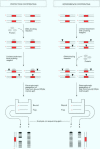
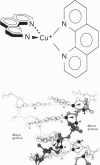
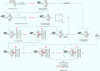
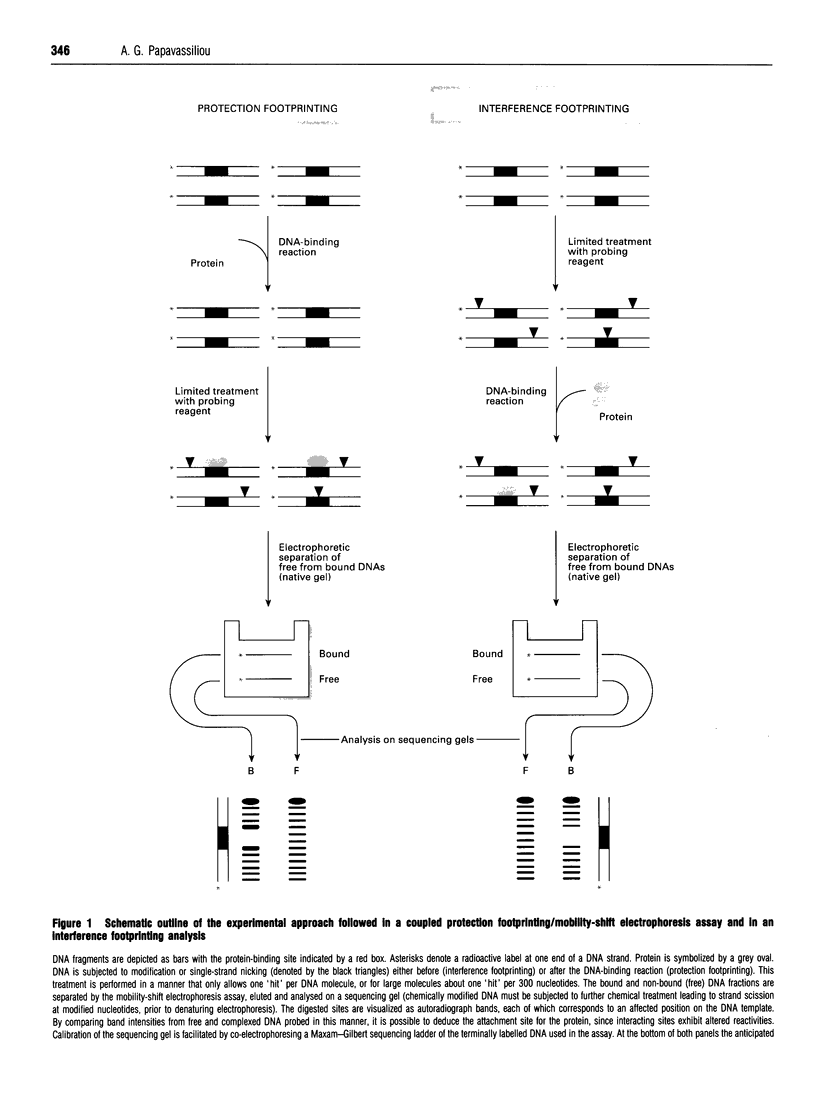
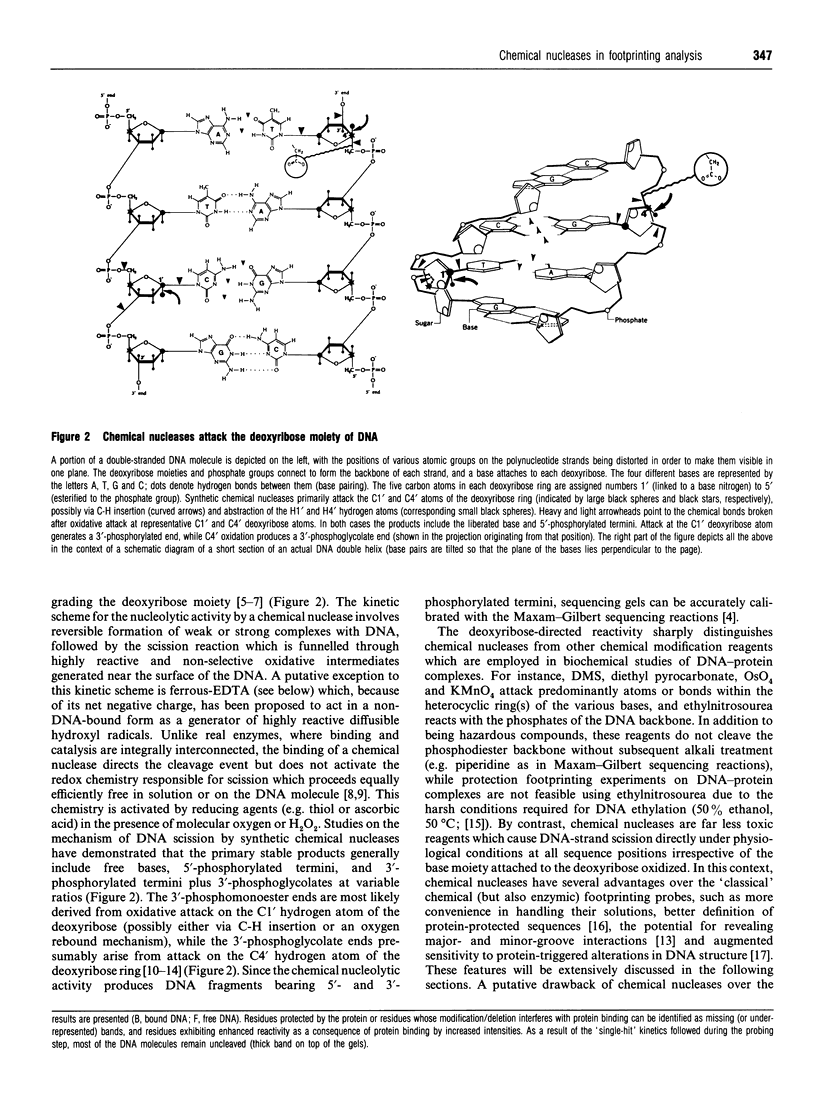
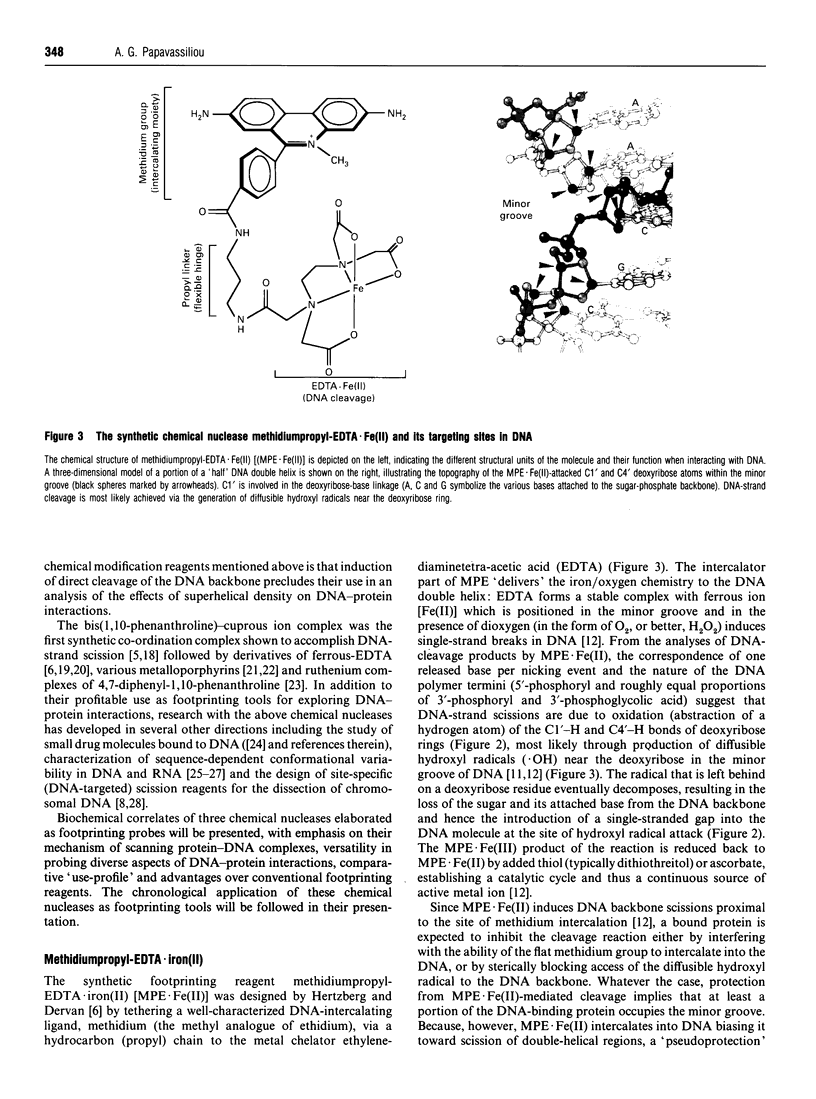
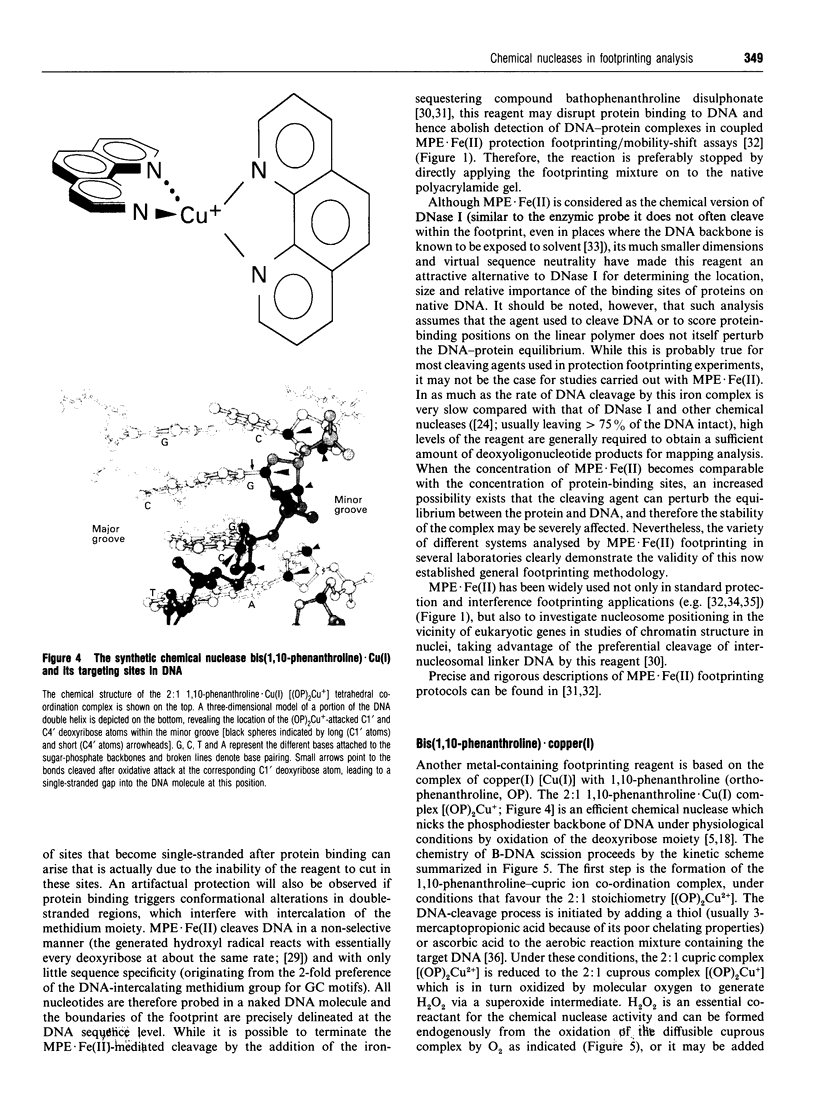
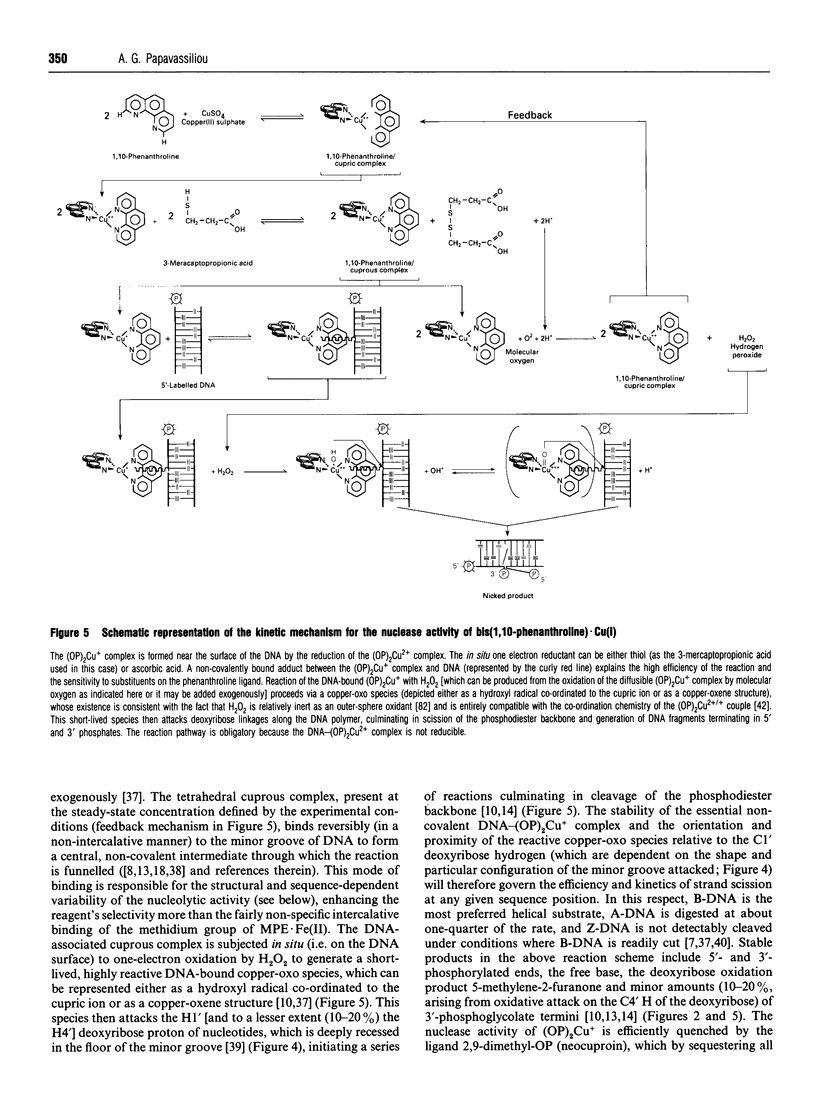
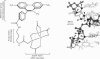Full text
PDF

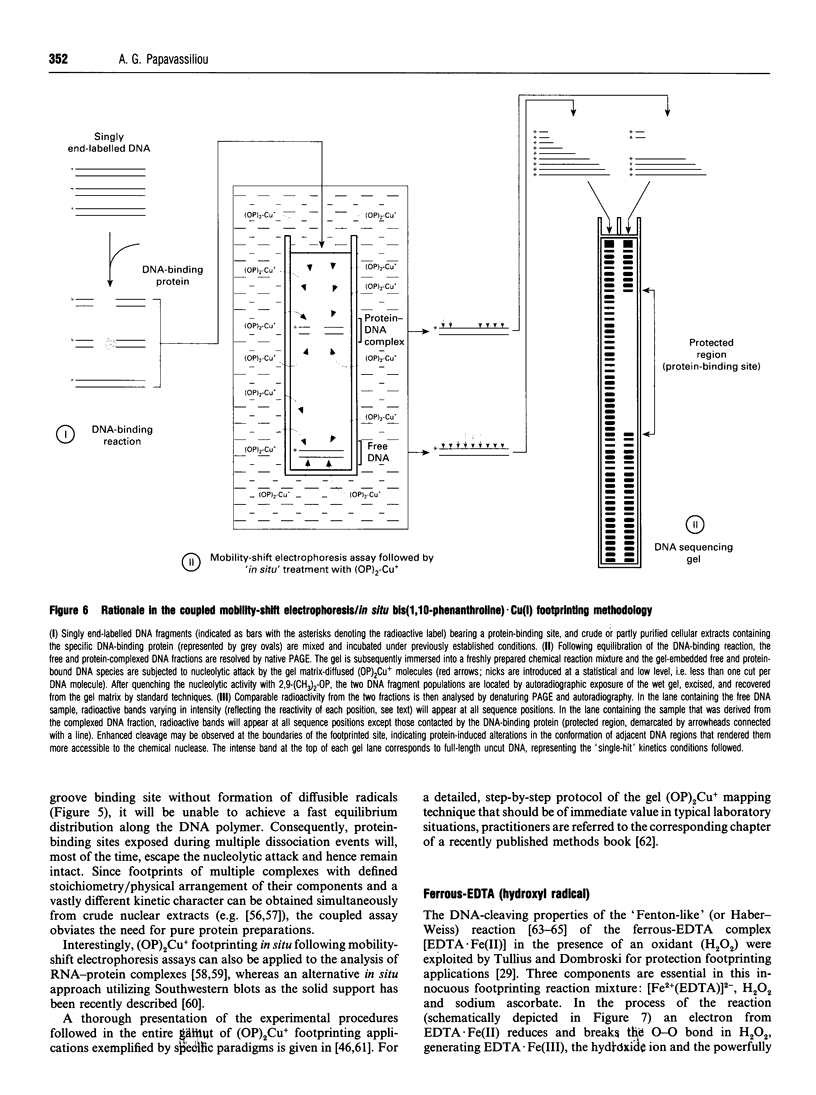
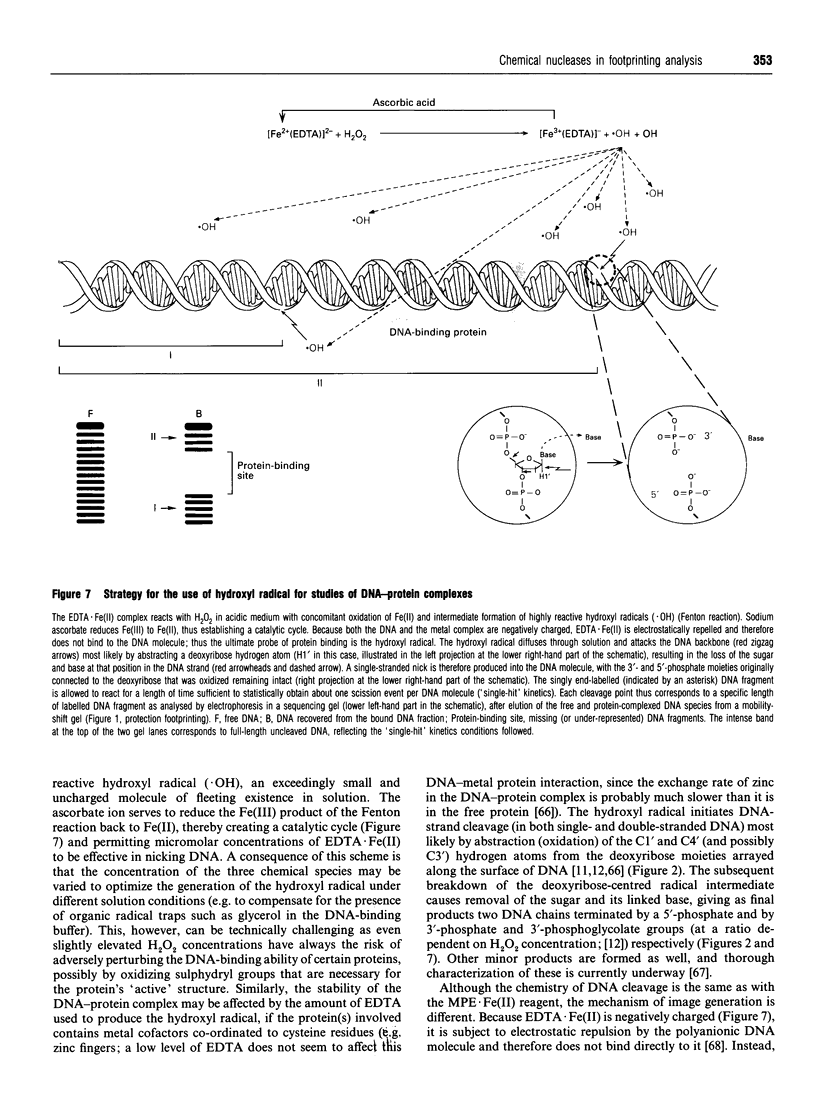

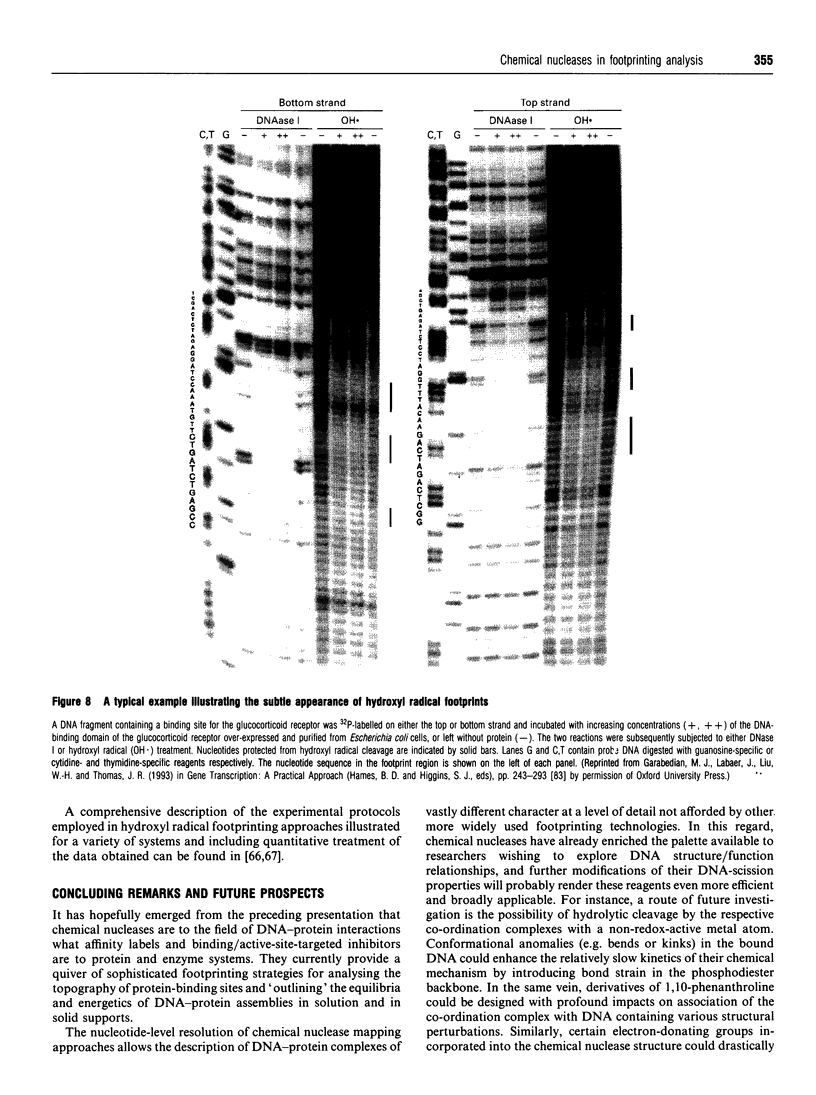


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barton J. K. Metals and DNA: molecular left-handed complements. Science. 1986 Aug 15;233(4765):727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Hertzberg R. P., Dervan P. B., Elgin S. C. Cleavage of chromatin with methidiumpropyl-EDTA . iron(II). Proc Natl Acad Sci U S A. 1983 Jun;80(11):3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry. 1990 Feb 13;29(6):1355–1361. doi: 10.1021/bi00458a001. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Gartenberg M. R., Shrader T. E. DNA bending in protein-DNA complexes. Methods Enzymol. 1991;208:118–146. doi: 10.1016/0076-6879(91)08011-6. [DOI] [PubMed] [Google Scholar]
