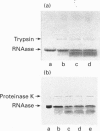
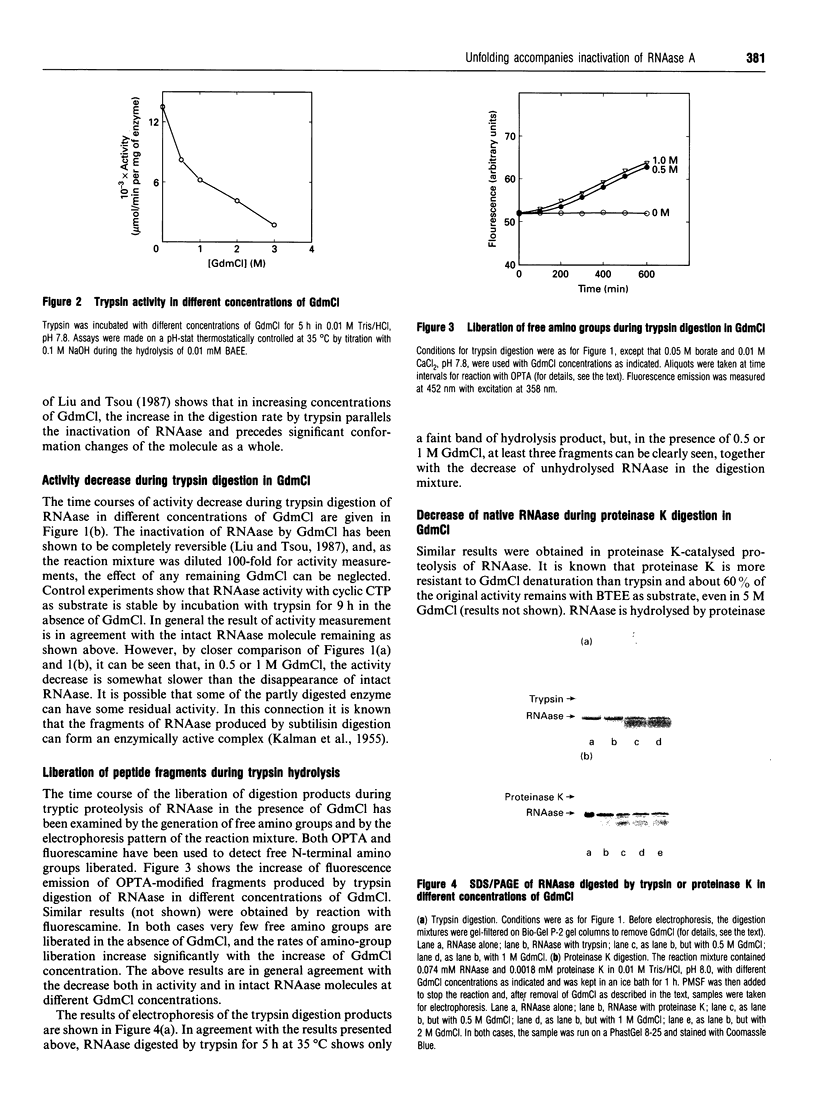
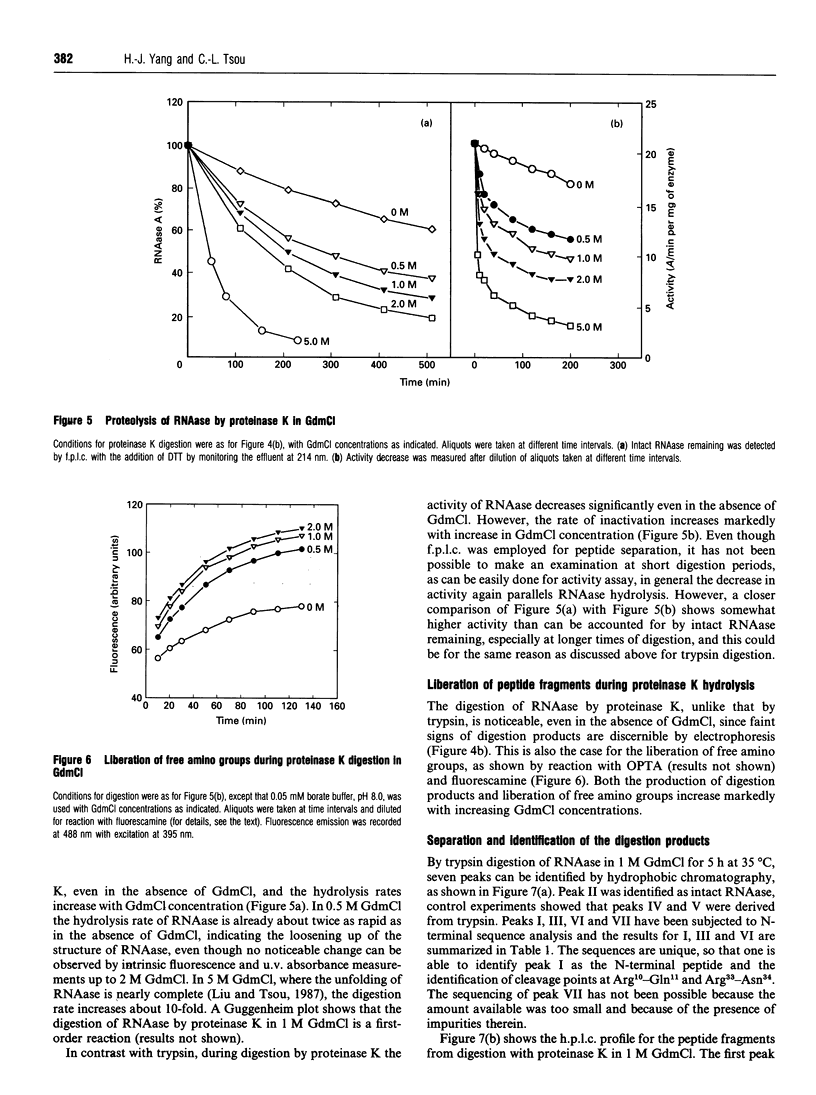
Abstract
Inactivation of pancreatic RNAase A occurs in guanidinium chloride (GdmCl) at low concentrations before the unfolding of the molecule as a whole can be detected [Liu and Tsou (1987) Biochim. Biophys. Acta 916, 455-464]. We have now shown that the rate of digestion of the RNAase molecule by either trypsin or proteinase K increases significantly at low concentrations of GdmCl where the enzyme is largely inactivated, but fluorescence and absorption measurements reveal no conformational changes. N-Terminal sequence analysis of the peptide fragments generated shows that proteolysis occurs primarily at or near the active site. The decrease in activity of RNAase at low concentrations of GdmCl is therefore due to partial unfolding of the molecule, particularly at the active site and not to an inhibition by the denaturant.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANFINSEN C. B., HARRINGTON W. F., HVIDT A., LINDERSTRØM-LANG K., OTTESEN M., SCHELLMAN J. Studies on the structural basis of ribonuclease activity. Biochim Biophys Acta. 1955 May;17(1):141–142. doi: 10.1016/0006-3002(55)90331-0. [DOI] [PubMed] [Google Scholar]
- Arnone M. I., Birolo L., Giamberini M., Cubellis M. V., Nitti G., Sannia G., Marino G. Limited proteolysis as a probe of conformational changes in aspartate aminotransferase from Sulfolobus solfataricus. Eur J Biochem. 1992 Mar 15;204(3):1183–1189. doi: 10.1111/j.1432-1033.1992.tb16745.x. [DOI] [PubMed] [Google Scholar]
- Benz F. W., Roberts G. C. Nuclear magnetic resonance studies of the unfolding of pancreatic ribonuclease. II. Unfolding by urea and guanidine hydrochloride. J Mol Biol. 1975 Jan 25;91(3):367–387. doi: 10.1016/0022-2836(75)90386-1. [DOI] [PubMed] [Google Scholar]
- Betton J. M., Desmadril M., Yon J. M. Detection of intermediates in the unfolding transition of phosphoglycerate kinase using limited proteolysis. Biochemistry. 1989 Jun 27;28(13):5421–5428. doi: 10.1021/bi00439a016. [DOI] [PubMed] [Google Scholar]
