Abstract
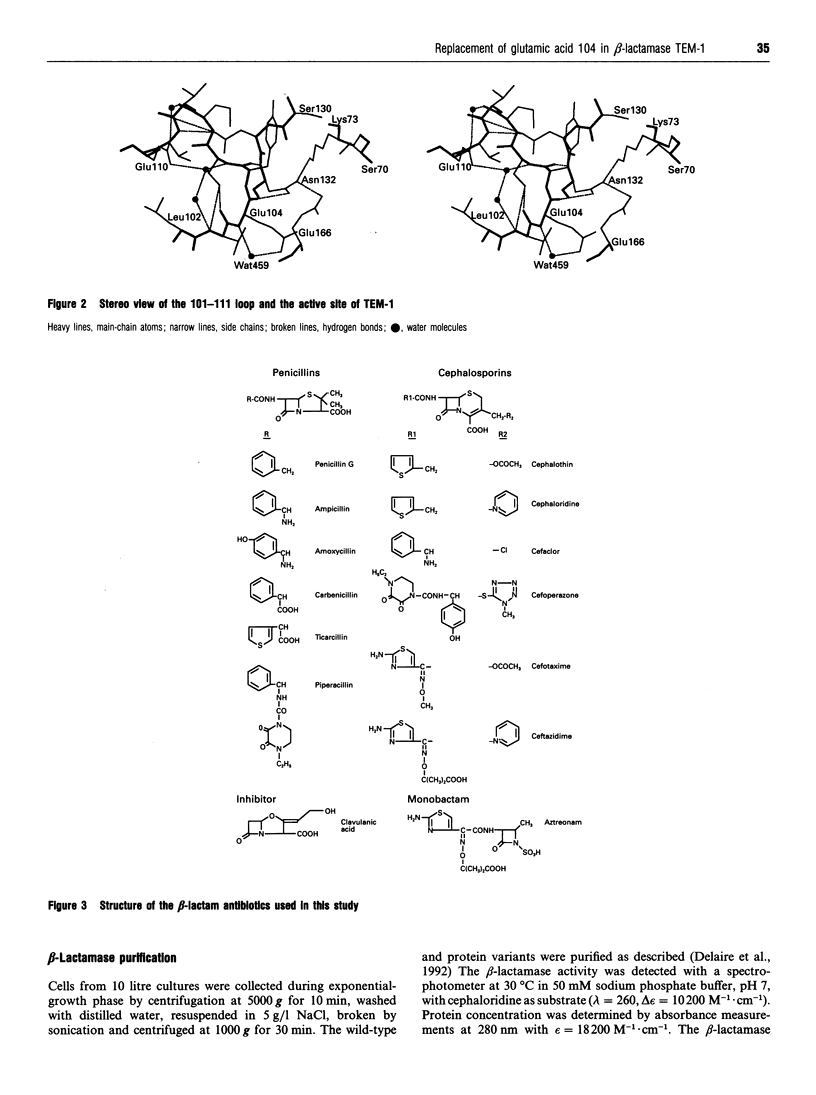
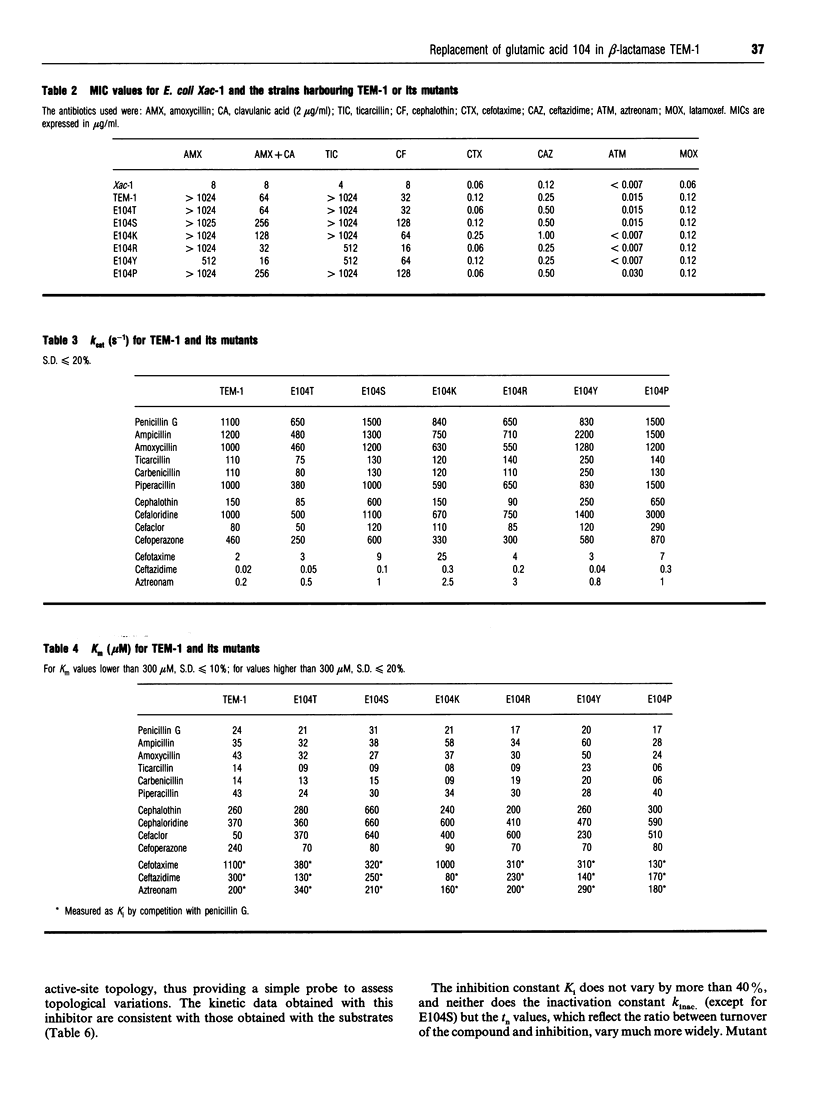
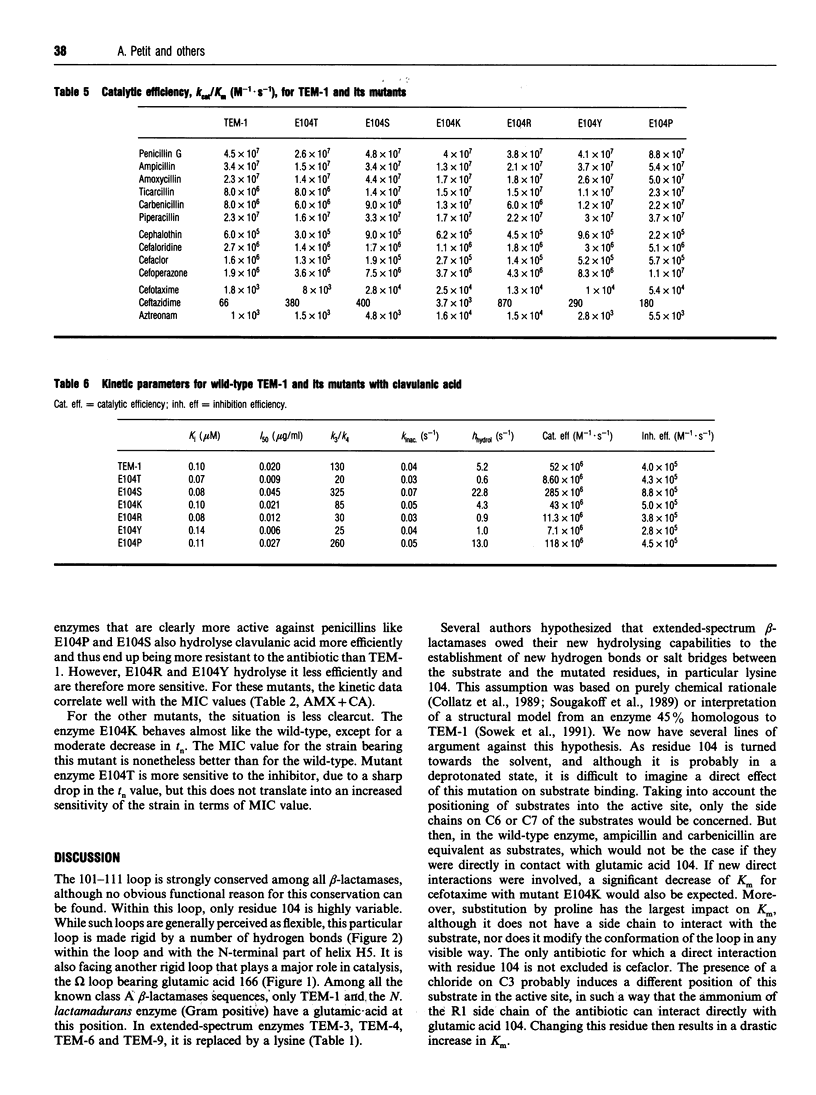
Residue 104 is frequently mutated from a glutamic acid to a lysine in the extended-spectrum TEM beta-lactamases responsible for the resistance to third-generation cephalosporins in clinical Gram negative strains. Among class A beta-lactamases, it is the most variable residue within a highly conserved loop which delineates one side of the active site of the enzymes. To investigate the role of this residue in the extended-spectrum phenotype, it has been replaced by serine, threonine, lysine, arginine, tyrosine and proline. All these substitutions yield active enzymes, with no drastic changes in kinetic properties compared with the wild-type enzyme, except with cefaclor, but an overall improved affinity for second- and third-generation cephalosporins. Only mutant E104K exhibits a significant ability to hydrolyse cefotaxime. Molecular modelling shows that the substitutions have generally no impact on the conformation of the 101-111 loop as the side chains of residues at position 104 are all turned towards the solvent. Unexpectedly, the E104P mutant turns out to be the most efficient enzyme. All our results argue in favour of an indirect role for this residue 104 in the substrate specificity of the class A beta-lactamases. This residue contributes to the precise positioning of residues 130-132 which are involved in substrate binding and catalysis. Changing residue 104 could also modify slightly the local electrostatic potential in this part of the active site. The limited kinetic impact of the mutations at this position have to be analysed in the context of the microbiological problem of resistance to third-generation cephalosporins. Although mutation E104K improves the ability of the enzyme to hydrolyse these compounds, it is not sufficient to confer true resistance, and is always found in clinical isolates associated with at least one mutation at another part of the active site. It is the combined effect of the two mutations that synergistically enhances the hydrolytic capability of the enzyme towards third-generation cephalosporins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Coulson A. F., Frère J. M., Ghuysen J. M., Joris B., Forsman M., Levesque R. C., Tiraby G., Waley S. G. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991 May 15;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélémy M., Péduzzi J., Bernard H., Tancrède C., Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum beta-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992 Jul 13;1122(1):15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- Bush K. Characterization of beta-lactamases. Antimicrob Agents Chemother. 1989 Mar;33(3):259–263. doi: 10.1128/aac.33.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanal C., Poupart M. C., Sirot D., Labia R., Sirot J., Cluzel R. Nucleotide sequences of CAZ-2, CAZ-6, and CAZ-7 beta-lactamase genes. Antimicrob Agents Chemother. 1992 Sep;36(9):1817–1820. doi: 10.1128/aac.36.9.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collatz E., Tran Van Nhieu G., Billot-Klein D., Williamson R., Gutmann L. Substitution of serine for arginine in position 162 of TEM-type beta-lactamases extends the substrate profile of mutant enzymes, TEM-7 and TEM-101, to ceftazidime and aztreonam. Gene. 1989 May 30;78(2):349–354. doi: 10.1016/0378-1119(89)90237-0. [DOI] [PubMed] [Google Scholar]
- Delaire M., Labia R., Samama J. P., Masson J. M. Site-directed mutagenesis at the active site of Escherichia coli TEM-1 beta-lactamase. Suicide inhibitor-resistant mutants reveal the role of arginine 244 and methionine 69 in catalysis. J Biol Chem. 1992 Oct 15;267(29):20600–20606. [PubMed] [Google Scholar]
- Herzberg O., Kapadia G., Blanco B., Smith T. S., Coulson A. Structural basis for the inactivation of the P54 mutant of beta-lactamase from Staphylococcus aureus PC1. Biochemistry. 1991 Oct 1;30(39):9503–9509. doi: 10.1021/bi00103a017. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A., Medeiros A. A. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991 Sep;35(9):1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsch C., Mourey L., Masson J. M., Samama J. P. Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution. Proteins. 1993 Aug;16(4):364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- Labia R., Andrillon J., Le Goffic F. Computerized microacidimetric determination of beta lactamase Michaelis-Menten constants. FEBS Lett. 1973 Jun 15;33(1):42–44. doi: 10.1016/0014-5793(73)80154-1. [DOI] [PubMed] [Google Scholar]
- Labia R., Barthélémy M. L'enzymogramme des beta-lactamases: adaptation en cel de la méthode iodométrique. Ann Microbiol (Paris) 1979 Oct;130B(3):295–304. [PubMed] [Google Scholar]
- Lenfant F., Labia R., Masson J. M. Probing the active site of beta-lactamase R-TEM1 by informational suppression. Biochimie. 1990 Jun-Jul;72(6-7):495–503. doi: 10.1016/0300-9084(90)90073-p. [DOI] [PubMed] [Google Scholar]
- Lenfant F., Petit A., Labia R., Maveyraud L., Samama J. P., Masson J. M. Site-directed mutagenesis of beta-lactamase TEM-1. Investigating the potential role of specific residues on the activity of Pseudomonas-specific enzymes. Eur J Biochem. 1993 Nov 1;217(3):939–946. doi: 10.1111/j.1432-1033.1993.tb18324.x. [DOI] [PubMed] [Google Scholar]
- Livrelli V., Peduzzi J., Joly B. Sequence and molecular characterization of the ROB-1 beta-lactamase gene from Pasteurella haemolytica. Antimicrob Agents Chemother. 1991 Feb;35(2):242–251. doi: 10.1128/aac.35.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabilat C., Courvalin P. Development of "oligotyping" for characterization and molecular epidemiology of TEM beta-lactamases in members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1990 Nov;34(11):2210–2216. doi: 10.1128/aac.34.11.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne A., Misselyn-Bauduin A. M., Joris B., Erpicum T., Granier B., Frère J. M. The diversity of the catalytic properties of class A beta-lactamases. Biochem J. 1990 Jan 1;265(1):131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Quinn J. P., Miyashiro D., Patel M., Bush K., Singer S. B., Graves D., Palzkill T., Arvin A. M. Outbreak of ceftazidime resistance due to a novel extended-spectrum beta-lactamase in isolates from cancer patients. Antimicrob Agents Chemother. 