Abstract
We present an in silico approach for drug discovery, dubbed connectivity enhanced structure activity relationship (ceSAR). Building on the landmark LINCS library of transcriptional signatures of drug-like molecules and gene knockdowns, ceSAR combines cheminformatic techniques with signature concordance analysis to connect small molecules and their targets and further assess their biophysical compatibility using molecular docking. Candidate compounds are first ranked in a target structure–independent manner, using chemical similarity to LINCS analogs that exhibit transcriptomic concordance with a target gene knockdown. Top candidates are subsequently rescored using docking simulations and machine learning–based consensus of the two approaches. Using extensive benchmarking, we show that ceSAR greatly reduces false-positive rates, while cutting run times by multiple orders of magnitude and further democratizing drug discovery pipelines. We further demonstrate the utility of ceSAR by identifying and experimentally validating inhibitors of BCL2A1, an important antiapoptotic target in melanoma and preterm birth–associated inflammation.
Drug discovery can be accelerated by combining transcriptomic signature connectivity analysis with targeted docking simulations.
INTRODUCTION
Accelerating drug discovery and repurposing are paramount for the development of targeted treatment options for precision medicine and the ability to respond to public health crises, such as the COVID-19 pandemic. Systematic efforts for drug discovery rely on high-throughput in vitro or ex vivo screening approaches, often in conjunction with in silico screening of small-molecule libraries resulting in a large number of candidate compounds targeting the druggable part of the genome (1–3). Parallel advances in pharmacogenomics and large-scale candidate drug profiling in cell lines and other model systems, such as Connectivity Map (4), NCI60 (5) and Cancer Cell Line Encyclopedia (6), or Genomics of Drug Sensitivity in Cancer (7), have further revolutionized drug discovery, target and mode of action prediction, and repurposing (8). Transcriptional signature connectivity analysis has been used for drug target discovery and to identify drugs that may reverse a signature of a disease state or have the same mode of action because of the similarity of their signatures (4, 9–13).
With the goal of connecting drugs and their targets, the Library of Integrated Network-based Cellular Signatures (LINCS) consortium has compiled a library of transcriptional signatures for over 15,000 drug-like molecules and ~4400 gene knockdowns (KDs), as well as over 2000 overexpression constructs in multiple cell lines (12, 13). As a result, LINCS transcriptional signatures can be used to directly correlate downstream transcriptional responses induced by chemical perturbations with those induced by loss or gain of function of the target protein. Thus, LINCS enables direct exploration of drug-gene relationships on previously unattainable scales by connecting substantial subsets of both the drug-like universe of small molecules and druggable genome (13–15). While showing great promise as a unique big data resource for pharmacogenomics, we posit that systematic benchmarking and integrative methods are required to establish the extent to which LINCS can be used to identify compounds that directly target specific proteins.
Signature concordance–based identification of putative inhibitors has the advantage of not requiring the target protein structure. On the other hand, because of the nature of signal transduction pathways, similar downstream transcriptional signatures may result from the loss of function of multiple upstream proteins in signaling cascades or pathways converging on the same transcriptional targets, such as signaling cascades involving multiple kinases and phosphorylation events between a growth receptor and a transcription factor in many types of cancer (16). Thus, the analysis of concordance between signatures of small molecules and the target gene KD is likely to identify candidate molecules that serve as pathway inhibitors and not necessarily direct inhibitors of the target protein.
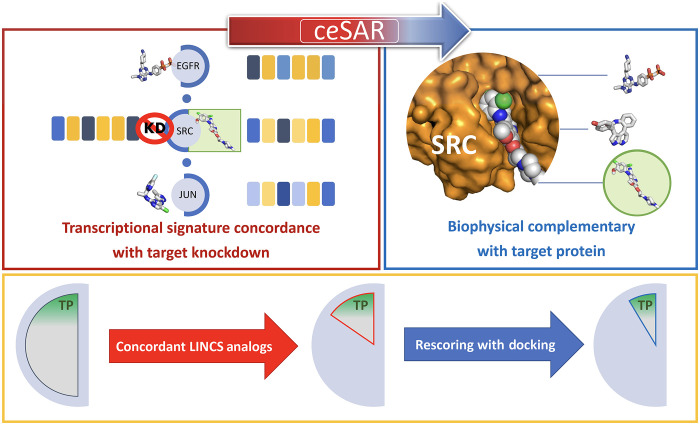
For example, a signature connectivity analysis to nominate putative inhibitors of SRC may identify compounds targeting the epidermal growth factor receptor (EGFR)–SRC–JUN signaling cascade as “concordant,” i.e., positively correlated with the SRC KD signature, although only one of them targets SRC directly (Fig. 1). To achieve a higher specificity to a given target in a pathway, the predicted binding affinity to the target protein may be used to complement the signature connectivity–based approach and identify consensus candidates. In this context, in silico docking techniques have been widely used to computationally predict binding affinities between small molecules and their structurally resolved targets, often coupled with structure activity relationship (SAR) analysis to reduce high positive rates in drug screening by considering a family of moieties sharing chemical similarity and to further increase specificity for validated hits (17).
Fig. 1. The overall principle of the ceSAR approach.
Candidate molecules are first ranked by their chemical similarity to concordant LINCS analogs, i.e., drug-like molecules with transcriptional signatures concordant to a signature of the target gene KD (red box), and subsequently reranked by docking simulations to assess their biophysical complementarity with the target protein (blue box). By combining signature concordance and biophysical complementarity, the library of candidate compounds is reduced to a small subset enriched for true positives (TP) for further validation (yellow box). Here, a fictitious SRC KD signature consists of six genes, with blue indicating down-regulated and yellow indicating up-regulated genes. Signatures of three compounds targeting the EGFR-SRC-JUN cascade are concordant with that of SRC KD, but only the actual SRC inhibitor (green circle) is found to fit the binding pocket by docking.
Here, we introduce an integrative approach for drug discovery, dubbed connectivity enhanced SAR (ceSAR), that combines the principles of transcriptional signature concordance and biophysical complementarity. Capitalizing on the LINCS library of transcriptional signatures, ceSAR integrates drug and target transcriptional signature connectivity analysis with cheminformatic and virtual screening approaches. A library of candidate compounds is reduced via fast chemical similarity search to identify those candidates that are structural analogs to a concordant LINCS small molecule. For a target gene, concordant LINCS small molecules are identified as those compounds whose signatures are concordant with a gene KD signature. The resulting small subset of candidate compounds can be subsequently rescored in conjunction with docking, using consensus ranking and machine learning (ML)–based models to combine these two complementary approaches.
Building on this overall principle, we aim to accelerate drug discovery by (i) demonstrating that the success rate in identifying candidate inhibitors of specific protein targets can be improved through the integration of transcriptional signature connectivity analysis with a targeted molecular docking; (ii) extending the signature connectivity analysis to an arbitrary set of user-provided candidate molecules that were not profiled as part of LINCS; and (iii) introducing an ultrafast method to compute chemical similarity and identify concordant LINCS analogs as a basis for efficient ranking of candidate molecules. By integrating these advances, ceSAR is shown to substantially reduce false-positive rates while greatly lowering the overall computational cost of virtual screening.
RESULTS
Benchmarking ceSAR
We systematically evaluate the performance of ceSAR and compare it with the results of AutoDock (18) and MTiOpenScreen (19) docking methods, using a diverse subset of targets from the DUD-E benchmark, which is widely used in the field to assess the performance of docking and virtual screening methods (20). For each target, DUD-E contains a library of compounds to be scored, on average (see table S2) comprising about 550 known binders, i.e., true positives, and 20,000 carefully selected drug-like decoy molecules classified as true negatives. Thus, DUD-E benchmark provides the ground truth to determine the success rate of virtual screening methods and assess their ability to discriminate between the known binders and decoys (20). Here, we used a subset of 20 targets from the DUD-E benchmark that had gene KDs available within LINCS. For direct comparison and evaluation of consensus methods, we performed docking simulations for DUD-E libraries using AutoDock v. 4.2 and the original target conformations and binding sites from the DUD-E benchmark. For a subset of DUD-E targets and for the original DUD compound libraries, we also performed docking simulations using the MTiOpenScreen web server. The results of the latter are included in the Supplementary Materials.
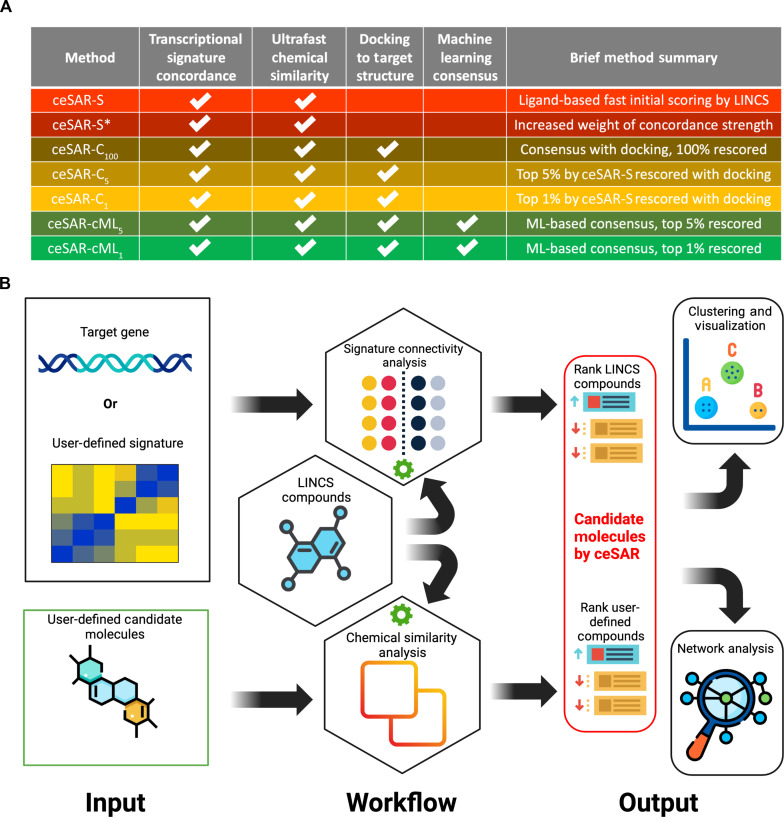
Signature concordance, chemical similarity, and docking derived features can be combined in multiple different ways. Therefore, we systematically evaluated several forms of ceSAR that are defined in Fig. 2, including the ultrafast ligand-based and consensus-based approaches. In the simplest and structure-independent form of the method, which is referred to as ceSAR-S (for signature-based), candidate molecules are ranked using a chemical similarity score, which is the Tanimoto coefficient (21) for the closest concordant LINCS analog. Here, concordant is defined as having a significantly positively correlated signature with a target gene KD signature (22). We also consider an alternative form of the method, referred to as ceSAR-S*, that combines signature concordance and chemical similarity to the analogs using the Fisher consensus.