- CROOK E. M., MATHIAS A. P., RABIN B. R. Spectrophotometric assay of bovine pancreatic ribonuclease by the use of cytidine 2':3'-phosphate. Biochem J. 1960 Feb;74:234–238. doi: 10.1042/bj0740234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton T. E. Protein folding. Biochem J. 1990 Aug 15;270(1):1–16. doi: 10.1042/bj2700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin J. R., Hare P. E. Chromatographic analysis of amino acids and primary amines with o-phthalaldehyde detection. Anal Biochem. 1977 Jul;81(1):151–156. doi: 10.1016/0003-2697(77)90608-x. [DOI] [PubMed] [Google Scholar]
- Dolgikh D. A., Gilmanshin R. I., Brazhnikov E. V., Bychkova V. E., Semisotnov G. V., Venyaminov SYu, Ptitsyn O. B. Alpha-Lactalbumin: compact state with fluctuating tertiary structure? FEBS Lett. 1981 Dec 28;136(2):311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- Haynie D. T., Freire E. Structural energetics of the molten globule state. Proteins. 1993 Jun;16(2):115–140. doi: 10.1002/prot.340160202. [DOI] [PubMed] [Google Scholar]
- KALMAN S. M., LINDERSTRØM-LANG K., OTTESEN M., RICHARDS F. M. Degradation of ribonuclease by subtilisin. Biochim Biophys Acta. 1955 Feb;16(2):297–299. doi: 10.1016/0006-3002(55)90224-9. [DOI] [PubMed] [Google Scholar]
- Kelly S. M., Duncan D., Price N. C. Unfolding and refolding of the NAD(+)-dependent isocitrate dehydrogenase from yeast. Int J Biol Macromol. 1993 Apr;15(2):75–79. doi: 10.1016/0141-8130(93)90001-3. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Lin Y. Z., Liang S. J., Zhou J. M., Tsou C. L., Wu P. Q., Zhou Z. K. Comparison of inactivation and conformational changes of D-glyceraldehyde-3-phosphate dehydrogenase during thermal denaturation. Biochim Biophys Acta. 1990 Apr 19;1038(2):247–252. doi: 10.1016/0167-4838(90)90212-x. [DOI] [PubMed] [Google Scholar]
- Liu W., Tsou C. L. Activity change during unfolding of bovine pancreatic ribonuclease A in guanidine. Biochim Biophys Acta. 1987 Dec 18;916(3):455–464. doi: 10.1016/0167-4838(87)90192-0. [DOI] [PubMed] [Google Scholar]
- Mast A. E., Enghild J. J., Salvesen G. Conformation of the reactive site loop of alpha 1-proteinase inhibitor probed by limited proteolysis. Biochemistry. 1992 Mar 17;31(10):2720–2728. doi: 10.1021/bi00125a012. [DOI] [PubMed] [Google Scholar]