- Dervan P. B. Design of sequence-specific DNA-binding molecules. Science. 1986 Apr 25;232(4749):464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Dixon W. J., Hayes J. J., Levin J. R., Weidner M. F., Dombroski B. A., Tullius T. D. Hydroxyl radical footprinting. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Flanagan W. M., Papavassiliou A. G., Rice M., Hecht L. B., Silverstein S., Wagner E. K. Analysis of the herpes simplex virus type 1 promoter controlling the expression of UL38, a true late gene involved in capsid assembly. J Virol. 1991 Feb;65(2):769–786. doi: 10.1128/jvi.65.2.769-786.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A. D., Mattaj I. W., Rio D. C. RNA-protein interactions. Cell. 1991 Dec 20;67(6):1041–1046. doi: 10.1016/0092-8674(91)90282-4. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry. 1989 Nov 28;28(24):9521–9527. doi: 10.1021/bi00450a041. [DOI] [PubMed] [Google Scholar]
- Henner W. D., Grunberg S. M., Haseltine W. A. Enzyme action at 3' termini of ionizing radiation-induced DNA strand breaks. J Biol Chem. 1983 Dec 25;258(24):15198–15205. [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Jezewska M. J., Bujalowski W., Lohman T. M. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of DNA is highly specific for duplex DNA. Biochemistry. 1989 Jul 25;28(15):6161–6164. doi: 10.1021/bi00441a006. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kakkis E., Calame K. A plasmacytoma-specific factor binds the c-myc promoter region. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7031–7035. doi: 10.1073/pnas.84.20.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Kuwabara M., Yoon C., Goyne T., Thederahn T., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper ion: reaction with CGCGAATTCGCG and its complexes with netropsin and EcoRI. Biochemistry. 1986 Nov 18;25(23):7401–7408. doi: 10.1021/bi00371a023. [DOI] [PubMed] [Google Scholar]
- Landolfi N. F., Yin X. M., Capra J. D., Tucker P. W. Protection analysis (or "footprinting") of specific protein-DNA complexes in crude nuclear extracts using methidiumpropyl-EDTA-iron (II). Biotechniques. 1989 May;7(5):500–504. [PubMed] [Google Scholar]
- Law R., Kuwabara M. D., Briskin M., Fasel N., Hermanson G., Sigman D. S., Wall R. Protein-binding site at the immunoglobulin mu membrane polyadenylylation signal: possible role in transcription termination. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9160–9164. doi: 10.1073/pnas.84.24.9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Marshall L. E., Graham D. R., Reich K. A., Sigman D. S. Cleavage of deoxyribonucleic acid by the 1,10-phenanthroline-cuprous complex. Hydrogen peroxide requirement and primary and secondary structure specificity. Biochemistry. 