Abstract
Endothelial cells play an essential role in inflammation through synthesis and secretion of chemoattractant cytokines and expression of adhesion molecules required for inflammatory cell attachment and infiltration. The mechanisms by which endothelial cells control the pro-inflammatory response depend on the type of inflammatory stimuli, endothelial cell origin, and tissue involved. In the present study, we investigated the role of the transcription factor c-Myc in inflammation using a conditional knockout mouse model in which Myc is specifically deleted in the endothelium. At a systemic level, circulating monocytes, the chemokine CCL7, and the extracellular-matrix protein osteopontin were significantly increased in endothelial c-Myc knockout (EC-Myc KO) mice, whereas the cytokine TNFSF11 was downregulated. Using an experimental model of steatohepatitis, we investigated the involvement of endothelial c-Myc in diet-induced inflammation. EC-Myc KO animals displayed enhanced pro-inflammatory response, characterized by increased expression of pro-inflammatory cytokines and leukocyte infiltration, and worsened liver fibrosis. Transcriptome analysis identified enhanced expression of genes associated with inflammation, fibrosis, and hepatocellular carcinoma in EC-Myc KO mice relative to control (CT) animals after short-exposure to high-fat diet. Analysis of a single-cell RNA-sequencing dataset of human cirrhotic livers indicated downregulation of MYC in endothelial cells relative to healthy controls. In summary, our results suggest a protective role of endothelial c-Myc in diet-induced liver inflammation and fibrosis. Targeting c-Myc and its downstream pathways in the endothelium may constitute a potential strategy for the treatment of inflammatory disease.
Keywords: c-Myc, endothelial cells, fibrosis, inflammation
1 ∣. INTRODUCTION
Endothelial cells play an essential role in the regulation of inflammation. As the innermost layer of blood vessels, endothelial cells form an anti-inflammatory and anticoagulatory surface under normal circumstances and function as a selective barrier between blood and other tissues.1,2 However, during infection, hemodynamic stress, hyperlipidemia, and/or hyperglycemia, endothelial cells undergo pro-inflammatory changes that lead to impairment of endothelial function and hyperactivation. Enhanced expression of adhesion molecules and increased permeability promote the binding of circulating leukocytes to the endothelium and infiltration into tissues. Activated endothelial cells also secrete pro-inflammatory cytokines and chemokines into the surrounding microenvironment, further contributing to the migration and recruitment of inflammatory cells.3,4
The endothelial pro-inflammatory response is largely regulated at transcriptional level and by mechanisms determined by the type of inflammatory stimuli, endothelial cell origin, and target tissue.4-6 The transcriptional factor c-Myc is a “master regulator” of the genome that contributes to the regulation of essential biological processes including proliferation, differentiation, metabolism, and cell fate.7-9 Deregulation of c-Myc expression has been extensively studied in cancer.9,10 However, little is known about the role of c-Myc in the endothelium. To date, most studies have investigated the role of endothelial c-Myc in embryonic development and tumor angiogenesis.11-13 We previously reported that c-Myc plays an essential role in endothelial cells by controlling not only proliferation and angiogenesis, but also the expression of inflammatory mediators, suggesting a potential role in regulating inflammation.14 c-Myc is downstream of major pro-inflammatory pathways and has been shown to regulate the expression of different inflammatory genes.15-19 However, the specific role of endothelial c-Myc in inflammation has not been fully explored. In the present work, we investigated the contribution of endothelial c-Myc to inflammation in vivo.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Animals
All animal experiments were approved by the University of Miami Animal Care and Use Committee according to the National Institutes of Health guidelines for the care and use of laboratory animals. Knockout mice carrying deletion of c-Myc specifically in endothelial cells were generated by crossing mouse lines Cdh5(PAC)-CreERT2 (B6.CBA-Tg(Cdh5-cre/ERT2)1Rha,20 obtained from Cancer Research Technology Limited (Ximbio) (London, UK) through material transfer agreement) and Mycflox/flox (B6.129S6-Myctm2Fwa/Mmjax,21 obtained from Jackson Laboratory (Bar Harbor, ME, USA), Stock #032046-JAX). Controls (CTs) consisted of littermates with identical floxed Myc allele but lacking Cre. In order to confirm localization of Cre activity, CT and EC-Myc KO animals were crossed with mT/mG Cre reporter mouse line (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB–tdTomato–EGFP)Luo/J,22 obtained from The Jackson Laboratory, Stock #007676). Both male and female littermates were used in the study for most of the analyses performed as indicated throughout the text. Deletion of c-Myc in endothelial cells was induced at 4–5 weeks of age by daily intraperitoneal injections of 2 mg tamoxifen (#13258, Cayman Chemical, Ann Arbor, MI, USA) for five consecutive days. For diet challenge studies, two weeks after the induction of knockout, animals were exposed to a low-fat control- (CTD) or high-fat diet (HFD) containing 1.25% cholesterol (#TD.08485 and #TD.02028, respectively, Teklad, Indianapolis, IN, USA). Experiments were performed for short- (5 weeks) or long-term (22 weeks) for molecular and pathological analysis, respectively. All animals were housed on a 12-h light/dark cycle with free access to food and water. At endpoint, animals were sedated by intraperitoneal injection of a ketamine-xylazine anesthetic mixture (100 mg and 10 mg/kg body weight, respectively). Blood samples were collected by cardiac puncture and separated for complete blood cell count and serum/plasma analysis. All organs were collected and snap frozen in liquid nitrogen or fixed in 10% formalin for molecular and pathology assessment, respectively.
2.2 ∣. Complete blood cell count and flow cytometry analysis
Blood samples were collected by cardiac puncture in EDTA-coated tubes and immediately submitted to the University of Miami Comparative Pathology Lab for complete blood cell count (CBC) (five males and one female per experimental group). Bone marrow was harvested by flushing femurs with phosphate-buffered saline and processed for analysis of cellular composition by fluorescence-activated cell sorting (five males and five females per experimental group). For immunophenotyping, hematopoietic cells were labeled with antibody cocktails for long-term hematopoietic stem cells (Lin− Sca-1+ c-Kit+ Flk2− CD90low), short-term hematopoietic stem cells (Lin− Sca-1+ c-Kit+ Flk2+ CD90low), multipotent progenitors (Lin− Sca-1+ c-Kit+ Flk2+ CD90−), granulocyte-monocyte progenitors (Lin− Il7r− c-Kit+ Sca-1− CD34+ CD16/32high), common myeloid progenitors (Lin− IL7r− c-Kit+ Sca-1− CD34+ FCRgLow), megakaryocyte-erythroid progenitors (Lin− Il7r− c-Kit+ Sca-1− CD34− CD16/32low), lymphoid progenitors (Lin− Thy1.1− Il7r+ c-Kit+ Sca-1+), granulocytes (Cd11b+ Ly6G+), erythroid cells (Ly6G− Ter119+), B lymphocytes (Ly6G− B220+ CD19+), monocytes/macrophages (CD11b+ F4-80+), T lymphocytes (CD3+ CD4+ CD8−/CD3+ CD4− CD8+), and natural killer cells (CD49b+ Nk1.1+). Lin denotes lineage specific markers (CD3, CD45R/B220, CD11b, Ly-6G (Gr-1), and Ter119). Staining was performed according to manufacturer's instructions and data were acquired using a BD LSR-II analyzer (BD Biosciences, San Jose, CA, USA). Results were analyzed using FlowJo software (BD Biosciences) according to the gating strategy in Figure S1. A complete list of the antibodies (Invitrogen, Waltham, MA, USA) tested is provided in Supporting Information.