- Seshadri K., Balaji P. V., Rao V. S., Vishveshwara S. Computer modelling studies of ribonuclease A-pyrimidine nucleotide complexes. J Biomol Struct Dyn. 1993 Oct;11(2):395–415. doi: 10.1080/07391102.1993.10508734. [DOI] [PubMed] [Google Scholar]
- Tsou C. L. Conformational flexibility of enzyme active sites. Science. 1993 Oct 15;262(5132):380–381. doi: 10.1126/science.8211158. [DOI] [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. Early folding intermediate of ribonuclease A. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8197–8201. doi: 10.1073/pnas.87.21.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N., Semisotnov G. V., Pain R. H., Ptitsyn O. B. 'All-or-none' mechanism of the molten globule unfolding. FEBS Lett. 1992 Dec 7;314(1):89–92. doi: 10.1016/0014-5793(92)81468-2. [DOI] [PubMed] [Google Scholar]
- Wilson J. E. The use of monoclonal antibodies and limited proteolysis in elucidation of structure-function relationships in proteins. Methods Biochem Anal. 1991;35:207–250. doi: 10.1002/9780470110560.ch4. [DOI] [PubMed] [Google Scholar]
- Xie G. F., Tsou C. L. Conformational and activity changes during guanidine denaturation of D-glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1987 Jan 5;911(1):19–24. doi: 10.1016/0167-4838(87)90265-2. [DOI] [PubMed] [Google Scholar]
- Yao Q. Z., Tian M., Tsou C. L. Comparison of the rates of inactivation and conformational changes of creatine kinase during urea denaturation. Biochemistry. 1984 Jun 5;23(12):2740–2744. doi: 10.1021/bi00307a032. [DOI] [PubMed] [Google Scholar]
- Zhang Y. L., Zhou J. M., Tsou C. L. Inactivation precedes conformation change during thermal denaturation of adenylate kinase. Biochim Biophys Acta. 1993 Jun 24;1164(1):61–67. doi: 10.1016/0167-4838(93)90112-5. [DOI] [PubMed] [Google Scholar]
- Zhou H. M., Zhang X. H., Yin Y., Tsou C. L. Conformational changes at the active site of creatine kinase at low concentrations of guanidinium chloride. Biochem J. 1993 Apr 1;291(Pt 1):103–107. doi: 10.1042/bj2910103. [DOI] [PMC free article] [PubMed] [Google Scholar]