1981 Jan 20;20(2):244–250. doi: 10.1021/bi00505a003. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavassiliou A. G. 1,10-Phenanthroline-copper ion nuclease footprinting of DNA-protein complexes in situ following mobility-shift electrophoresis assays. Methods Mol Biol. 1994;30:43–78. doi: 10.1385/0-89603-256-6:43. [DOI] [PubMed] [Google Scholar]
- Papavassiliou A. G. In situ (OP)2-Cu+ mapping of electrophoretically resolved RNA-protein complexes. Anal Biochem. 1993 Oct;214(1):331–334. doi: 10.1006/abio.1993.1498. [DOI] [PubMed] [Google Scholar]
- Papavassiliou A. G. Localisation of DNA-protein contact points by DMS resistance of complexes resolved in gel retardation assays. Nucleic Acids Res. 1993 Feb 11;21(3):757–758. doi: 10.1093/nar/21.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavassiliou A. G., Silverstein S. J. Interaction of cell and virus proteins with DNA sequences encompassing the promoter/regulatory and leader regions of the herpes simplex virus thymidine kinase gene. J Biol Chem. 1990 Jun 5;265(16):9402–9412. [PubMed] [Google Scholar]
- Pope L. E., Sigman D. S. Secondary structure specificity of the nuclease activity of the 1,10-phenanthroline-copper complex. Proc Natl Acad Sci U S A. 1984 Jan;81(1):3–7. doi: 10.1073/pnas.81.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope L. M., Reich K. A., Graham D. R., Sigman D. S. Products of DNA cleavage by the 1,10-phenanthroline-copper complex. Inhibitors of Escherichia coli DNA polymerase I. J Biol Chem. 1982 Oct 25;257(20):12121–12128. [PubMed] [Google Scholar]
- Revzin A., Ceglarek J. A., Garner M. M. Comparison of nucleic acid-protein interactions in solution and in polyacrylamide gels. Anal Biochem. 1986 Feb 15;153(1):172–177. doi: 10.1016/0003-2697(86)90077-1. [DOI] [PubMed] [Google Scholar]
- Rush J. D., Maskos Z., Koppenol W. H. Distinction between hydroxyl radical and ferryl species. Methods Enzymol. 1990;186:148–156. doi: 10.1016/0076-6879(90)86104-4. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman D. S. Chemical nucleases. Biochemistry. 1990 Oct 2;29(39):9097–9105. doi: 10.1021/bi00491a001. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Chen C. H. Chemical nucleases: new reagents in molecular biology. Annu Rev Biochem. 1990;59:207–236. doi: 10.1146/annurev.bi.59.070190.001231. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Graham D. R., D'Aurora V., Stern A. M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline . cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J Biol Chem. 1979 Dec 25;254(24):12269–12272. [PubMed] [Google Scholar]