2.3 ∣. Lipid analysis
Lipid analysis of plasma and liver samples was outsourced to the Mouse Metabolic Phenotyping Center at University of California Davis using assays for lipid extraction from livers (#D3301), triglycerides (#D3404), and cholesterol (#D3405).
2.4 ∣. Inflammatory cytokine analysis
Cytokine levels were determined by multiplex analysis of serum samples collected three months after induction of c-Myc deletion using a customed designed panel (#LXSAMSM, outsourced to R&D Systems, Minneapolis, MN, USA) (only males were tested). In experimental diet studies, cytokines were measured in liver lysates using a commercially available mouse cytokine array panel (#MD31, outsourced to Eve Technologies, Calgary, AB, Canada). Liver lysates were prepared in RIPA buffer supplemented with protease and phosphatase inhibitor cocktail (#R0278 and #MSSAFE, respectively, Sigma-Aldrich, St. Louis, MO, USA). Both males and females were tested.
2.5 ∣. RNA sequencing analysis
Liver RNA extraction and sequencing were performed by Beijing Genomics Institute (Shenzhen, Guangdong, China). RNA quality was tested before sequencing on BGI DNBseq platform. The raw sequencing reads were aligned to GRCm38 mouse reference genome using a web server BioJupies.23 Genes exhibiting at least 0.5 count per million in at least 25% of the samples were selected for further analysis in R (version 3.6.1). Differentially expressed genes were identified using DESeq2 (version 1.26.0) and genes with a p value <.05 and the absolute fold change >1.5 were considered significant.24 Venn diagram analysis was performed with Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html). Gene ontology analysis was performed using EnrichR.25 Pathway and functional analysis were conducted with Ingenuity Pathway Analysis (Qiagen, Hilden, Germany).26 For each experimental group, four males and three females were analyzed. Gene expression data have been uploaded into the Gene Expression Omnibus database under accession number GSE176573. A complete R-script to reproduce the analysis is available at the authors' GitHub page (https://github.com/cdorodrigs/AtherogenicPrj).
2.6 ∣. Single-cell RNA sequencing analysis
Using the GSE136103 dataset, cells were integrated, clustered, and dimensionally resolved using the R package Seurat as described previously.27,28 Donors consisted of five healthy individuals (four males and one female) and five cirrhotic patients (three males and two females). Authors of the original article provided tSNE embeddings of all non-parenchymal cells as well as the endothelial cell cluster.29 A complete R-script to reproduce the analysis is available at the lead authors' GitHub page (https://github.com/cdorodrigs/AtherogenicPrj).
2.7 ∣. Histology analysis
Formalin fixed liver lobes were embedded in paraffin. Sections (4 μm) were stained with hematoxylin and eosin (H&E) for foci count and picrosirius red for fibrosis assessment. All slides used for analysis were scanned at 20× using a virtual slide scanner (VS120, Olympus, Tokyo, Japan). Image analysis was performed blindly by two independent observers using ImageJ software. Inflammation was assessed on H&E-stained slides by counting the total number of inflammatory foci and normalization to total tissue area. Results represent the average of normalized counts in four lobes. The degree of inflammation was estimated based on a scoring system including no inflammation (score 0, no foci/mm2 tissue), mild inflammation (score 1, <2 foci/mm2 tissue), moderate inflammation (score 2, 2–8 foci/mm2 tissue), and severe inflammation (score 3, more than 8 foci/mm2 tissue). Fibrosis was assessed by estimation of the percentage of area positive for picrosirius red. Positive staining was measured using a color deconvolution plugin in ImageJ in which the red channel image was selected for quantification. For immunohistochemistry, paraffin-embedded tissue sections were stained with an anti-myeloperoxidase (MPO) antibody (#ab208670, Abcam, Cambridge, UK) and an anti-CD68 antibody (#ab125212, Abcam) to identify neutrophils and macrophages, respectively. Twenty randomly selected areas of tissue sections were photographed using a Zeiss Axioscope (Zeiss, Göttingen, Germany) at 10× magnification with five images for each lobe. For quantification, MPO staining was assessed by counting the number of positive cells in each image, and CD68 staining was analyzed by estimation of the percentage of positively stained area using ImageJ color deconvolution plugin in which the brown channel image was selected for quantification.
2.8 ∣. Isolation of endothelial cells
Animal was anesthetized with a ketamine-xylazine solution. The abdomen was opened and the intestine was gently moved to the side to expose the inferior vena cava and portal vein. A catheter connected to a perfusion pump was inserted into the inferior vena cava and the portal vein was cut for outflow drainage. The liver was first perfused with a warm (37°C) Hanks solution supplemented with 0.5 mM EGTA until the liver became pale, and was then perfused with 50 ml of 0.05% collagenase type IV (#LS004188, Worthington, Lakewood, NJ, USA) prepared in Hanks solution supplemented with 3 mM calcium chloride for about 10 min. After perfusion, the liver was harvested, transferred to a sterile Petri dish containing Hanks solution, and gently pressed with the flat end of a 3 ml syringe plunger to release cells. Cell suspension was passed through a 70 μm strainer and centrifuged at 60 g for 2 min to pellet hepatocytes. The supernatant containing non-parenchymal cells was then centrifuged at 500 g for 5 min. The resulting pellet was resuspended in 20 ml of 17.6% Optiprep solution (prepared in Hanks solution) and 20 ml of 8.2% Optiprep solution (prepared in Hanks solution) was carefully layered on top of the cell suspension. The gradient was centrifuged at 1400 g for 30 min with minimal acceleration and deceleration. Cells at the interface were collected, washed three times with Hanks solution, and labeled with an anti-mouse biotinylated CD31 antibody (#558737, BD Biosciences). CD31-labeled endothelial cells were isolated by magnetic sorting using CELLection™ Biotin Binder Kit (#11533D, Invitrogen) according to the manufacturer's instructions.