Fig. 2. The hierarchy of ceSAR methods.
Dependencies and notation for the hierarchy of ceSAR methods introduced and benchmarked in this work (A) and the overall ceSAR-S workflow (B). Note that the methods highlighted in red in (A) do not depend on protein structure. ceSAR-S (and ceSAR-S*) workflow shown in (B) is implemented in sig2lead.net and stand-alone sig2lead application that combine signature connectivity analysis for LINCS compounds with chemical similarity analysis for user defined compounds. The latter is scored on the basis of their chemical similarity to concordant LINCS analogs. Note that sig2lead allows for user defined loss-of-function transcriptional signatures when a LINCS KD signature is not available.
To combine signature connectivity with docking, we consider a simple form of consensus, referred to as ceSAR-C, which defines the combined rank of candidate molecules as the geometric average of signature connectivity and docking-based ranks. If docking is performed for all compounds in the library, then the method is referred to as ceSAR-C100. When the library is first reduced using ceSAR-S to the top 5 or 1% of the library, consensus forms of the method are referred to as ceSAR-C5 and ceSAR-C1, respectively. The corresponding ML consensus approaches are referred to as ceSAR-cML1 and ceSAR-cML5.
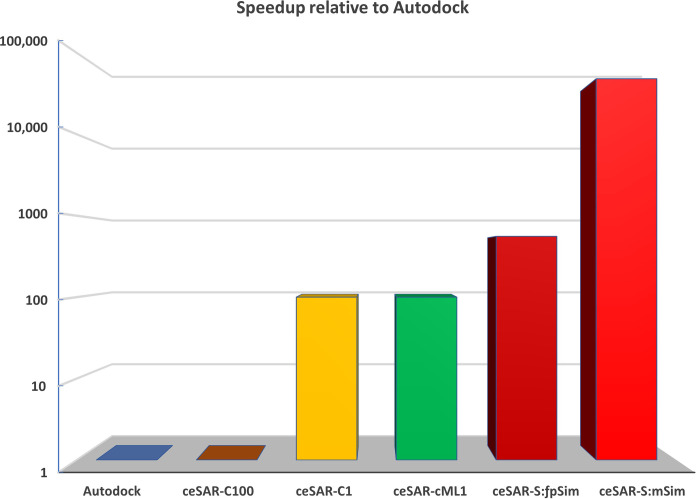
We first compare ceSAR and docking methods in terms of computing time. Figure 3 shows that ceSAR-S with the ultrafast minSim (minority Sim) algorithm to compute Jaccard similarity (here Tanimoto coefficient) is about 50,000 times faster than docking. Thus, ligand-based and structure-independent ceSAR-S has a negligible computational cost relative to docking simulations and can be executed within minutes on a personal laptop for a typical DUD-E library of about 20,000 candidate molecules. On the other hand, ceSAR-C1 and ceSAR-cML1 reduce the computational time 100-fold compared to docking, as only the top 1% of the library, reduced by using ceSAR-S, needs to be screened by docking. Thus, performing targeted docking for a small subset of the entire library selected by the structure-independent and fast ceSAR-S still greatly reduces the computational cost. We next evaluate whether these gains in speed come at a cost in terms of accuracy.
Fig. 3. Average speedup (in logarithmic scale) on 20 DUD-E targets for methods relative to AutoDock.
ceSAR The consensus approaches ceSAR-C1 (yellow) and ceSAR-cML1 (green) reduce the run time by 100× compared with docking. Structure-independent ceSAR-S reduces the run time by ~560× when using the fpSim function that represents current methods (see Materials and Methods) to compute the chemical similarity (ceSAR-S:fpSim, dark red) and by ~48,000× when using the ultrafast minSim (ceSAR-S:mSim, red) algorithm introduced in this work.
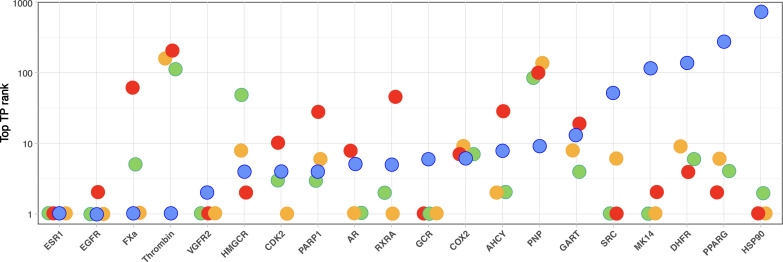
The success of virtual drug screening is critically dependent on the ability to retain at least some true binders as the library is reduced to a small subset amenable to experimental validation. Therefore, we start the evaluation of ceSAR methods using the top true-positive rank as an important and easy to interpret metric of success (Fig. 4). While AutoDock performs well on four targets [ESR1, EGFR, Factor Xa (FXa), and Thrombin], nominating a true binder as the top-ranking candidate, it also fails to identify at least one true positive in the top 100 compounds for four targets [mitogen-activated protein kinase 14 (MK14), dihydrofolate reductase (DHFR), peroxisome proliferator–activated receptor (PPARG), and heat shock protein 90 (HSP90)]. Signature concordance–based ceSAR-S nominates a true positive as the top-ranking candidate for five targets [ESR1, vascular endothelial growth factor receptor 2 (VGFR2), glucocorticoid receptor (GCR), SRC, and HSP90], while failing to identify at least one true positive in the top 100 compounds for only two targets [Thrombin and purine nucleoside phosphorylase (PNP)].
Fig. 4. Top true-positive ranks for 20 DUD-E targets.
Results for AutoDock (blue), ceSAR-S (red), ceSAR-C1 (yellow), and ceSAR-cML1 (green) consensus approaches. Note complementarity of signature connectivity and docking approaches, with ceSAR working well when docking fails for the last five targets (SRC, MK14, DHFR, PPARG, and HSP90), while docking working well when ceSAR fails (Thrombin and PNP). Note also that the consensus methods are more robust and outperform both AutoDock and ceSAR-S in terms of the number of targets with a true positive as the top-ranking candidate or within the top 10 candidates. CDK2, cyclin-dependent kinase 2; COX2, cyclooxygenase 2.
ceSAR-S performs well on targets on which AutoDock fails, whereas AutoDock performs well when ceSAR-S fails. Thus, the two approaches are complementary, enabling the ceSAR consensus methods to perform better relative to individual methods. Both, ceSAR-C1 and ceSAR-cML1 are very robust, failing to identify at least one true positive in the top 10 compounds for only two and three targets, respectively. The number of “catastrophic failures” without a single true positive in the top 100 compounds is further reduced to only two targets (Thrombin and PNP) for ceSAR-C1 and one target for ceSAR-cML1 (Thrombin).
Even the simple ceSAR-C1 consensus method selects a true positive as the top-ranking candidate for 10 targets. While real-life applications typically require multiple compounds to be tested, the DUD-E benchmark shows that combining signature connectivity and docking yields a 50% success rate with just one compound to be tested experimentally, as opposed to a 25% success rate for ceSAR-S and 20% for AutoDock. When combining all methods evaluated here, one can further increase the sensitivity and achieve a 100% success rate on the DUD-E benchmark by selecting for further experimental validation the union of top 10 candidates nominated by each of the four methods.
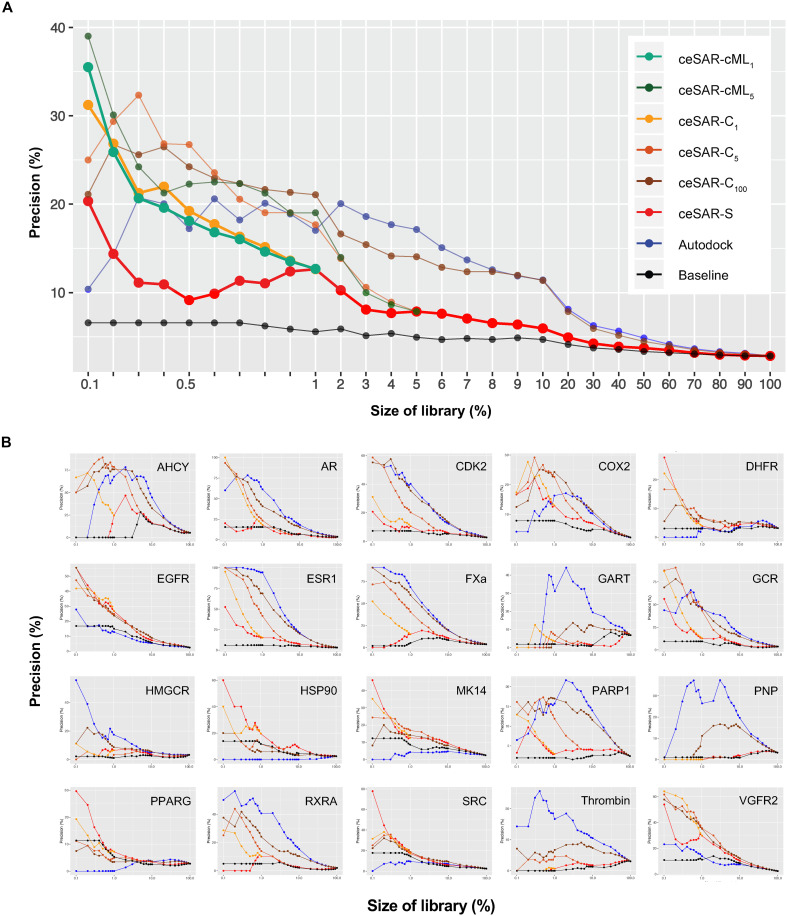
The performance of ceSAR methods is further summarized in Fig. 5, using the median and individual precision curves for 20 targets. Precision curves show the percentage of true binders upon reducing the DUD-E compound libraries to small subsets amenable to further validation. Note that the precision, or positive predictive value, defined as PPV = TP/(TP + FP), where TP denotes the number of true-positive predictions and FP denotes the number of false-positive predictions, captures the fraction of true binders as the library is reduced. Thus, PPV measures the likelihood of successfully identifying an inhibitor through experimental validation of a library subset.