- Sigman D. S., Kuwabara M. D., Chen C. H., Bruice T. W. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 1991;208:414–433. doi: 10.1016/0076-6879(91)08022-a. [DOI] [PubMed] [Google Scholar]
- Spassky A., Rimsky S., Buc H., Busby S. Correlation between the conformation of Escherichia coli -10 hexamer sequences and promoter strength: use of orthophenanthroline cuprous complex as a structural index. EMBO J. 1988 Jun;7(6):1871–1879. doi: 10.1002/j.1460-2075.1988.tb03020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky A., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper ion. Conformational analysis and footprinting of the lac operon. Biochemistry. 1985 Dec 31;24(27):8050–8056. doi: 10.1021/bi00348a032. [DOI] [PubMed] [Google Scholar]
- Spassky A. Visualization of the movement of the Escherichia coli RNA polymerase along the lac UV5 promoter during the initiation of the transcription. J Mol Biol. 1986 Mar 5;188(1):99–103. doi: 10.1016/0022-2836(86)90484-5. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Thederahn T., Spassky A., Kuwabara M. D., Sigman D. S. Chemical nuclease activity of 5-phenyl-1,10-phenanthroline-copper ion detects intermediates in transcription initiation by E. Coli RNA polymerase. Biochem Biophys Res Commun. 1990 Apr 30;168(2):756–762. doi: 10.1016/0006-291x(90)92386-e. [DOI] [PubMed] [Google Scholar]