2.9 ∣. Real-time PCR
Total RNA was extracted from CD31-isolated cells with miRNeasy Tissue/Cells Advanced Kit (#217684, Qiagen). RNA concentration and quality were determined using a NanoDrop 2000 spectrophotometer (ThermoFisher, Waltham, MA, USA). cDNA synthesis was performed using High-Capacity cDNA Reverse Transcription Kit (#4368814, Applied Biosystems, Waltham, MA, USA) according to the manufacturer's instructions. Real-time PCR was performed on 7500 Real-Time PCR System (Applied Biosystems) with Taqman gene expression master mix (#4369514, Applied Biosystems) and probes (Hprt, Mm01545399_m1; Myc, Mm00487804_m1; Cdkn1a, Mm04205640_g1). Target gene expression was normalized by Hprt. Fold changes were calculated relative to CT group by the ΔΔCt method.
2.10 ∣. Statistical analysis
All statistical analysis was performed using SigmaPlot software and p values <.05 were considered significant. Unless otherwise noted, data are presented as mean ± standard deviation and individual animals are represented by dots in all graphs. Sample numbers are indicated in figure legends. For comparison between two groups, we performed Student t-test considering two-tailed p-values. Mann–Whitney U test and Welch's t-test were used as alternatives when samples did not pass normality test or equal variance test, respectively. For comparison among four groups, we performed two-way analysis of variance followed by Holm-Sidak post hoc analysis.
3 ∣. RESULTS
3.1 ∣. Endothelial c-Myc knockout promotes systemic inflammation
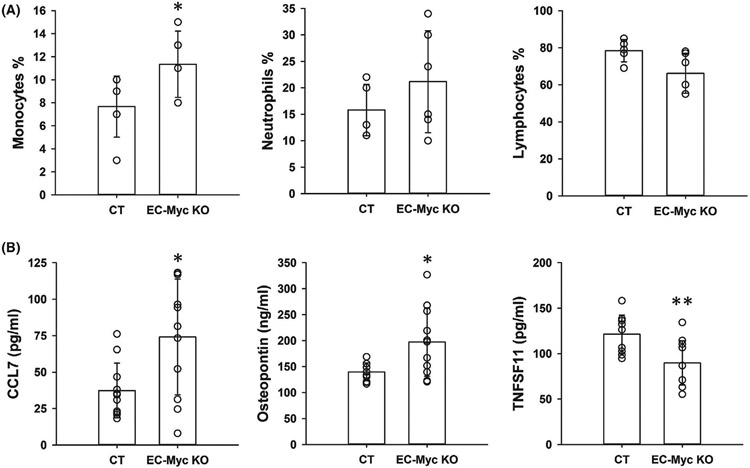
We previously reported that c-Myc knockdown in human umbilical vein endothelial cells (HUVECs) induced expression of cytokines and adhesion molecules involved in inflammatory cell adhesion and infiltration. In addition, c-Myc knockdown enhanced TNF-α-induced inflammation.14 To investigate the impact of endothelial c-Myc loss in inflammation in vivo, we generated a mouse model in which c-Myc is conditionally deleted specifically in the endothelium (Figure S2). CBC analysis showed a significant increase in monocyte fraction in EC-Myc KO mice by 47.7% relative to CT (11.33 ± 2.80 vs. 7.67 ± 2.66%, p = .045). No significant changes were observed in neutrophil and lymphocyte populations (Figure 1A). Complete CBC analysis is presented in Table S1. We performed flow cytometry analysis of bone marrow cells to determine whether endothelial c-Myc deletion caused alterations in hematopoiesis and did not find significant differences between CT and EC-Myc KO groups (Table S2).
FIGURE 1.
Endothelial c-Myc knockout promotes systemic inflammation. (A) Monocyte, neutrophil, and lymphocyte fraction of total white blood cells analyzed from CT and EC-Myc KO complete blood count (n = 6). (B) Multiplex analysis of selected inflammatory mediators in serum significantly altered by endothelial c-Myc knockout (n = 11). CT, control; EC-Myc KO, endothelial c-Myc knockout. *p < .05, **p < .01
We performed multiplex analysis of inflammation markers in serum from CT and EC-Myc KO animals using a custom-designed panel. The results showed that loss of endothelial c-Myc caused a significant increase in the circulating levels of the cytokine CCL7 and the extracellular matrix protein osteopontin by 100% (74 ± 40 vs. 37 ± 19 pg/ml, p = .012) and 41% (197 414 ± 65 967 vs. 139 986 ± 17 031 pg/ml, p = .011), respectively, whereas TNFSF11 was significantly decreased by 26% (90 ± 24 vs. 121 ± 21 pg/ml, p = .004) (Figure 1B). A summary of all inflammation targets analyzed is presented in Table S3. These findings suggest that endothelial c-Myc is involved in the regulation of systemic inflammation.
3.2 ∣. Transcriptome analysis of endothelial c-Myc knockout livers after exposure to HFD stress suggests enhanced inflammatory response
To further investigate the role of endothelial c-Myc in the context of inflammation, we challenged CT and EC-Myc KO mice with a HFD used in studies of nonalcoholic steatohepatitis (NASH). This diet promotes hypercholesterolemia and accumulation of cholesterol in the liver, triggering inflammation and fibrosis.30 As expected, animals developed an increase in liver weight and cholesterol content in both plasma and liver (Table S4).