Fig. 5. Precision curves for 20 DUD-E targets.
The median (A) and individual (B) precision curves for 20 DUD-E targets as a function of the library size. AutoDock is compared with ceSAR-S and consensus approaches, ceSAR-C1, ceSAR-C5,and ceSAR-C100, and ML-based ceSAR-cML1 and ceSAR-cML5, and with a simple baseline method (Baseline) that ignores signature connectivity and accounts for compositional biases in LINCS compound library.
AutoDock is successful in eliminating the most unlikely binders using biophysical small molecule–target protein complementarity and the predicted binding energies, leading to initial success and higher precision at the level of 5 to 10% of the original library size. However, docking struggles to correctly rank true positives and the remaining more challenging true negatives, resulting in a drop of accuracy as the size of the library is reduced further.
On the other hand, despite its negligible computational cost, ceSAR-S yields precision of 10% or more as the library is reduced to the top 2% or less, while outperforming AutoDock at the furthest library reduction, for which it yields a median precision of about 20% compared to 10% for docking. Note that DUD-E target libraries comprise, on average, ~20,000 candidate compounds, so reducing the library size to less than 1% is desirable to reduce the number of compounds for testing, reflecting real-life challenges. In addition, note that ceSAR-S improves consistently upon baseline, which ignores signature concordance while identifying the closest LINCS analogs of DUD-E compounds. Comparison with this defined baseline allows one to account for biases in LINCS coverage of different compound classes and to assess the signal due to signature concordance, which is represented by the surface area between the red and black curves.
The consensus forms of ceSAR lead to further improvements, outperforming AutoDock for library sizes below 0.5%. At the most reduced library size, the median precision reaches about 30, 35, and 40% for ceSAR-C1, ceSAR-cML1, and ceSAR-cML5, respectively. Thus, consensus ceSAR methods yield between three- and fourfold improvement over docking. ceSAR-C1 and ceSAR-cML1 consensus methods, which use docking for the top 1% of the library ranked by ceSAR-S, have the most significantly different distributions over 20 targets with respect to the baseline, as well as to individual AutoDock and ceSAR-S methods, as measured by the Kullback-Leibler divergence measure (fig. S14), while offering a substantial speedup since only a small fraction (1%) of the library needs to be rescored using consensus with docking. We conclude that ceSAR-C1 and ceSAR-cML1 consensus methods provide the best trade-off between speed and accuracy on the DUD-E benchmark, while the performance of consensus-based methods is robust with respect to the choice of the top library subset for integration with docking.
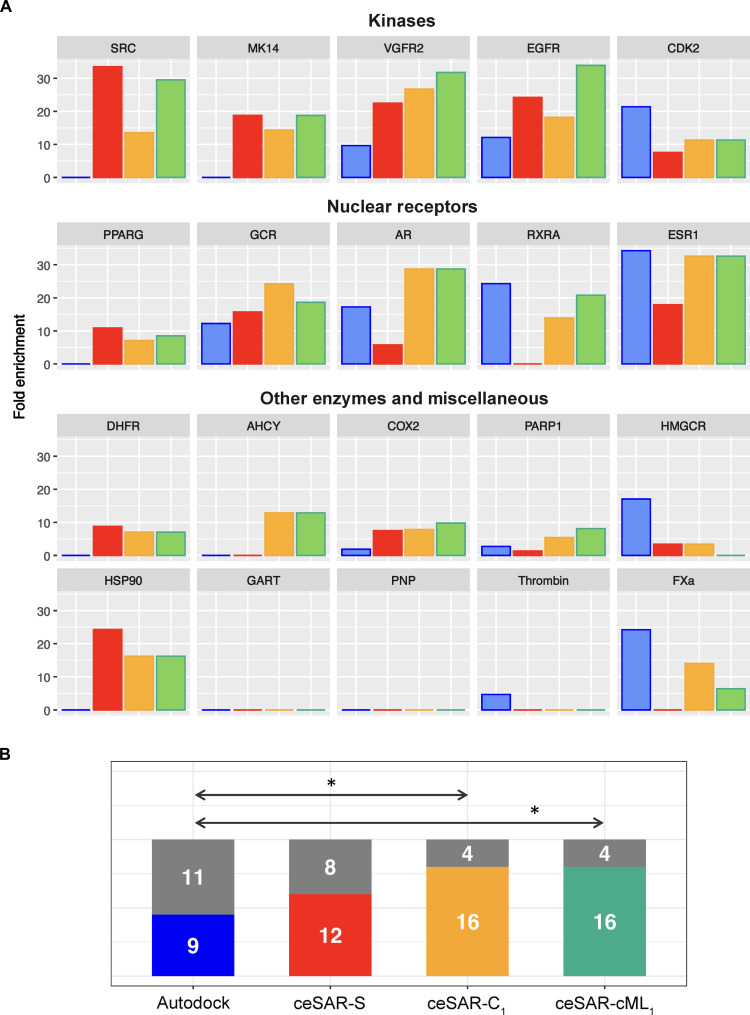
Another metric of success in virtual screening is the enrichment into true positives as the library is reduced. Enrichment factor is defined as the ratio of true-positive fraction in a subset of the library versus the initial true-positive fraction in the entire library. Since for DUD-E compound libraries, the initial true-positive fraction is about 2.5% on average, at the furthest library reduction, AutoDock, ceSAR-S, ceSAR-C1, and ceSAR-cML1 yield about 4-, 8-, 12-, and 14-fold enrichment on average, respectively (Fig. 6). However, there are considerable differences in the level of success for different targets, as also illustrated by individual precision curves in Fig. 5B, as well as receiver operating characteristic and precision-recall curves in fig. S18.
Fig. 6. Enrichment factors for DUD-E targets at 0.1% library.
(A) Fold enrichment into true positives, defined as the ratio of true-positive fraction for full versus reduced library for AutoDock (blue) versus ceSAR-S (red) and consensus methods ceSAR-C1 (yellow) and ceSAR-cML1 (green). (B) The number of targets with ≥5-fold enrichment versus limited or no enrichment (gray) at 0.1% library reduction. Note that ceSAR approaches are more robust and consensus methods ceSAR-C1 and ceSAR-cML1 significantly outperform docking (P = 0.02), reducing the number of targets with limited or no enrichment to four while greatly reducing the computational cost. Statistically significant differences are indicated by arrows with asterisks (*) in (B).
ceSAR performs very well on kinases and moderately well on nuclear receptors, while docking alone is less robust and fails for some targets in those classes. While the degree of success varies considerably for other enzymes and miscellaneous targets (DUD-E definitions), the results are more robust for the two consensus methods, underscoring the complementarity of signature connectivity and docking-based scores. This is further highlighted in Fig. 6B by comparing the number of targets with at least fivefold enrichment as opposed to the number of targets with limited or no enrichment. The consensus ceSAR methods fail to enrich substantially for only four targets and ceSAR-S for eight targets, as compared with 11 such failures for AutoDock. A more detailed analysis of enrichment factors is included in table S7.
Overall, multiple metrics of success show that ceSAR is more robust in comparison with docking, which performs well on some targets while also failing completely for multiple targets in terms of enrichment at the extreme library reductions or top true-positive rank. Meanwhile, when combined with the initial reduction of compound libraries by ceSAR-S, targeted docking provides a complementary principle to rescore the candidate compounds identified through signature connectivity, resulting in increased performance of ceSAR consensus approaches.
To further investigate the sources of success and limitations of the method, we tested AutoDock Vina as an alternative docking method for rescoring a small subset of the library identified by ceSAR-S. In a direct comparison of AutoDock and AutoDock-based ceSAR results from this work with those reported in a recent evaluation of DOCK 3.7 and AutoDock Vina on DUD-E benchmark (23) included in table S7, AutoDock Vina slightly outperforms AutoDock in terms of the adjusted log area under the curve while achieving substantially higher median enrichment at 1% library size, although further enrichment is strongly target dependent, similar to AutoDock.
We used the MTiOpenScreen docking server that uses AutoDock Vina to compare the results with AutoDock. On a subset of eight DUD-E targets included in fig. S23, the consensus ceSAR-C1 with AutoDock Vina yields median precision of about 41 at 0.1% library reduction, compared with about 39% for ceSAR-C1 with AutoDock on the same subset of targets, indicating that both docking methods can discriminate false positives that remain after the initial, signature concordance–based library reduction. Similar trends are observed on the original DUD benchmark, using the MTiOpenScreen docking server. Together, these results demonstrate robustness across docking programs and different candidate compound libraries.
Identification of BCL2A1 inhibitors using ceSAR
Although such an approach is less efficient, ceSAR can be extended to incorporate signature connectivity–based rescoring after using docking to reduce the library size. Note that this is different from the consensus approaches considered above in that the order of library reduction is reversed. On the DUD-E benchmark, first, reducing the library to the top 1% using AutoDock and then rescoring using a simple consensus of AutoDock and ceSAR-S yielded median precision of about 27% at the furthest library reduction, i.e., better than individual methods but slightly worse than ceSAR-C1. The AutoDock first form of the combined approach is tested here to identify specific inhibitors of an important antiapoptotic target, namely, BCL2A1 (A1). A1 has been implicated in a number of diseases, ranging from inflammation in preterm birth (24) to chemotherapeutic resistance in melanoma (25). To date, very few small-molecule inhibitors specific to A1 have been identified.
Most antiapoptotic proteins prevent apoptosis by binding and sequestration of proapoptotic proteins, achieved via binding to their “BH3” domain (26). A major success in targeting this family was the development of a Bcl-2 inhibitor ABT-737 (27), which was modified to a bioavailable version ABT-263 or navitoclax. Unfortunately, ABT-263 also bound Bcl-xL, whose role in promoting platelet survival leads to thrombocytopenia in humans (28). This observation spurned the development of ABT-199, which showed specificity for Bcl-2 without inhibiting Bcl-xL (29, 30). Thus, despite their structural similarity, it is possible to selectively target individual Bcl-2 family members.