1992 Sep;36(9):1991–1996. doi: 10.1128/aac.36.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Normark S. Sensitivity of Escherichia coli to various beta-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic beta-lactamases: a quantitative predictive treatment. Mol Microbiol. 1987 Jul;1(1):29–36. doi: 10.1111/j.1365-2958.1987.tb00523.x. [DOI] [PubMed] [Google Scholar]
- Sakurai Y., Tsukamoto K., Sawai T. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J Bacteriol. 1991 Nov;173(21):7038–7041. doi: 10.1128/jb.173.21.7038-7041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. C. beta-Lactamases of gram-negative bacteria: new challenges for new drugs. Clin Infect Dis. 1992 May;14(5):1089–1099. doi: 10.1093/clinids/14.5.1089. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sougakoff W., Petit A., Goussard S., Sirot D., Bure A., Courvalin P. Characterization of the plasmid genes blaT-4 and blaT-5 which encode the broad-spectrum beta-lactamases TEM-4 and TEM-5 in enterobacteriaceae. Gene. 1989 May 30;78(2):339–348. doi: 10.1016/0378-1119(89)90236-9. [DOI] [PubMed] [Google Scholar]
- Sowek J. A., Singer S. B., Ohringer S., Malley M. F., Dougherty T. J., Gougoutas J. Z., Bush K. Substitution of lysine at position 104 or 240 of TEM-1pTZ18R beta-lactamase enhances the effect of serine-164 substitution on hydrolysis or affinity for cephalosporins and the monobactam aztreonam. Biochemistry. 1991 Apr 2;30(13):3179–3188. doi: 10.1021/bi00227a004. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., Sutoh K., James M. N. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 A resolution. Nature. 1992 Oct 22;359(6397):700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedel G., Mabilat C., Goussard S., Picard B., Fournier G., Gilly L., Paul G., Philippon A. Two variants of transferrable extended-spectrum TEM-beta-lactamase successively isolated from a clinical Escherichia coli isolate. FEMS Microbiol Lett. 1992 Jun 1;72(2):161–166. doi: 10.1016/0378-1097(92)90522-p. [DOI] [PubMed] [Google Scholar]