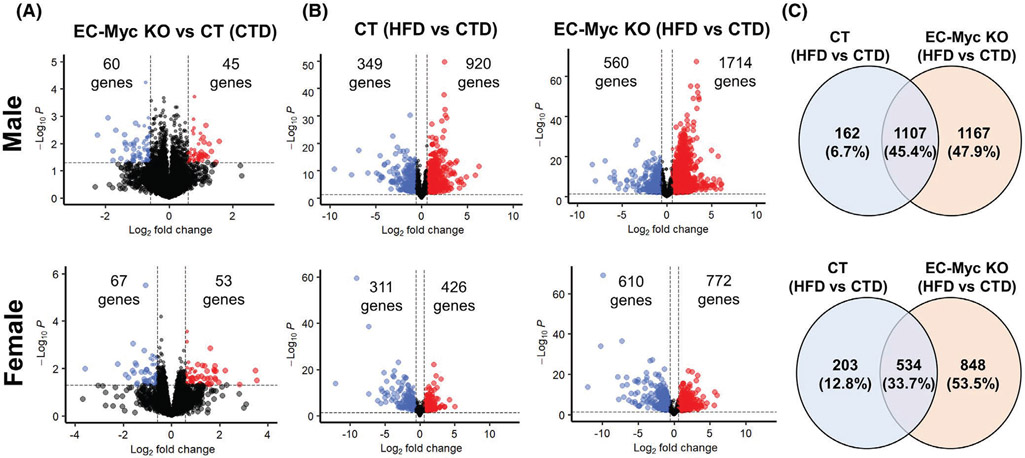
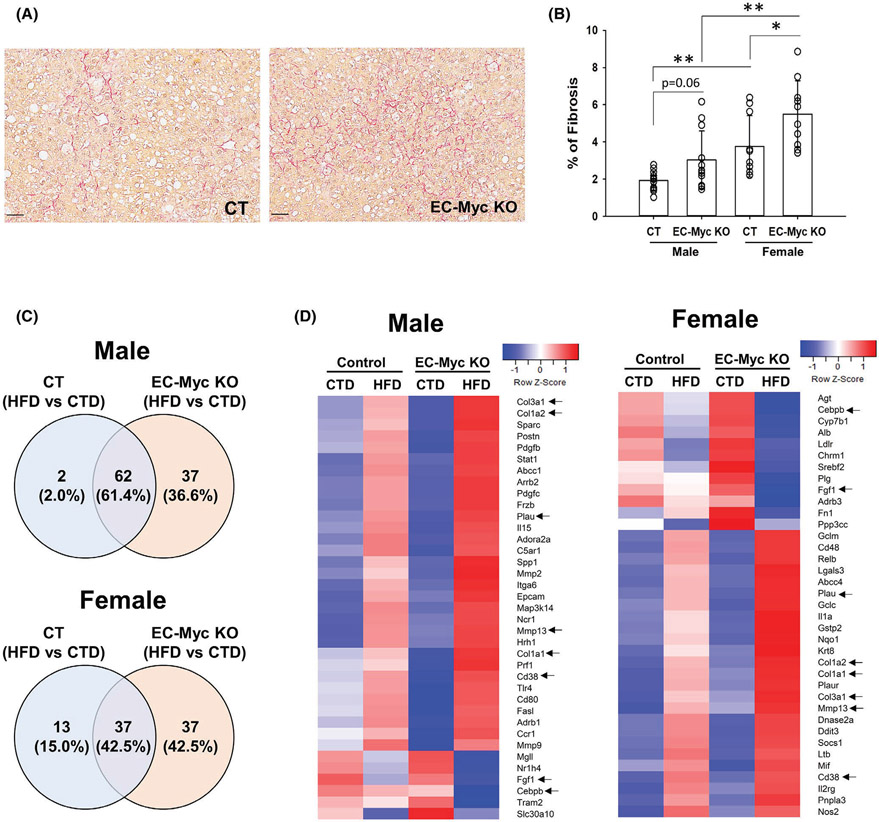
We performed transcriptome analysis on livers from CT and EC-Myc KO mice after short-term exposure to CTD and HFD. Loss of endothelial c-Myc under CTD had a modest impact in gene expression profile (Figure 2A). Exposure to HFD promoted significant changes in gene expression profile of both CT and EC-Myc KO mice, and the response was more robust in EC-Myc KO group compared to CT (2274 vs. 1269 genes in males and 1382 vs. 737 genes in females) (Figure 2B). Venn diagram analysis showed high similarity between CT and EC-Myc KO mice in response to HFD, with a total of 1107 and 534 common targets in male and female animals, respectively. However, a large subset of genes was exclusive to EC-Myc KO group relative to CT (1167 vs. 162 genes in males and 848 vs. 203 genes in females) (Figure 2C).
FIGURE 2.
Differential gene expression of CT and EC-Myc KO mice after short-term exposure to HFD. (A) Volcano plots indicating the number of genes significantly altered in EC-Myc KO mice relative to CT under CTD. (B) Volcano plots indicating the number of genes significantly altered by HFD exposure in CT and EC-Myc KO livers. (C) Venn diagrams indicating the number of genes significantly altered by HFD exposure in CT and EC-Myc KO livers. The top panels represent results in males (n = 4) and the bottom panels represent results in females (n = 3). CT, control; CTD, low-fat control diet; EC-Myc KO, endothelial c-Myc knockout; HFD, high-fat diet
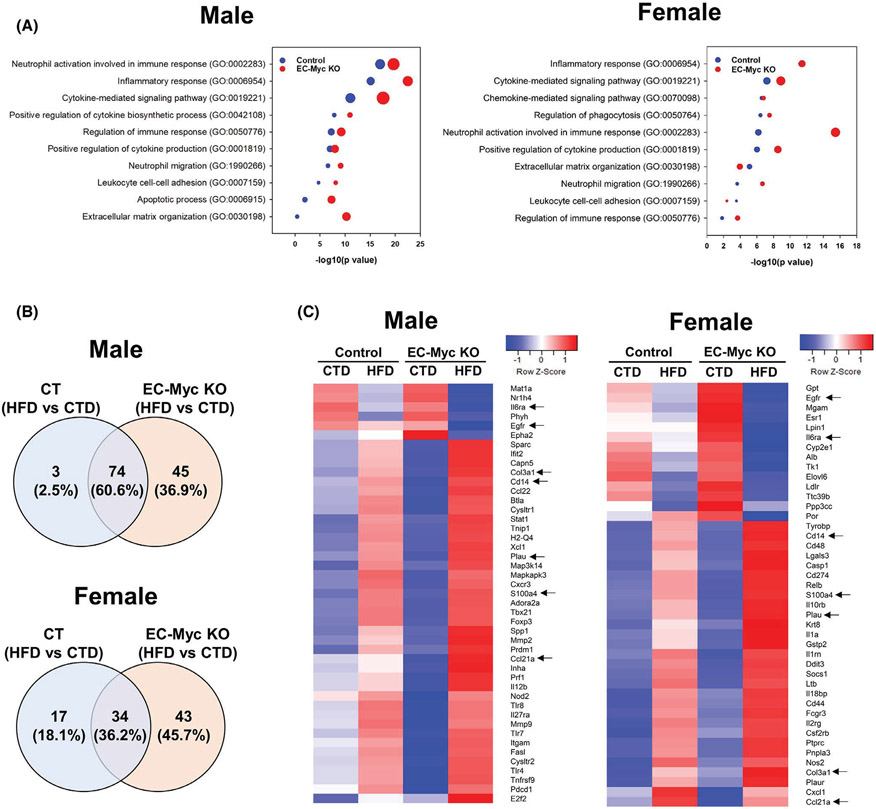
Gene ontology analysis of differentially expressed genes showed that inflammation-associated pathways were among the top biological processes upregulated in both CT and EC-Myc KO livers in response to HFD. However, for most of the pathways, the number of gene targets and the degree of significance were both higher in EC-Myc KO group (Figure 3A). We compared the list of differentially expressed liver inflammation-related genes in each experimental group after exposure to HFD using Venn diagram analysis. Although both experimental groups shared similarities in the context of inflammation (74 and 34 common genes in males and females, respectively), EC-Myc KO mice had a much larger list of exclusive targets than CT (45 vs. 3 genes in males and 43 vs. 17 genes in females) (Figure 3B). Interestingly, although male and female EC-Myc KO animals shared some exclusive gene targets after HFD exposure such as Plau, S100a4 and Il6ra, we observed a sex-specific response in inflammatory gene expression (Figure 3C).