To address this, we screened a compound library of 90,087 drug-like small molecules using AutoDock v. 4.2. The top 300 compounds found by docking were clustered, and representatives of each cluster were tested in vitro using a differential scanning fluorimetry (DSF) thermal shift assay to detect compound binding to BCL2A1, and a fluorescence polarization (FP) competition assay to test for inhibition of Noxa BH3 domain binding to BCL2A1 (Fig. 7A). The top 20 compounds identified by the combination of DSF and FP assays are listed in Table 1; 15 of the top 20 hits showed Ki values in the low micromolar range.
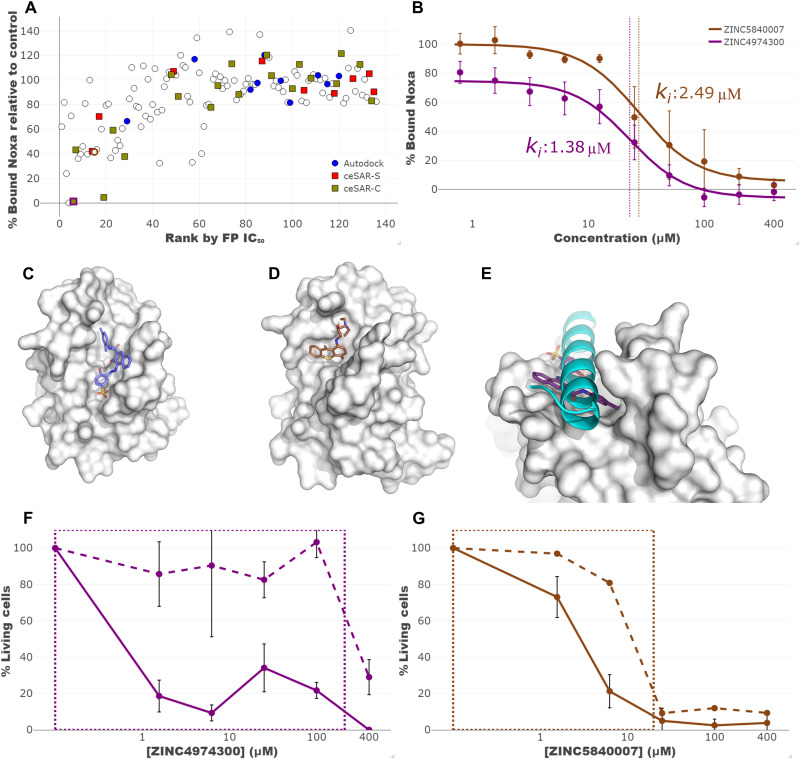
Fig. 7. Experimental validation of in silico candidate compounds targeting BCL2A1.
A total of 116 compounds, identified initially by AutoDock and rescored using ceSAR, along with 23 structural analogs were tested experimentally by FP and DSF. These 139 compounds are shown in (A), with top 20 candidates ranked by AutoDock, ceSAR-S, and ceSAR-C highlighted in blue, red, and yellow, respectively. At a single high dose, several compounds showed inhibition of the BCL2A1-Noxa interaction, including some of the most promising candidates nominated by the consensus ceSAR-C approach. Dose-response FP curves for two candidate compounds, with IC50 values indicated by vertical lines, are shown in (B). These compounds were predicted to bind within the BH3 peptide binding pocket (C and D) in a manner that would drive competitive inhibition of BH3 binding (E). These compounds were demonstrated to induce the death of wild-type, but not bax−/−bak−/− activated T cells, with a box displaying the potential therapeutic window in (F and G).
Table 1. Top 20 compounds based on experimental validation using consensus of DSF and FP.
Compounds are listed in rank order based on the geometric mean of ranks obtained using two complementary experimental assays: the absolute value of Tm change determined by DSF and high-dose degree of inhibition determined by FP. Note that the top three compounds by experimental consensus were identified within the top 10 ceSAR-C consensus ranks. Highlighted in bold are two compounds that induced cell death in an activated T cell death assay, consistent with inhibition of the prosurvival BCL2A1 protein. NA, not applicable.
| ZINC ID | Experimental rank | AutoDock rank | ceSAR-S rank | ceSAR-C rank | FP IC50 (μM) | Ki (μM) |
|---|---|---|---|---|---|---|
| ZINC01717014 | 1 | 67 | 2 | 5 | 146 | 7.39 |
| ZINC05930871 | 2 | 3 | 80 | 10 | 70.1 | 3.55 |
| ZINC04974300 | 3 | 6 | 23 | 6 | 27.3 | 1.38 |
| ZINC04934733 | 4 | 64 | 51 | 75 | >400 | >20.3 |
| ZINC04804154 | 5 | 91 | 57 | 90 | 254 | 12.9 |
| ZINC04803984 | 6 | 102 | 77 | 104 | 31.8 | 1.61 |
| ZINC01679491 | 7 | 44 | 46 | 52 | >400 | >20.3 |
| ZINC01673413 | 8 | 37 | 40 | 41 | 4.5 | 0.23 |
| ZINC04758328 | 9 | 72 | 61 | 82 | 68.9 | 3.49 |
| ZINC01592019 | 10 | 103 | 72 | 100 | 299.4 | 15.2 |
| ZINC01593456 | 11 | 89 | 19 | 48 | 48.5 | 2.46 |
| ZINC05839997 | 12 | 43 | 70 | 70 | >400 | >20.3 |
| ZINC01650974 | 13 | 100 | 25 | 62 | 28.3 | 1.43 |
| ZINC01600320 | 14 | 22 | 14 | 12 | 21.3 | 1.08 |
| ZINC03194765 | 15 | 79 | 10 | 25 | 55.4 | 2.81 |
| ZINC04366919 | 16 | 29 | 78 | 56 | 103 | 5.22 |
| ZINC04409853 | 17 | 63 | 49 | 71 | 387 | 19.6 |
| ZINC05840007 | 18 | 53 | 76 | 79 | 49.1 | 2.49 |
| ZINC04367231 | 19 | 87 | 63 | 91 | NA | NA |
| ZINC00344612 | 20 | 31 | 45 | 39 | >400 | >20.3 |
ceSAR-S was subsequently applied to rescore the subset of compounds selected for experimental validation. As before, ceSAR-S and docking results are combined into consensus ceSAR-C ranking using the geometric mean of AutoDock and ceSAR-S ranks. Compounds were classified as inhibitors for the sake of benchmarking the AutoDock first ceSAR method if they caused a thermal shift upon addition to the BCL2A1-Noxa reaction and had a median inhibitory concentration (IC50), as defined by dose-response FP, of 400 μM or less, which corresponded to a Ki of 20 μm or less. As shown in fig. S24, as the set of compounds is reduced to a subset, both ceSAR-S and ceSAR-C yield a higher precision and enrichment into experimentally validated inhibitors relative to AutoDock alone. These results provide support for the hypothesis that a set of putative weak binders identified experimentally (and guided by the initial virtual screening) can be successfully reduced to an enriched subset using ceSAR-S rescoring in conjunction with docking and thus reduce the number of compounds for more stringent validation.
Given the role of Bcl2A1 in the survival of T lymphocytes (31), we used an in vitro T cell survival assay to determine specificity of Bcl2A1 inhibition in a biologically relevant system. In addition, we took advantage of Bax/Bak-deficient T cells as these cells lack the ability to undergo apoptosis via the Bcl2-regulated pathway (32) and serve to control for off-target toxicity. Two compounds demonstrated a therapeutic window in the low micromolar range in which they induced the death of activated T cells while failing to kill T cells derived from bax−/−bak−/− mice, indicating specific inhibition without off-target effects in this concentration range (Fig. 7, F and G). More results of experimental validation, including detailed results of screening by FP and DSF, are included in figs. S25 to S27 and tables S8 and S9.
Thus, rescoring candidate compounds obtained using docking simulations can yield further enrichment into true positives and limit the compounds that need to be tested experimentally. Conversely, observed enrichment into true positives for an important and challenging target illustrates how a set of experimentally identified weak binders can be used to seed signature connectivity–based ceSAR search with the goal of identifying additional candidate compounds.
DISCUSSION
Accelerating drug discovery and repurposing are paramount for advancing personalized precision medicine. In this contribution, we introduce ceSAR, an efficient in silico method to accelerate drug discovery and repurposing. ceSAR improves upon existing approaches by (i) extending the application of transcriptional signature connectivity analysis to an arbitrary set of candidate compounds that are not included in LINCS; (ii) enabling ultrafast chemical similarity search for concordant LINCS analogs using the minSim algorithm introduced here; and (iii) combining small-molecule docking simulations with signature connectivity analysis to increase the specificity for a target protein. By reducing false-positive rates while also greatly reducing the computational cost of virtual screening, ceSAR addresses two major limitations of current virtual screening approaches.
Over the past two decades, transcriptional and other drug activity profiles have been increasingly used in drug design, mode of action identification, and SAR analyses (4, 12, 33, 34). For example, identifying targets for small molecules, thus identifying these molecules as potential inhibitors, can be facilitated by comparing bioactivity profiles or transcriptional signatures of a compound to known inhibitors (33). Another example is the use of the connectivity map approach to connecting gene expression profiles of disease states, such as drug-resistant forms of cancer, with discordant drug signatures, allowing one to identify drugs that can potentially be used to reverse the disease signature (4, 12, 15).
In contrast to these previous efforts, ceSAR directly connects the transcriptional signatures of small molecules with the signature of a gene KD to identify antagonists of a specific target rather than a potential pathway inhibitor. To that end, ceSAR combines signature connectivity analysis with atomistic docking simulations and predicted binding energies to improve specificity. We would like to note that overexpression signatures could be used to identify agonists of a specific target, which is the subject of a future study.
ceSAR capitalizes on the LINCS transcriptional signature database that made available a large library of both small-molecule perturbation signatures and genetic perturbation signatures, thus covering a substantial subspace of the drug-like chemical universe and druggable subset of the genome in one or multiple biological contexts (12, 13). While building on LINCS, the signature connectivity–based ceSAR-S method integrates chemical similarity and signature connectivity analyses to increase overall success rates and expand virtual screening and SAR analysis to other suitable libraries of compounds, including those identified in high-throughput experimental screening.