- Tullius T. D. DNA footprinting with hydroxyl radical. Nature. 1988 Apr 14;332(6165):663–664. doi: 10.1038/332663a0. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- UDENFRIEND S., CLARK C. T., AXELROD J., BRODIE B. B. Ascorbic acid in aromatic hydroxylation. I. A model system for aromatic hydroxylation. J Biol Chem. 1954 Jun;208(2):731–739. [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Roeder R. G., Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988 Sep 9;241(4871):1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- Veal J. M., Merchant K., Rill R. L. The influence of reducing agent and 1,10-phenanthroline concentration on DNA cleavage by phenanthroline + copper. Nucleic Acids Res. 1991 Jun 25;19(12):3383–3388. doi: 10.1093/nar/19.12.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal J. M., Rill R. L. Sequence specificity of DNA cleavage by bis(1,10-phenanthroline)copper(I). Biochemistry. 1988 Mar 22;27(6):1822–1827. doi: 10.1021/bi00406a004. [DOI] [PubMed] [Google Scholar]
- Veal J. M., Rill R. L. Sequence specificity of DNA cleavage by bis(1,10-phenanthroline)copper(I): effects of single base pair transitions on the cleavage of preferred pyrimidine-purine-pyrimidine triplets. Biochemistry. 1989 Apr 18;28(8):3243–3250. doi: 10.1021/bi00434a019. [DOI] [PubMed] [Google Scholar]
- Ward B., Skorobogaty A., Dabrowiak J. C. DNA cleavage specificity of a group of cationic metalloporphyrins. Biochemistry. 1986 Nov 4;25(22):6875–6883. doi: 10.1021/bi00370a021. [DOI] [PubMed] [Google Scholar]
- Yoon C., Kuwabara M. D., Law R., Wall R., Sigman D. S. Sequence-dependent variability of DNA structure. Influence of flanking sequences and fragment length on digestion by conformationally sensitive nucleases. J Biol Chem. 1988 Jun 15;263(17):8458–8463. [PubMed] [Google Scholar]
- Yoon C., Kuwabara M. D., Spassky A., Sigman D. S. Sequence specificity of the deoxyribonuclease activity of 1,10-phenanthroline-copper ion. Biochemistry. 1990 Feb 27;29(8):2116–2121. doi: 10.1021/bi00460a022. [DOI] [PubMed] [Google Scholar]
- del Angel R. M., Papavassiliou A. G., Fernández-Tomás C., Silverstein S. J., Racaniello V. R. Cell proteins bind to multiple sites within the 5' untranslated region of poliovirus RNA. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8299–8303. doi: 10.1073/pnas.86.21.8299. [DOI] [PMC free article] [PubMed] [Google Scholar]