FIGURE 3.
Transcriptome analysis of inflammatory pathways in CT and EC-Myc KO liver after short-term exposure to HFD. (A) Gene ontology analysis of inflammation-related biological processes upregulated in male (left panel) and female (right panel) mice by HFD exposure in CT and EC-Myc KO liver. Dot size indicates the number of genes. (B) Venn diagrams indicating the number of inflammation-related genes in male (top panel) and female (bottom panel) mice significantly altered by HFD in CT and EC-Myc KO liver. (C) Heatmaps showing inflammation-related genes in male (left panel) and female (right panel) mice significantly altered exclusively in EC-Myc KO livers after HFD exposure (n = 3–4). Genes marked with black arrows are present in both male and female animals. CT, control; CTD, low-fat control diet; EC-Myc KO, endothelial c-Myc knockout; HFD, high-fat diet
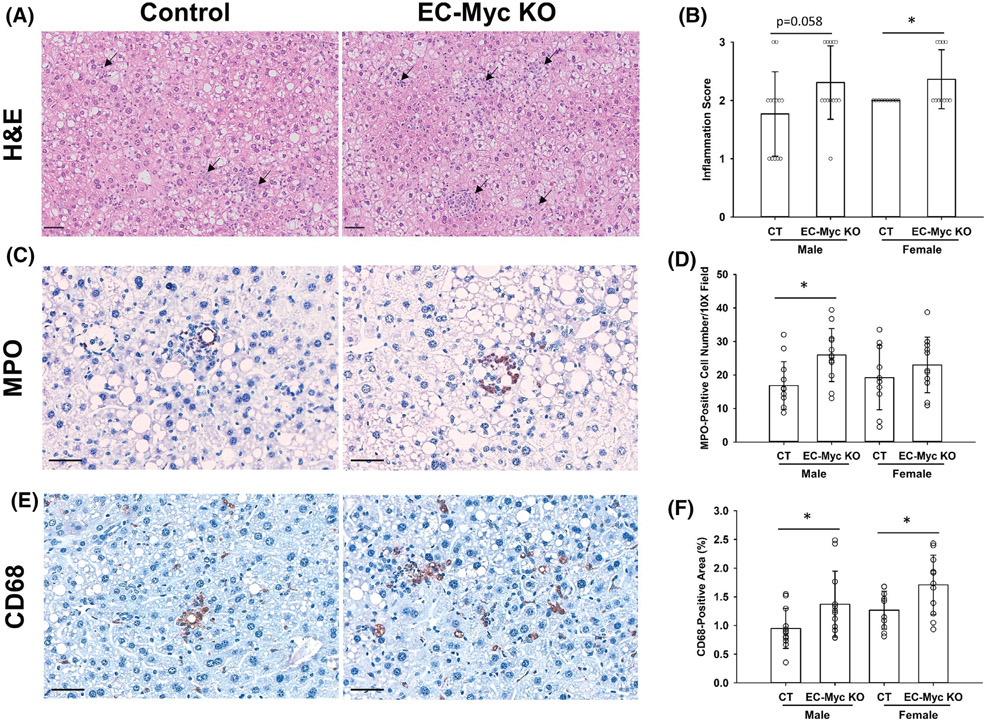
3.3 ∣. Endothelial c-Myc loss exacerbates diet-induced liver inflammation
Our transcriptome findings suggested that EC-Myc KO mice developed a stronger pro-inflammatory response to HFD diet. Therefore, to investigate a pathophysiological correlate, we performed histology analysis of liver sections to assess the degree of inflammation after long-term diet exposure (Figure 4A). EC-Myc KO mice showed a higher degree of inflammation than CT, with most animals showing moderate or severe inflammation (Score 2 and 3). Although both male and female knockout mice showed similar trends, results were significant only in females (Figure 4B). To identify the type of infiltrated inflammatory cells involved, we performed immunohistochemistry on liver sections for MPO and CD68, markers of neutrophils and macrophages, respectively (Figure 4C,E). EC-Myc KO mice showed increased neutrophil infiltration in males compared to CT, while macrophage infiltration was increased in both male and female EC-Myc KO mice (Figure 4D,F).
FIGURE 4.
Loss of endothelial c-Myc enhances HFD-induced leukocyte infiltration in the liver. (A) Representative images of H&E-stained liver sections of CT and EC-Myc KO livers after long-term exposure to HFD showing inflammatory foci (black arrows). (B) Analysis of inflammation degree. Results represent the score value for each individual animal. (C) Representative images of immunohistochemical staining for MPO on liver sections of CT and EC-Myc KO mice. (D) Quantification of MPO staining. Results represent the number of MPO-positive cells per 10× field. (E) Representative images of immunohistochemical staining for CD68 on liver sections of CT and EC-Myc KO mice. (F) Quantification of CD68 staining. Results represent the percentage of positively stained area relative to total tissue area (n = 10–13). Scale bar, 50 μm. CT, control; EC-Myc KO, endothelial c-Myc knockout; H&E, hematoxylin and eosin; HFD, high-fat diet; MPO, myeloperoxidase. *p < .05
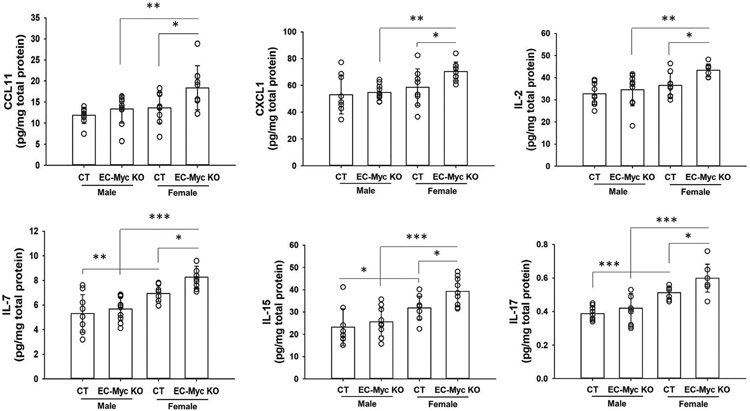
We next performed multiplex analysis of inflammation markers in liver lysates from CT and EC-Myc KO mice exposed to HFD for a short term to identify potential mediators responsible for the increased leukocyte infiltration. Multiple chemokines and cytokines were increased in EC-Myc KO liver compared to CT, and the most significant changes were observed in EC-Myc KO females (Figure 5). A summary of all cytokines analyzed is presented in Table S5.