Docking-based in silico screening, on the other hand, relies on shape, electrostatics and other interaction-based complementarity between putative inhibitors and target proteins, requiring structural information about the target and its relevant conformational states that may be unavailable (35). The results of docking simulations may also be sensitive to the choice of the target’s conformation, empirical force fields, docking programs, and sampling depth (4, 12, 15). In addition, traditional virtual screening approaches require substantial computing resources, so reducing the scope of docking simulations to a more targeted set of candidates, by applying ceSAR-S as the initial filter, is highly desirable.
ceSAR-S ranks candidate molecules based on their chemical similarity to concordant LINCS analogs, which involves computing pairwise similarities between user-provided candidate compounds and LINCS compounds. To address this computational bottleneck, we developed an efficient solution for computing the Tanimoto coefficient and retrieving concordant LINCS analogs with sparse binary fingerprints used here for fast chemical similarity search. The algorithm, dubbed minSim, reduces the computation to minority states and optimally exploits the sparse nature of binary fingerprints without using approximate techniques, such as those based on hashing (36, 37). As shown in table S1, for the retrieval from the LINCS library for different DUD-E datasets, minSim provides between 60- and 150-fold speedup compared to fpSim function, which represents current approaches for exact chemical similarity search (38). Note that these speedups are consistent with the observed levels of sparsity in the LINCS dataset, while reflecting the varying degree of sparsity in DUD-E datasets of query molecules.
Overall, the average central processing unit (CPU) time per DUD-E target required to perform ceSAR-S search was about 3.7 CPU minutes on a laptop computer with two Intel i5-4200U @ 1.6-GHz cores. For comparison, the average CPU time per target required to perform AutoDock simulations, using the search depth and grid sizes defined in the Supplementary Materials, was on the order of 3000 CPU hours on a computational cluster consisting mostly of 16 Intel (R) Xeon (R) CPU E5-2667 v3 @ 3.20-GHz core nodes. Thus, ceSAR-S reduced run times by roughly 50,000-fold compared to AutoDock. This marked speedup makes it possible to perform in silico enrichment on large chemical libraries on a personal computer, without the need to use a high-end computing platform.
Despite its negligible computational cost, the connectivity-based and target structure–independent ceSAR-S method achieves twofold improvement over AutoDock in terms of the median precision at the furthest library reductions. The initial ceSAR-S search can be subsequently combined with docking simulations to achieve higher specificity for a target by filtering out different subsets of likely false positives: those that fail to induce signatures concordant with a KD of the target gene versus those that are deemed as incongruent with the target protein binding site. The complementarity of these two principles is demonstrated in figs. S1 to S4.
ceSAR-C1 and ceSAR-cML1 consensus methods yield three- to fourfold increases in median precision and enrichment of true binders compared to docking alone, while still greatly reducing the overall computational cost compared to docking alone. In terms of the top true-positive rank, ceSAR-C1 yields a 50% success rate with just one (top ranking) compound to be tested experimentally, as opposed to a 20% success rate for ceSAR-S or AutoDock. The consensus approach is also shown to be more robust, as docking and ceSAR-S fail on different targets. As a result, the fraction of targets with limited or no enrichment at 0.1% library size is 55% for AutoDock, 40% for ceSAR-S, and only 20% for consensus ceSAR-C1 and ceSAR-cML1 methods. Likewise, the number of targets for which none of the true positives is ranked among the top 100 candidates is reduced from four for AutoDock to two for ceSAR-S and ceSAR-C1 and one for ceSAR-cML1.
Note that the success of ceSAR is not due to overrepresentation of known binders from DUD-E datasets among the LINCS compounds. As can be seen from table S2, both true binders and decoys from DUD-E have a similar overlap with the LINCS library, as indicated by similar distributions of Tanimoto coefficients for the closest LINCS analogs for both subsets. Furthermore, the baseline approach that ignores signature connectivity and simply uses the Tanimoto coefficient to the closest LINCS analog, irrespective of its “concordance” with the target KDs, performs poorly as demonstrated in figs. S12 to S14 and S17.
AutoDock outperforms signature connectivity enhanced ceSAR methods in the case of three targets: HMGCR, Thrombin, and PNP. None of these three targets have robust coverage of close LINCS analogs of true binders in their respective DUD-E datasets (Fig. 8A and table S2); furthermore, they are characterized by relatively weak concordance between LINCS small-molecule and KD signatures (fig. S9). These failures underscore current limitations of ceSAR, which are expected to be gradually alleviated as LINCS-like resources grow, while suggesting criteria that can be used to predict the likelihood of success.
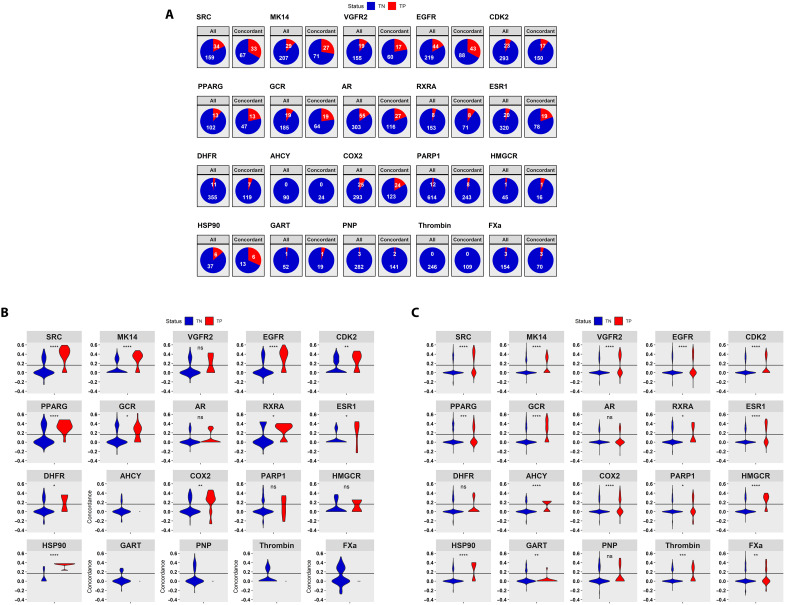
Fig. 8. Performance of ceSAR-S on compounds directly represented in LINCS versus those with LINCS analogs.
Enrichment into true positives by signature concordance for compounds directly included in LINCS is shown in (A); distributions of concordance scores for true positives (TP) (red) versus true negatives (TN) (blue) included directly in LINCS are shown in (B); and distributions of concordance scores for those with LINCS analogs at Tanimoto coefficient of 0.8 or more are included in (C). ns, not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
In this context, it is instructive to highlight again the importance of expanding the applicability of signature connectivity analysis beyond compounds directly included in LINCS. Namely, although LINCS provides transcriptional signatures for a large set of molecules, broadly covering the drug-like universe, not all classes of drugs are well represented. To analyze these biases systematically, for each target, we first consider the product of DUD-E and LINCS libraries, which is defined as a subset of DUD-E compounds that have a LINCS counterpart with the Tanimoto coefficient of 1.0.
In Fig. 8A, the per-target fractions of all true positives and true negatives (decoys) included in LINCS are contrasted with the fraction of true positives versus true negatives retained in the subset of concordant LINCS compounds. For most targets, a clear enrichment into true positives is observed and can be further explained by the shift toward higher values of concordance scores for true positives relative to true negatives that are directly represented in LINCS, as shown in Fig. 8B. This is further supported by the analysis of concordance scores for true positives and true negatives in table S2. However, for six targets (AHCY, HMGCR, GART, PNP, Thrombin, and FXa), the total number of true positives included in LINCS is three or less, limiting our ability to use signature connectivity directly and assess the performance.
To address this limitation, ceSAR effectively transfers signature connectivity–based signal from concordant LINCS analogs to candidate compounds by considering their chemical similarity. As shown in Fig. 8C, for relatively close analogs with Tanimoto coefficient of >0.8, this principle allows one to extend the signature concordance filter to all targets considered here. Statistically significantly higher concordance values for true positives are observed for all but three targets. Results at Tanimoto of 0.9, which are included in fig. S10, are consistent with these trends as well. Together, these results support the assumption that despite possible caveats, including off-target effects both at the protein and short hairpin RNA levels, compensatory mechanisms due to protein isoforms, or regulatory feedback loops, a small-molecule inhibitor of a protein target is likely to phenocopy KD of the same target at a statistically significant level of transcriptional signature concordance and that this signal can be effectively captured using LINCS data in conjunction with chemical similarity–based transfer of concordance scores.
Note that some classes of targets and their antagonists, including kinase inhibitors, are already well represented in LINCS, contributing to high accuracy of ceSAR on the five kinases included in our evaluation, for which the ultrafast ceSAR-S outperforms docking in terms of all metrics considered here (figs. S17 and S18). These results indicate that overall results may improve substantially as databases become more representative of the total drug-like chemical universe. In this regard, iLINCS (22), which provides the foundation for this work and the source of high-quality signatures and their precomputed concordance values, is expected to grow over time, contributing to increased accuracy of ceSAR.
While the current evaluation of ceSAR is limited to gene KD signatures generated by the LINCS project to use consistently harmonized data for benchmarking, the method can be applied to gene KD/loss-of-function signatures generated by the user as well. These signatures can be readily generated using either public or private gene expression data with standard bioinformatics pipelines. By enabling user-provided loss-of-function signatures as part of the sig2lead.net server and the stand-alone sig2lead app implementation of ceSAR-S (see also Fig. 2), the utility of the method is extended beyond the set of available LINCS KD signatures.
Last, in the past several years, there has been a flurry of deep learning methods, such as DeepVS (39) or Deep Docking (40) that aim to learn from databases of known active and inactive compounds, including DUD-E, and are reported to outperform docking methods in cross-validation (39, 41). However, despite optimistic claims of improved performance, challenges of overcoming biases in training sets and achieving robust generalization remain (42–44). In this work, molecular descriptors and ligand-protein features are not used directly. Instead, ligand fingerprint representation is simply used to identify LINCS analogs with the goal of transferring the signal captured by the concordance of small-molecule and target KD signatures. Hence, even in the ML-based consensus form of ceSAR that combines a very limited number of ligand and protein structure-independent features, the method does not pose a substantial risk of overfitting. Conversely, signature concordance and chemical similarity–based features evaluated here may contribute to future development of improved deep learning methods for virtual screening.
