrophobic region of the dimer interface, with their side chains packed against those of residues Ala422' and Val421' in the second subunit. A series of mutant glutathione reductases was constructed in which the identities of the residues at positions 421 and 422 were changed. Mutations were designed so as to present like charges (mutants Val421-->Glu:Ala422-->Glu and Val421-->Lys:Ala422-->Lys) or opposite charges (mutant Val421-->Lys:Ala422-->Glu) across the dimer interface to assess the role of electrostatic interactions in dimer stability. A fourth mutant (Val421-->His:Ala422-->His) was also constructed to investigate the effects of introducing a potentially protonatable bulky side chain into a crowded region of the dimer interface. In all cases, an active dimeric enzyme was found to be assembled but each mutant protein was thermally destabilized. A detailed steady-state kinetic analysis indicated that each mutant enzyme no longer displayed the Ping Pong kinetic behaviour associated with the wild-type enzyme but exhibited what was best described as a random bireactant ternary complex mechanism. This leads, depending on the chosen substrate concentration, to apparent sigmoidal, hyperbolic or complex kinetic behaviour. These experiments, together with others reported previously, indicate that simple mutagenic changes in regions distant from the active site can lead to dramatic switches in steady-state kinetic mechanism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akerboom T. P., Bilzer M., Sies H. The relationship of biliary glutathione disulfide efflux and intracellular glutathione disulfide content in perfused rat liver. J Biol Chem. 1982 Apr 25;257(8):4248–4252. [PubMed] [Google Scholar]
- Berry A., Scrutton N. S., Perham R. N. Switching kinetic mechanism and putative proton donor by directed mutagenesis of glutathione reductase. Biochemistry. 1989 Feb 7;28(3):1264–1269. doi: 10.1021/bi00429a047. [DOI] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Deonarain M. P., Berry A., Scrutton N. S., Perham R. N. Alternative proton donors/acceptors in the catalytic mechanism of the glutathione reductase of Escherichia coli: the role of histidine-439 and tyrosine-99. Biochemistry. 1989 Dec 12;28(25):9602–9607. doi: 10.1021/bi00451a008. [DOI] [PubMed] [Google Scholar]
- Ferdinand W. The interpretation of non-hyperbolic rate curves for two-substrate enzymes. A possible mechanism for phosphofructokinase. Biochem J. 1966 Jan;98(1):278–283. doi: 10.1042/bj0980278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer S., Perham R. N. Glutathione reductase from Escherichia coli: cloning and sequence analysis of the gene and relationship to other flavoprotein disulfide oxidoreductases. Biochemistry. 1986 May 6;25(9):2736–2742. doi: 10.1021/bi00357a069. [DOI] [PubMed] [Google Scholar]
- Henderson G. B., Murgolo N. J., Kuriyan J., Osapay K., Kominos D., Berry A., Scrutton N. S., Hinchliffe N. W., Perham R. N., Cerami A. Engineering the substrate specificity of glutathione reductase toward that of trypanothione reduction. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8769–8773. doi: 10.1073/pnas.88.19.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leistler B., Perham R. N. Solubilizing buried domains of proteins: a self-assembling interface domain from glutathione reductase. Biochemistry. 1994 Mar 15;33(10):2773–2781. doi: 10.1021/bi00176a005. [DOI] [PubMed] [Google Scholar]
- Mittl P. R., Berry A., Scrutton N. S., Perham R. N., Schulz G. E. Anatomy of an engineered NAD-binding site. Protein Sci. 1994 Sep;3(9):1504–1514. doi: 10.1002/pro.5560030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittl P. R., Berry A., Scrutton N. S., Perham R. N., Schulz G. E. Structural differences between wild-type NADP-dependent glutathione reductase from Escherichia coli and a redesigned NAD-dependent mutant. J Mol Biol. 1993 May 20;231(2):191–195. doi: 10.1006/jmbi.1993.1274. [DOI] [PubMed] [Google Scholar]
- Mittl P. R., Schulz G. E. Structure of glutathione reductase from Escherichia coli at 1.86 A resolution: comparison with the enzyme from human erythrocytes. Protein Sci. 1994 May;3(5):799–809. doi: 10.1002/pro.5560030509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordhoff A., Bücheler U. S., Werner D., Schirmer R. H. Folding of the four domains and dimerization are impaired by the Gly446-->Glu exchange in human glutathione reductase. Implications for the design of antiparasitic drugs. Biochemistry. 1993 Apr 20;32(15):4060–4066. doi: 10.1021/bi00066a029. [DOI] [PubMed] [Google Scholar]
- ORNSTEIN L. DISC ELECTROPHORESIS. I. BACKGROUND AND THEORY. Ann N Y Acad Sci. 1964 Dec 28;121:321–349. doi: 10.1111/j.1749-6632.1964.tb14207.x. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Deonarain M. P., Perham R. N. Active site complementation in engineered heterodimers of Escherichia coli glutathione reductase created in vivo. Proc Biol Sci. 1990 Dec 22;242(1305):217–224. doi: 10.1098/rspb.1990.0127. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Engineering of an intersubunit disulphide bridge in glutathione reductase from Escherichia coli. FEBS Lett. 1988 Dec 5;241(1-2):46–50. doi: 10.1016/0014-5793(88)81028-7. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Purification and characterization of glutathione reductase encoded by a cloned and over-expressed gene in Escherichia coli. Biochem J. 1987 Aug 1;245(3):875–880. doi: 10.1042/bj2450875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Deonarain M. P., Berry A., Perham R. N. Cooperativity induced by a single mutation at the subunit interface of a dimeric enzyme: glutathione reductase. Science. 1992 Nov 13;258(5085):1140–1143. doi: 10.1126/science.1439821. [DOI] [PubMed] [Google Scholar]