FIGURE 5.
Endothelial c-Myc knockout increases inflammatory cytokines in the liver after HFD exposure. Multiplex analysis of inflammatory cytokines in liver lysates from CT and EC-Myc KO mice after short-term exposure to HFD (n = 9–10). CT, control; EC-Myc KO, endothelial c-Myc knockout; HFD, high-fat diet. *p < .05, **p < .01, ***p < .001
3.4 ∣. Endothelial c-Myc knockout enhances diet-induced liver fibrosis
Chronic inflammation plays a central role in the development of liver fibrosis. To assess the degree of fibrosis, we performed picrosirius red staining of liver tissue sections from CT and EC-Myc KO animals after long-term exposure to HFD (Figure 6A). Quantification of staining confirmed an increase in fibrosis in EC-Myc KO males (3.03 ± 1.56 vs. 1.92 ± 0.52% stained area, p = .06) and females (5.50 ± 1.79 vs. 3.76 ± 1.65% of stained area, p = .01) relative to CT animals. Females developed higher degree of fibrosis than males in both experimental groups (Figure 6B).
FIGURE 6.
Endothelial c-Myc deletion exacerbates HFD-induced liver fibrosis. (A) Representative images of picrosirius red-stained liver sections of CT and EC-Myc KO livers after long-term exposure to HFD. Scale bar, 50 μm. (B) Quantification of fibrosis. Results represent the percentage of positively stained area relative to total tissue area (n = 8–13). (C) Venn diagrams indicating the number of fibrosis-related genes in male (top panel) and female (bottom panel) mice significantly altered in control and EC-Myc KO livers after short-term exposure to HFD (n = 3–4). (D) Heatmaps showing fibrosis-related genes significantly altered in male (left panel) and female (right panel) mice exclusively in EC-Myc KO livers after short-term exposure to HFD (n = 3–4). Genes marked with black arrows are present in both male and female animals. CT, control; CTD, low-fat control diet; EC-Myc KO, endothelial c-Myc knockout; HFD, high-fat diet. *p < .05, **p < .01
Ingenuity pathway analysis of our transcriptome data indicated that HFD exposure altered the expression of liver fibrosis-related genes in both CT and EC-Myc KO animals (Figure S3A). Venn diagram analysis of liver fibrosis-related genes revealed a much larger list of exclusive targets in EC-Myc KO group compared to CT in both male (37 vs. 2 genes) and female (37 vs. 13 genes) mice (Figure 6C). Similar to what we found for inflammation-related genes, male and female EC-Myc KO animals showed a sex-specific gene list, while sharing some fibrosis-related targets such as the metalloprotease gene Mmp13, as well as the collagen genes Col1a1, Col1a2 and Col3a1 (Figure 6D).
3.5 ∣. Endothelial c-Myc loss promotes liver cancer-related gene expression changes
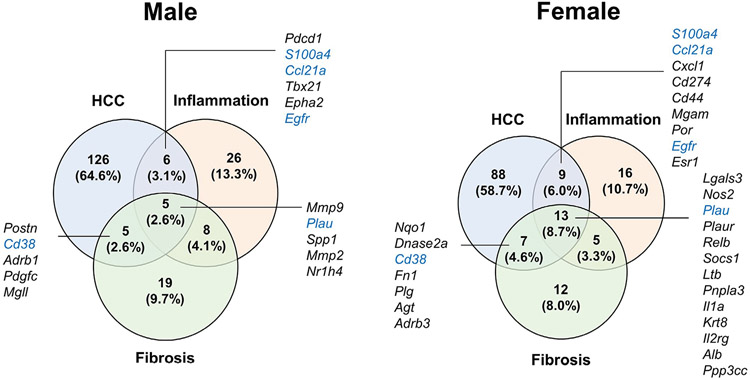
Ingenuity pathway analysis of our transcriptome dataset also identified top hepatotoxicity functions affected by HFD exposure. Among the top five functions identified, liver damage, inflammation, and fibrosis were present in both CT and EC-Myc KO mice, although the EC-Myc KO group showed a larger number of affected genes (Figure S3A). Interestingly, we found that EC-Myc KO animals specifically showed functions associated with deregulated proliferation: liver hyperplasia/hyperproliferation in males (p = 1.16E-18 vs. 9.94E-05 in CT) and hepatocellular carcinoma (HCC) in females (p = 2.95E-15 vs. 3.69E-06 in CT). These are highly relevant findings as liver inflammation and fibrosis have been shown to increase the risk of liver cancer.31 We compared all experimental groups regarding the list of HCC-related genes altered by HFD exposure and found a large subset of genes were exclusive to the EC-Myc KO group in both male (142 genes) and female (117 genes) mice (Figure S3B). Herein, we once again observed target differences between male and female EC-Myc KO mice despite some shared targets. Among the induced genes exclusive to the EC-Myc KO group, several were associated with cell cycle regulation and promotion of hyperproliferation, such as Dlgap5, Kif20b, and Ube2c. Other genes have been linked to the remodeling of the tumor microenvironment, cell migration, and invasion, such as Vcan and Vim. Some of the gene targets were also related to liver inflammation and fibrosis functions, indicating possible association with the development of HCC (Figure 7 and Table 1).
FIGURE 7.
Analysis of HCC-related genes in EC-Myc KO liver after short-exposure to HFD. Venn diagrams indicating the number of inflammation, fibrosis, and HCC-related genes in male (left panel) and female (right panel) mice significantly altered by HFD exclusively in EC-Myc KO livers (n = 3–4). Genes marked in blue are present in both male and female animals. EC-Myc KO, endothelial c-Myc knockout; HCC, hepatocellular carcinoma; HFD, high-fat diet
TABLE 1.