The performance of ceSAR adds substantially to the utility of LINCS as a big data resource for pharmacogenomics and provides a strong rationale for continued large-scale transcriptional profiling of drug-like molecules and druggable parts of the genome. We anticipate that with further advances in the CRISPR technology, more accurate gene signatures will be obtained, leading to increased performance of the approach. To that end, we note that improving the quality of signatures available via iLINCS through more rigorous quality control and benchmarking has led to substantial improvements in the performance of ceSAR (fig. S21). At the same time, continued advances in determining three-dimensional structures of proteins and their complexes using cryo–electron microscopy and other techniques, including artificial intelligence–based protein structure prediction, will further expand the protein targetable space, adding importance to accelerating the speed of virtual screening approaches.
Through the integration of signature connectivity analysis, fast exact chemical similarity search, and virtual screening approaches, ceSAR greatly reduces false-positive rates and improves accuracy while reducing run times by multiple orders of magnitude. Thus, ceSAR provides a fast, robust, and accurate platform for drug discovery and repurposing that has a potential to further democratize drug discovery pipelines and accelerate advances in personalized precision medicine as illustrated in this work by the discovery of specific inhibitors of BCL2A1.
MATERIALS AND METHODS
ceSAR candidate molecule ranking
For a library of small molecules, Q, and a target gene t with at least one KD transcriptional signature available in LINCS, t ∈ L, the simplest form of ceSAR considered here, referred to as ceSAR-S, ranks candidate compounds by identifying their closest chemical analogs in the LINCS library of transcriptionally profiled chemical perturbagens, k ∈ L, that induce signatures concordant with those of the target KDs. Specifically, for a target gene t, with at least one KD transcriptional signature available in LINCS, t ∈ L, and a library of small molecules to be ranked, Q, the following similarity score is computed for each q ∈ Q as a basis for ranking
where σ(q, k) is the Tanimoto coefficient (21) between compounds q and k ∈ L represented as binary fingerprints and, thus, a real number between 0 and 1, while (k, t) is the maximum concordance over all cell lines for t, and cell line, concentration, exposure time LINCS tuples for k, Lk, between the signatures of chemical perturbagen k and genetic KDs of t
Conceptually, taking the maximum value of a signature concordance score over all cell lines and concentrations for chemical perturbagens follows the assumption that genetic and chemically induced loss of function may result in the most pronounced signatures and their concordance in some unknown biological contexts, as represented by different cell lines included in LINCS. Extreme Pearson correlation–based concordance measure used by iLINCS is adopted here (22), and the threshold for significant concordance is set to c0 = 0.162, which corresponds to Bonferroni-corrected P value of 0.05 (22). The performance of the method is robust with respect to the choice of this threshold as demonstrated in figs. S19 to S21.
By increasing the similarity threshold, s0 ∈ [0,1], one can reduce the initial library to an enriched subset, while the concordance value is used to break ties, in particular resulting in concordance-based ranking of candidate molecules with direct LINCS analogs, i.e., those with the score s(q) = 1. Thus, ceSAR-S uses the signature concordance primarily as an initial filter, while considering effectively only very close analogs when reducing the library to a small subset for further reranking and validation.
We also consider an alternative form of the method, referred to as ceSAR-S*, that finds the closest concordant analog in the LINCS library for each compound and then ranks the compounds by combining signature concordance and chemical similarity to the analogs using the Fisher consensus. As discussed in the Supplementary Materials, while ceSAR-S* improves somewhat the performance in terms of separation of true positives and decoys, it achieves a lower precision at the furthest library reduction, as compounds with strongly concordant distant analogs, including potential pathway inhibitors, are also retained in this case.
ML-based consensus
We also consider more complex, ML-based models to combine signature connectivity–related features, including the strength of concordance, with docking-generated features, including the predicted binding energy. Two alternative feature representations, with 11 or 13 features derived from signature concordance, chemical similarity, and docking predictions, are used (see the Supplementary Materials). Two random forest (11 features and 13 features) and two neural network (11 features and 13 features) models are trained 20 times using leave-one-target cross-validation, with 1 of the 20 DUD-E targets kept as the test set each time. The training is performed with a balanced approach in which all true positives are used for each DUD-E library, while the negative examples are sampled from two subsets of true negatives: one from the top 5% (ceSAR-S ranking) and the other from the remaining 95% of the library. For accuracy assessment, only results on targets not included in the training are used, with the predictions generated by, first, using ceSAR-S to reduce to 5 or 1% of the library for ceSAR-cML5 or ceSAR-cML1, respectively, and then applying the four ML models. Each of these four ML models generates a probability of being a true positive, and Fisher’s consensus approach is used to combine the individual probabilities, representing the evidence of being a true positive versus true negative assigned by each of the models, to provide a meta-consensus ML reranking of candidates selected by ceSAR-S.
Sparse binary fingerprints for chemical similarity search
Binary fingerprints are widely used in cheminformatics for efficient chemical similarity searches and SAR analyses (45–48). In this approximation, small molecules are represented as binary vectors indicating the presence of substructures, subgraphs, pharmacophores, or chemical groups (45, 46). Here, we use the 1024-bit atom-pair fingerprint representation (45, 49), as generated by the ChemmineR package (38), which leads to a sparse binary vector representation of LINCS compounds. As shown in fig. S5, very few of the fingerprint features have a relatively balanced split between ones and zeros across the LINCS compounds. In addition, all LINCS compounds have less than 120 ones in their respective fingerprints with a length of 1024 bits, with a median of about 50 ones.
Fast exact chemical similarity search using minSim
Consider now a search for analogs of a query compound q ∈ Q against database compounds k ∈ L using binary fingerprints described above. The formula for the Tanimoto coefficient, σ(q, k), which, for two binary fingerprints q and k, is defined as the ratio of the number of positions with ones in both q and k versus the number of positions with ones in either q or k, can be written in the following form
where m(q) and m(k) are the number of ones that can be precomputed for all database molecules, while sim(q, k) is the number of ones in common for q and k.
Note that the computation of sim(q, k) can be limited to only those columns in the binary fingerprint where q is in the minority state, which is assumed to be 1. Furthermore, using preprocessing of the reference dataset of compounds (here, LINCS library), one can optimally exploit the sparsity in each column by precomputing indices of database compounds in the minority state at each column, as illustrated in fig. S7. Namely, the following list of database vectors ki is precomputed for each column j in the fingerprint
The minSim algorithm computes all Tanimoto coefficients for a query molecule q by updating integer counters sim(q, k), which are set to zero for all k at the beginning of the search, in a simple loop over minority columns in q and minority lists in each minority column:
We posit that minSim optimally exploits the sparse nature of binary fingerprints by considering only those fingerprint columns (positions) where the query molecule q is in the minority state and by using precomputed lists of all database compounds k that are in the minority states at these positions. The implementation of the algorithm in R is included in fig. S8. Note also that minSim computes the exact Jaccard similarity, without using approximate techniques, such as those based on hashing (36–38).
Statistical analysis
The median difference of the precision and top true-positive rank between benchmarked methods was assessed using the two-sided Wilcoxon test. The difference of the whole distribution of the precision values at different sizes of the reduced libraries between methods was assessed by using the Kullback-Leibler divergence measure and the chi-square test.
BCL2A1 protein purification
BCL2A1 (A1) protein, residues 1 to 152, P104K, C113S, was expressed in the BL21 strain of Escherichia coli in an H596 vector with a hexa-His-MBP tag provided by A. Evdokimov. All purification steps were performed in 20 mM tris (pH 7.0) and 500 mM NaCl. Cells were induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside overnight, pelleted, and lysed via sonicator. After cell lysis, cell debris was pelleted out, and the supernatant was filtered and run through Ni-affinity chromatography. Protein-containing fractions were pooled, and the tag was removed with TEV protease rocking at room temperature. The cleaved proteins were run through a subtractive Ni–nitrilotriacetic acid column and lastly through an S75 size exclusion column.
Thermal shift assay
A total of 100 μM compounds were applied to purified A1 at 4.4 μM in triplicate. Sypro Orange dye was added at a final dilution of 1:1000 to protein- and compound-containing wells. An Applied Biosystems StepOnePlus was used to perform DSF by elevating the temperature from 20° to 99°C, measuring fluorescence at every half degree. Melting temperature (Tm) was recorded as the maximum of the first derivative, indicating that half of the protein population was unfolded. Compounds observed to induce a change in Tm that was greater than three SDs compared to the control were included for further validation.
FP assay
FP was subsequently used to test for specificity of binding by displacement of fluorescein isothiocyanate (FITC)–labeled mouse NoxaB peptide (Peptide 2.0). FP assays were performed in two steps: single-point high-concentration compounds and dose response of FP hits. BCL2A1 was added at 3 to 100 μM of each compound in 20 mM tris (pH 7), 500 mM NaCl, and 0.005% Tween 20 buffer. After adding 375 nM labeled Noxa, 96-well plates were incubated overnight at 20°C in the dark to achieve equilibrium before FP was measured with a BioTek Synergy H2. Autofluorescent and fluorescent quenching compounds were corrected via ratiometric correction as previously described (50). Compounds that showed a substantial shift in polarization, along with those identified in the thermal shift assays, had dose response measured via FP.
Dose-response curves
Dose-response curves were measured in triplicate by adding 3 μM BCL2A1 to a serial twofold (and, for further validation, 1.33-fold) dilution series of each compound ranging from 400 μM to 781 nM in 20 mM tris (pH 7), 500 mM NaCl, and 0.005% Tween 20 buffer, following the same FP protocol described above. Dose-response curves were fitted with the four-paramater logistic equation using SigmaPlot 12.5 (Systat Software Inc. San Jose, CA) to determine IC50 values. To optimize reproducibility and signal-to-noise ratio in the FP data, we used relatively high concentrations of FITC-Noxa, which resulted in elevated IC50 values. Fitted IC50 values were converted to Ki values using the Cheng-Prusoff equation (51)
where Ki is the inhibition constant of the compound, IC50 is the concentration of compound in the FP assay that results in 50% maximal binding, [A] is the concentration of FITC-Noxa peptide, and KD is the dissociation constant for peptide binding to BCL2A1. The published KD (20 nM) for BCL2A1 binding to mouse NoxaA (52) was used as the value for the affinity when calculating Ki values.