Genes exclusively altered in EC-Myc KO liver by short-term HFD exposure and their association with inflammation, fibrosis, and hepatocellular carcinoma
| Gene | Male |
Female |
Function |
||||
|---|---|---|---|---|---|---|---|
| Fold change | p value | Fold change | p value | Inflammation | Fibrosis | HCC | |
| Mmp13 | 4.78 | 8.96E-04 | 6.87 | 1.09E-03 | √ | ||
| Cd14 | 3.34 | 3.83E-08 | 1.81 | 5.94E-03 | √ | ||
| Gna15 | 2.94 | 1.23E-04 | 2.74 | 2.44E-03 | √ | ||
| Dlgap5 | 2.90 | 7.90E-04 | 3.02 | 3.40E-03 | √ | ||
| Plau | 2.89 | 2.48E-05 | 3.02 | 2.04E-05 | √ | √ | √ |
| Kif23 | 2.76 | 1.15E-05 | 2.16 | 3.83E-03 | √ | ||
| Col3a1 | 2.64 | 8.08E-04 | 1.70 | 4.57E-03 | √ | √ | |
| Col1a1 | 2.58 | 5.30E-03 | 1.88 | 7.72E-03 | √ | ||
| Kif20b | 2.50 | 1.22E-04 | 3.38 | 2.58E-04 | √ | ||
| Kifc1 | 2.46 | 1.45E-03 | 3.58 | 1.01E-03 | √ | ||
| Vcan | 2.41 | 3.96E-03 | 2.92 | 4.47E-03 | √ | ||
| Clec1b | 2.28 | 3.33E-18 | 1.87 | 2.50E-03 | √ | ||
| Ube2c | 2.20 | 7.18E-04 | 3.43 | 6.28E-04 | √ | ||
| S100a4 | 2.19 | 5.65E-03 | 2.74 | 5.06E-04 | √ | √ | |
| Cxcl14 | 2.12 | 5.23E-04 | 2.03 | 6.08E-03 | √ | ||
| Rad54l | 2.10 | 3.80E-03 | 1.85 | 8.95E-03 | √ | ||
| Tmem176a | 1.90 | 2.12E-12 | 1.54 | 1.15E-03 | √ | ||
| Gdpd1 | 1.87 | 5.57E-04 | 1.85 | 3.36E-03 | √ | ||
| Vim | 1.83 | 5.34E-05 | 1.51 | 6.74E-03 | √ | ||
| Anxa3 | 1.82 | 3.42E-06 | 1.77 | 1.79E-03 | √ | ||
| Col1a2 | 1.81 | 9.27E-03 | 1.57 | 9.75E-03 | √ | ||
| Anxa5 | 1.77 | 1.18E-05 | 1.52 | 7.25E-04 | √ | ||
| Aspm | 1.75 | 6.16E-03 | 2.23 | 5.56E-03 | √ | ||
| Ccdc80 | 1.74 | 4.05E-04 | 1.59 | 2.00E-04 | √ | ||
| Cd38 | 1.73 | 1.31E-11 | 1.64 | 3.58E-03 | √ | √ | |
| Ccl21a | 1.61 | 2.04E-02 | 2.19 | 6.75E-03 | √ | √ | |
| Nusap1 | 1.54 | 1.15E-02 | 2.39 | 2.29E-03 | √ | ||
| Klf13 | −1.53 | 6.01E-05 | −1.56 | 1.25E-04 | √ | ||
| Scap | −1.53 | 3.50E-07 | −1.64 | 5.72E-04 | √ | ||
| Hpd | −1.55 | 6.00E-05 | −2.08 | 9.28E-06 | √ | ||
| Cebpb | −1.56 | 4.18E-04 | −1.55 | 2.76E-04 | √ | ||
| Ceacam1 | −1.58 | 4.32E-15 | −1.54 | 2.93E-08 | √ | ||
| Fbp1 | −1.59 | 5.99E-06 | −2.01 | 4.22E-06 | √ | ||
| Acsm3 | −1.62 | 2.29E-10 | −1.52 | 1.72E-03 | √ | ||
| Fpgs | −1.65 | 2.71E-10 | −1.61 | 7.27E-05 | √ | ||
| Pfkfb2 | −1.66 | 5.94E-05 | −1.51 | 4.29E-03 | √ | ||
| Fgf1 | −1.70 | 7.62E-10 | −1.89 | 8.84E-08 | √ | ||
| Glul | −1.74 | 2.26E-06 | −2.13 | 7.67E-12 | √ | ||
| Pck1 | −1.92 | 7.39E-07 | −1.95 | 7.42E-08 | √ | ||
| Ar | −2.08 | 1.73E-03 | −2.44 | 2.69E-04 | √ | ||
| Egfr | −2.19 | 9.19E-08 | −1.96 | 8.72E-05 | √ | √ | |
| Il6ra | −2.23 | 9.81E-05 | −2.57 | 1.68E-12 | √ | ||
| Pim3 | −2.31 | 5.11E-07 | −1.63 | 1.46E-04 | √ | ||
Note: Results represent fold changes relative to CTD group. Genes exclusively altered in both EC-Myc KO male (n = 4) and female (n = 3) animals are shown. Abbreviations: CTD, low-fat control diet; EC-Myc KO, endothelial c-Myc knockout; HCC, hepatocellular carcinoma; HFD, high-fat diet.
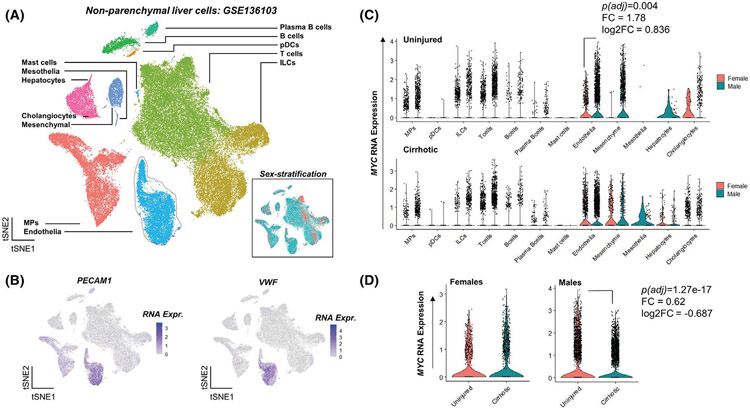
3.6 ∣. MYC expression is downregulated in endothelial cells from human cirrhotic livers
NASH is a progressive pathological condition characterized by inflammation and fibrosis, ultimately leading to cirrhosis. To determine the clinical relevance of our findings, we analyzed a single-cell RNA sequencing dataset of liver non-parenchymal cells from cirrhosis patients and healthy individuals.29 Clustering of 66 135 cells revealed 12 cell type-specific clusters including endothelia (Figure 8A). Classical endothelial markers PECAM1 and VWF were highly expressed in the endothelial cluster (Figure 8B). We analyzed MYC expression across all cell types and found that in healthy liver, MYC expression is 1.78-fold (adjusted p = .004) higher in male endothelial cells relative to females (Figure 8C). Interestingly, endothelial MYC expression was downregulated (−1.62-fold, adjusted p = 1.27E-17) in male cirrhotic liver compared to healthy individuals, indicating a possible role of endothelial MYC in cirrhosis (Figure 8D).