T cell death assay
T cells were isolated from C57BL/6 mice or LckCreBaxfl/fl/Bak−/− mice (32) using a pan–T cell isolation kit (Miltenyi Biotech) and stimulated with anti-CD3/CD28 for 24 hours. Single-cell suspensions from the spleen were generated by maceration through a 100-μm nylon mesh, followed by LympholyteM Ficoll gradient separation (CEDARLANE Labs). Purified cells were then cultured on anti-CD3–coated (3 μg/ml; coated overnight, BioLegend) six-well plates in the presence of soluble anti-CD28 (2 μg/ml; Bio X Cell) and interleukin-2 (IL-2) (10 ng/ml; R&D Systems Inc.) in RPMI medium (Life Technologies) for 24 hours at 37°C. Cells were then washed and cultured again in IL-2 (10 ng/ml) for 24 hours at 37°C. Cells were harvested and cultured for 24 hours on anti-CD3–coated 96-well plates at 500,000 cells per well with soluble anti-CD28 (2 μg/ml), IL-2 (10 ng/ml), 0.125 μg of purified anti-mouse FasL (BioLegend), and various concentrations of BCL2A1 inhibitor compounds ± polybrene (2 μg/ml; EMD Millipore). Live versus dead cells were enumerated by trypan blue staining using the TC20 automated cell counter (Bio-Rad Laboratories).
Acknowledgments
We would like to acknowledge Jetstream (https://jetstream-cloud.org/) cloud computing platform, which is used to host the sig2lead.net server, Cincinnati Children’s Hospital Medical Center computing cluster, and all members of the LINCS consortium.
Funding: This work was supported in part by the National Institutes of Health grants U54 HL127624, P30 ES006096, R01 MH107487, R01 CA122346, R01 GM128216, T32 CA236764, R01 CA237016, R21 HD090856, and UL1 TR001425, 2I01BX001110 BLR&D VA Merit award, University of Cincinnati Cancer Center Pilot Project award (to J.M.) and Cincinnati Children’s Innovation Fund award (to D.A.H. and A.B.H.).
Author contributions: A.W.T., J.R., and S.C. developed and benchmarked the method, contributed several sections of the manuscript, and implemented the sig2lead app. J.R., R.A., and B.S. developed and tested the minSim algorithm, B.S., M.P., M.F.-N., M.K., W.N., and M.M. developed the iLINCS platform and API interfaces for signature connectivity analysis used by sig2lead. A.W.T., S.H., A.B.H., and D.A.H. contributed the BCL2A1 experimental validation. A.L.M., M.F.C.-K., R.M., J.Y., S.B., W.N., W.S., N.N., and Y.Z. helped with the benchmarking and refinement of the method and the manuscript. J.M. conceived of the method and coordinated the study and writing of the manuscript.
Competing interests: A.W.T., D.A.H., A.B.H., and J.M. are coinventors on US patent #11,541,073 B2 (granted 3 January 2023) and on pending US patent applications #17/597,515 (filed 19 December 2022) and #18/083,617 (filed 19 December 2022), all of which were filed by Cincinnati Children’s Hospital Medical Center and are related to the present work. All other authors declare that they have no competing interests.
Data and materials availability: DUD-E datasets and target structures used here for benchmarking can be downloaded from http://dude.docking.org/subsets/dud38. The LINCS library of small molecules can be downloaded from the LINCS Data Portal (http://lincsportal.ccs.miami.edu/dcic-portal/), while its preprocessed counterpart for fast chemical similarity search and SAR analyses, as well as ML models trained as part of this work and scripts to combine ceSAR-S and docking scores into consensus predictions, can be downloaded from https://github.com/sig2lead. The gene KD and chemical perturbation LINCS signatures, as well as their precomputed concordance scores, are available through iLINCS and its API programmatic interfaces (www.ilincs.org). All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. ceSAR-S has been implemented as an R Shiny app, dubbed sig2lead, which is a public domain package that can be downloaded from https://github.com/sig2lead, and it is also available as a web server at http://sig2lead.net.
Supplementary Materials
This PDF file includes:
Sections S1 to S8
Figs. S1 to S27
Tables S1 to S9
References
REFERENCES AND NOTES
- 1.Seiler K. P., George G. A., Happ M. P., Bodycombe N. E., Carrinski H. A., Norton S., Brudz S., Sullivan J. P., Muhlich J., Serrano M., Ferraiolo P., Tolliday N. J., Schreiber S. L., Clemons P. A., ChemBank: A small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 36, D351–D359 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaulton A., Bellis L. J., Bento A. P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., Overington J. P., ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eder J., Sedrani R., Wiesmann C., The discovery of first-in-class drugs: Origins and evolution. Nat. Rev. Drug Discov. 13, 577–587 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J.-P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., Golub T. R., The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Shoemaker R. H., Monks A., Alley M. C., Scudiero D. A., Fine D. L., McLemore T. L., Abbott B. J., Paull K. D., Mayo J. G., Boyd M. R., Development of human tumor cell line panels for use in disease-oriented drug screening. Prog. Clin. Biol. Res. 276, 265–286 (1988). [PubMed] [Google Scholar]
- 6.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., Morais P., Meltzer J., Korejwa A., Jané-Valbuena J., Mapa F. A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I. H., Cheng J., Yu G. K., Yu J., Aspesi P., de Silva M., Jagtap K., Jones M. D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R. C., Liefeld T., MacConaill L., Winckler W., Reich M., Li N., Mesirov J. P., Gabriel S. B., Getz G., Ardlie K., Chan V., Myer V. E., Weber B. L., Porter J., Warmuth M., Finan P., Harris J. L., Meyerson M., Golub T. R., Morrissey M. P., Sellers W. R., Schlegel R., Garraway L. A., The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W., Soares J., Greninger P., Edelman E. J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J. A., Thompson I. R., Ramaswamy S., Futreal P. A., Haber D. A., Stratton M. R., Benes C., McDermott U., Garnett M. J., Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnamurthy N., Grimshaw A. A., Axson S. A., Choe S. H., Miller J. E., Drug repurposing: A systematic review on root causes, barriers and facilitators. BMC Health Serv. Res. 22, 970 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haverty P. M., Lin E., Tan J., Yu Y., Lam B., Lianoglou S., Neve R. M., Martin S., Settleman J., Yauch R. L., Bourgon R., Reproducible pharmacogenomic profiling of cancer cell line panels. Nature 533, 333–337 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Karaman B., Sippl W., Computational drug repurposing: Current trends. Curr. Med. Chem. 26, 5389–5409 (2019). [DOI] [PubMed] [Google Scholar]
- 11.El-Hachem N., Ba-Alawi W., Smith I., Mer A. S., Haibe-Kains B., Integrative cancer pharmacogenomics to establish drug mechanism of action: Drug repurposing. Pharmacogenomics 18, 1469–1472 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Subramanian A., Narayan R., Corsello S. M., Peck D. D., Natoli T. E., Lu X., Gould J., Davis J. F., Tubelli A. A., Asiedu J. K., Lahr D. L., Hirschman J. E., Liu Z., Donahue M., Julian B., Khan M., Wadden D., Smith I. C., Lam D., Liberzon A., Toder C., Bagul M., Orzechowski M., Enache O. M., Piccioni F., Johnson S. A., Lyons N. J., Berger A. H., Shamji A. F., Brooks A. N., Vrcic A., Flynn C., Rosains J., Takeda D. Y., Hu R., Davison D., Lamb J., Ardlie K., Hogstrom L., Greenside P., Gray N. S., Clemons P. A., Silver S., Wu X., Zhao W.-N., Read-Button W., Wu X., Haggarty S. J., Ronco L. V., Boehm J. S., Schreiber S. L., Doench J. G., Bittker J. A., Root D. E., Wong B., Golub T. R., A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keenan A. B., Jenkins S. L., Jagodnik K. M., Koplev S., He E., Torre D., Wang Z., Dohlman A. B., Silverstein M. C., Lachmann A., Kuleshov M. V., Ma’ayan A., Stathias V., Terryn R., Cooper D., Forlin M., Koleti A., Vidovic D., Chung C., Schürer S. C., Vasiliauskas J., Pilarczyk M., Shamsaei B., Fazel M., Ren Y., Niu W., Clark N. A., White S., Mahi N., Zhang L., Kouril M., Reichard J. F., Sivaganesan S., Medvedovic M., Meller J., Koch R. J., Birtwistle M. R., Iyengar R., Sobie E. A., Azeloglu E. U., Kaye J., Osterloh J., Haston K., Kalra J., Finkbiener S., Li J., Milani P., Adam M., Escalante-Chong R., Sachs K., Lenail A., Ramamoorthy D., Fraenkel E., Daigle G., Hussain U., Coye A., Rothstein J., Sareen D., Ornelas L., Banuelos M., Mandefro B., Ho R., Svendsen C. N., Lim R. G., Stocksdale J., Casale M. S., Thompson T. G., Wu J., Thompson L. M., Dardov V., Venkatraman V., Matlock A., Van Eyk J. E., Jaffe J. D., Papanastasiou M., Subramanian A., Golub T. R., Erickson S. D., Fallahi-Sichani M., Hafner M., Gray N. S., Lin J.-R., Mills C. E., Muhlich J. L., Niepel M., Shamu C. E., Williams E. H., Wrobel D., Sorger P. K., Heiser L. M., Gray J. W., Korkola J. E., Mills G. B., LaBarge M., Feiler H. S., Dane M. A., Bucher E., Nederlof M., Sudar D., Gross S., Kilburn D. F., Smith R., Devlin K., Margolis R., Derr L., Lee A., Pillai A., The library of integrated network-based cellular signatures NIH program: System-level cataloging of human cells response to perturbations. Cell Syst. 6, 13–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glicksberg B. S., Li L., Chen R., Dudley J., Chen B., Leveraging big data to transform drug discovery. Methods Mol. Biol. 1939, 91–118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musa A., Tripathi S., Dehmer M., Yli-Harja O., Kauffman S. A., Emmert-Streib F., Systems pharmacogenomic landscape of drug similarities from LINCS data: Drug association networks. Sci. Rep. 9, 7849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidović D., Koleti A., Schürer S. C., Large-scale integration of small molecule-induced genome-wide transcriptional responses, kinome-wide binding affinities and cell-growth inhibition profiles reveal global trends characterizing systems-level drug action. Front. Genet. 5, 10.3389/fgene.2014.00342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maia E. H. B., Assis L. C., de Oliveira T. A., da Silva A. M., Taranto A. G., Structure-based virtual screening: From classical to artificial intelligence. Front. Chem. 8, 343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forli S., Huey R., Pique M. E., Sanner M. F., Goodsell D. S., Olson A. J., Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 11, 905–919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labbé C. M., Rey J., Lagorce D., Vavruša M., Becot J., Sperandio O., Villoutreix B. O., Tufféry P., Miteva M. A., MTiOpenScreen: A web server for structure-based virtual screening. Nucleic Acids Res. 43, W448–W454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mysinger M. M., Carchia M., Irwin J. J., Shoichet B. K., Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 55, 6582–6594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajusz D., Rácz A., Héberger K., Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Chem. 7, 20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilarczyk M., Fazel-Najafabadi M., Kouril M., Shamsaei B., Vasiliauskas J., Niu W., Mahi N., Zhang L., Clark N. A., Ren Y., White S., Karim R., Xu H., Biesiada J., Bennett M. F., Davidson S. E., Reichard J. F., Roberts K., Stathias V., Koleti A., Vidovic D., Clarke D. J. B., Schürer S. C., Ma’ayan A., Meller J., Medvedovic M., Connecting omics signatures and revealing biological mechanisms with iLINCS. Nat. Commun. 13, 4678 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M., Shen C., Yang J., Wang Q., Huang N., Systematic investigation of docking failures in large-scale structure-based virtual screening. ACS Omega 7, 39417–39428 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Presicce P., Park C.-W., Senthamaraikannan P., Bhattacharyya S., Jackson C., Kong F., Rueda C. M., DeFranco E., Miller L. A., Hildeman D. A., Salomonis N., Chougnet C. A., Jobe A. H., Kallapur S. G., IL-1 signaling mediates intrauterine inflammation and chorio-decidua neutrophil recruitment and activation. JCI Insight 3, e98306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hind C. K., Carter M. J., Harris C. L., Chan H. T. C., James S., Cragg M. S., Role of the pro-survival molecule Bfl-1 in melanoma. Int. J. Biochem. Cell Biol. 59, 94–102 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Li K.-P., Shanmuganad S., Carroll K., Katz J. D., Jordan M. B., Hildeman D. A., Dying to protect: Cell death and the control of T cell homeostasis. Immunol. Rev. 277, 21–43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O’Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H., An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Wilson W. H., O’Connor O. A., Czuczman M. S., LaCasce A. S., Gerecitano J. F., Leonard J. P., Tulpule A., Dunleavy K., Xiong H., Chiu Y.-L., Cui Y., Busman T., Elmore S. W., Rosenberg S. H., Krivoshik A. P., Enschede S. H., Humerickhouse R. A., Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: A phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 11, 1149–1159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Souers A. J., Leverson J. D., Boghaert E. R., Ackler S. L., Catron N. D., Chen J., Dayton B. D., Ding H., Enschede S. H., Fairbrother W. J., Huang D. C. S., Hymowitz S. G., Jin S., Khaw S. L., Kovar P. J., Lam L. T., Lee J., Maecker H. L., Marsh K. C., Mason K. D., Mitten M. J., Nimmer P. M., Oleksijew A., Park C. H., Park C.-M., Phillips D. C., Roberts A. W., Sampath D., Seymour J. F., Smith M. L., Sullivan G. M., Tahir S. K., Tse C., Wendt M. D., Xiao Y., Xue J. C., Zhang H., Humerickhouse R. A., Rosenberg S. H., Elmore S. W., ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Sasaki H., Hirose T., Oura T., Otsuka R., Rosales I., Ma D., Lassiter G., Karadagi A., Tomosugi T., Dehnadi A., Matsunami M., Raju Paul S., Reeves P. M., Hanekamp I., Schwartz S., Colvin R. B., Lee H., Spitzer T. R., Cosimi A. B., Cippà P. E., Fehr T., Kawai T., Selective Bcl-2 inhibition promotes hematopoietic chimerism and allograft tolerance without myelosuppression in nonhuman primates. Sci. Transl. Med. 15, eadd5318 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H.-W., Park S.-J., Choi B. K., Kim H. H., Nam K.-O., Kwon B. S., 4-1BB promotes the survival of CD8+ T lymphocytes by increasing expression of Bcl-xL and Bfl-1. J. Immunol. 169, 4882–4888 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Tripathi P., Koss B., Opferman J. T., Hildeman D. A., Mcl-1 antagonizes Bax/Bak to promote effector CD4+ and CD8+ T-cell responses. Cell Death Differ. 20, 998–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng T., Li Q., Wang Y., Bryant S. H., Identifying compound-target associations by combining bioactivity profile similarity search and public databases mining. J. Chem. Inf. Model. 51, 2440–2448 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews H., Hanison J., Nirmalan N., “Omics”-informed drug and biomarker discovery: Opportunities, challenges and future perspectives. Proteomes 4, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dar K. B., Bhat A., Amin S., Hamid R., Anees S., Anjum S., Reshi B. A., Zargar M. A., Masood A., Ganie S. A., Modern computational strategies for designing drugs to curb human diseases: A prospect. Curr. Top. Med. Chem. 18, 2702–2719 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Rachkovskij D. A., Index structures for fast similarity search for binary vectors. Cybern. Syst. Anal. 53, 799–820 (2017). [Google Scholar]
- 37.J. Wang, H. T. Shen, J. Song, J. Ji, Hashing for similarity search: A survey. arXiv:1408.2927 [cs.DS] (13 August 2014).
- 38.González-Medina M., Jesús Naveja J., Sánchez-Cruz N., Medina-Franco J. L., Open chemoinformatic resources to explore the structure, properties and chemical space of molecules. RSC Adv. 7, 54153–54163 (2017). [Google Scholar]
- 39.Pereira J. C., Caffarena E. R., Dos Santos C. N., Boosting docking-based virtual screening with deep learning. J. Chem. Inf. Model. 56, 2495–2506 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Gentile F., Yaacoub J. C., Gleave J., Fernandez M., Ton A.-T., Ban F., Stern A., Cherkasov A., Artificial intelligence-enabled virtual screening of ultra-large chemical libraries with deep docking. Nat. Protoc. 17, 672–697 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Gentile F., Agrawal V., Hsing M., Ton A.-T., Ban F., Norinder U., Gleave M. E., Cherkasov A., Deep docking: A deep learning platform for augmentation of structure based drug discovery. ACS Cent. Sci. 6, 939–949 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sieg J., Flachsenberg F., Rarey M., In need of bias control: Evaluating chemical data for machine learning in structure-based virtual screening. J. Chem. Inf. Model. 59, 947–961 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Li Y., Fan Z., Rao J., Chen Z., Chu Q., Zheng M., Li X., An overview of recent advances and challenges in predicting compound-protein interaction (CPI). Med. Rev. 3, 465–486 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyu J., Irwin J. J., Shoichet B. K., Modeling the expansion of virtual screening libraries. Nat. Chem. Biol. 19, 712–718 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muegge I., Mukherjee P., An overview of molecular fingerprint similarity search in virtual screening. Expert Opin. Drug Discovery 11, 137–148 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Floris M., Manganaro A., Nicolotti O., Medda R., Mangiatordi G. F., Benfenati E., A generalizable definition of chemical similarity for read-across. J. Chem. 6, 39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willett P., Similarity searching using 2D structural fingerprints. Methods Mol. Biol. 672, 133–158 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Willett P., Similarity-based virtual screening using 2D fingerprints. Drug Discov. Today 11, 1046–1053 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Cereto-Massagué A., Ojeda M. J., Valls C., Mulero M., Garcia-Vallvé S., Pujadas G., Molecular fingerprint similarity search in virtual screening. Methods 71, 58–63 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Shapiro A. B., Walkup G. K., Keating T. A., Correction for interference by test samples in high-throughput assays. J. Biomol. Screen. 14, 1008–1016 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Yung-Chi C., Prusoff W. H., Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973). [DOI] [PubMed] [Google Scholar]
- 52.Smits C., Czabotar P. E., Hinds M. G., Day C. L., Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure 16, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Cao Y., Charisi A., Cheng L.-C., Jiang T., Girke T., ChemmineR: A compound mining framework for R. Bioinformatics 24, 1733–1734 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Pilarczyk, M. F. Najafabadi, M. Kouril, J. Vasiliauskas, W. Niu, B. Shamsaei, N. Mahi, L. Zhang, N. Clark, Y. Ren, S. White, R. Karim, H. Xu, J. Biesiada, M. F. Bennet, S. Davidson, J. F. Reichard, V. Stathias, A. Koleti, D. Vidovic, D. J. B. Clark, S. Schurer, A. Ma’ayan, J. Meller, M. Medvedovic, Connecting omics signatures of diseases, drugs, and mechanisms of actions with iLINCS. bioRxiv 826271 [Preprint] (2019). 10.1101/826271. [DOI]
- 55.J. Biesiada, A. Porollo, J. Meller, “On setting up and assessing docking simulations for virtual screening” in Rational Drug Design, Methods in Molecular Biology (Humana Press, 2012), pp. 1–16; http://link.springer.com/protocol/10.1007/978-1-62703-008-3_1. [DOI] [PubMed]
- 56.R Core Team, R: A language and environment for statistical computing (2017); www.R-project.org/.
- 57.D. Meyer, E. Dimitriadou, K. Hornik, A. Weingessel, F. Leisch, C.-C. Chang, C.-C. Lin, e1071: Misc functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien (2019).
- 58.Kuhn M., Building predictive models in R using the caret package. J. Stat. Softw. 28, 1–26 (2008).27774042 [Google Scholar]
- 59.O’Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., Hutchison G. R., Open Babel: An open chemical toolbox. J. Chem. 3, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.K. Horan, T. Girke, ChemmineOB: R interface to a subset of OpenBabel functionalities, version 1.18.0 (2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sections S1 to S8
Figs. S1 to S27
Tables S1 to S9
References