FIGURE 8.
MYC expression is downregulated in endothelial cells from human cirrhotic livers. (A) An integrated t-stochastic neighbor embedding (tSNE) plot of the GSE136103 dataset, showing cell type specific clusters of uninjured and cirrhotic livers (n = 5). Endothelial cluster in light blue is outlined. Inset shows the sample split based on sex. (B) tSNE plots showing the expression of classical endothelial markers PECAM1 and VWF to be high in the endothelial cluster. (C) Violin plots showing normalized RNA expression of MYC across all cell types separated based on sex in both uninjured and cirrhotic non-parenchymal liver cells. (D) Violin plots showing normalized RNA expression of MYC between uninjured and cirrhotic endothelial cells. A total of 66 135 cells are shown across 12 clusters. ILCs, innate lymphoid cells; MPs, mononuclear phagocytes; pDCs, plasmacytoid dendritic cells
4 ∣. DISCUSSION
In the present study, we show that c-Myc deficiency in endothelial cells promotes systemic inflammation and exacerbates HFD-induced liver inflammation and fibrosis in mice. These results are consistent with a previous report by our group showing that c-Myc knockdown in HUVECs increased the expression and secretion of inflammatory mediators as well as adhesion molecules involved in leukocyte adhesion.14 The current in vivo findings provide further support for an essential role of endothelial c-Myc in the regulation of systemic and tissue-specific inflammation.
Deregulated c-Myc expression has been correlated with inflammation in different experimental disease models, including chronic liver disease and cancer.15-19 However, the present study is the first to describe a specific role of c-Myc in endothelial-mediated inflammation. Interestingly, in non-endothelial cell types including cancer cells, induction of c-Myc has been shown to create a pro-inflammatory microenvironment that can lead to an influx of inflammatory cells, promoting tumor growth.15,16 Likewise, in a model of diabetic retinopathy, induction of c-Myc in Müller cells was associated with increased secretion of IL-1β, TNF-α, and IL-6, while c-Myc knockdown attenuated inflammation and disease progression.32 Although the use of different cell types may contribute to the apparently conflicting findings, it should also be appreciated that c-Myc regulates gene expression through transcriptional activation and repression by interacting with other DNA binding proteins in a context specific fashion. Therefore, any imbalance in c-Myc expression may compromise delicate homeostatic mechanisms.
Our gene expression profile indicates that EC-Myc KO animals develop a markedly stronger response to HFD exposure than CT mice. Inflammation was the top function identified in both experimental groups. However, EC-Myc KO animals displayed an exclusive list of genes, some of which have been previously reported to be associated with NASH. We were able to confirm that the enhanced pro-inflammatory profile in EC-Myc KO mice was also reflected at the protein level. The chemokines CCL11 and CXCL1, which were induced in EC-Myc KO animals exposed to HFD, have been previously reported to facilitate recruitment of eosinophils and neutrophils, resulting in liver inflammation.33-35 Among the interleukins significantly upregulated in EC-Myc KO mice, IL-17, a potent pro-inflammatory cytokine produced by Th17 cells, has been shown to stimulate other liver non-parenchymal cells to produce pro-inflammatory mediators that perpetuate liver inflammation.36 A pathological role for IL-17 has been described in different liver diseases including NASH.37-39
Chronic inflammation eventually triggers deregulation of repair mechanisms, leading to fibrosis.40 Several studies on non-endothelial cell types have correlated induction of c-Myc with fibrosis in different tissues.41-44 However, our results show that Myc deletion in endothelial cells exacerbates diet-induced liver fibrosis, suggesting a protective role. This difference could be simply interpreted as a cell-specific mechanism, but also attributed to different stages of injury and repair. As a stress sensor and an essential player in cell fate decisions, c-Myc upregulation is likely induced immediately after injury and ultimately promotes survival or death.45 In other scenarios in which repair is needed, induction of c-Myc sustains self-renewal.18,46 Interestingly, our findings differ from a recent report in which Gata4 is deleted specifically in liver sinusoidal endothelial cells (LSECs).47 Using gene set enrichment analysis, Winkler et al reported that liver fibrosis was promoted by activation of c-Myc-mediated Pdfgb expression in LSECs.47 One of the potential reasons for the apparent discrepancy is the use of different genetic models. We used a conditional approach to control the timing of endothelial Myc deletion in the endothelium, eliminating the impact of developmental defects.11,12 In our model, Myc deletion is driven by the Cdh5 promoter, resulting in c-Myc knockout in the entire vasculature. Conversely, Winkler et al used a non-inducible model in which Gata4 deletion is triggered during embryonic development, resulting in a more severe liver phenotype.48 This is not surprising as GATA4 has been shown to control liver development, and disruption of sinusoidal structure promotes Kupffer and stellate cell activation, ultimately triggering liver fibrosis.49,50 Our EC-Myc KO model showed significant pro-inflammatory changes at systemic level, but little impact in the liver unless animals were exposed to HFD. Since endothelial cells are highly heterogeneous across different tissues and engaged in organ-specific functions, regulation of gene expression in the entire endothelium or exclusively in LSECs is not expected to confer the same effects.5 In addition to the different models and knockout timing, there are significant differences in the diet and exposure time used in the Winkler study that could account for the divergent results. In our transcriptome analysis, one of the top genes induced by HFD in EC-Myc KO mice was S100a4, a member of the S100 calcium-binding protein family. In the liver, S100a4 is expressed by macrophages, and its expression level is increased in chronic liver disease and has been linked to fibrosis.51,52
In the context of diet-induced chronic liver disease, our analysis of a single-cell RNA sequencing dataset indicates that MYC is downregulated in liver endothelial cells from cirrhosis patients relative to healthy individuals, perhaps relating our findings to advanced fibrosis in human disease.29 Sex-stratification of the data indicated that MYC downregulation was significant only in males. However, one of the limitations in the analysis was the number of female donors relative to males.29 In our experimental model, we found that both male and female knockout mice show similar phenotypes after exposure to HFD. However, EC-Myc KO females showed an exacerbated response in most of pathological parameters analyzed, possibly due to different transcriptome changes induced by HFD. Although these sex-specific findings were not confirmed by the human data analysis, they are consistent with a recent meta-analysis study reporting that although women have a lower risk of steatosis, female patients with nonalcoholic fatty liver diseases have a higher risk of advanced fibrosis.53
Chronic liver disease, including NASH, is a risk factor for the development of hepatocellular carcinoma (HCC). Of relevance to this, our work identified a large number of genes previously reported in HCC exlusively in EC-Myc KO mice after short-exposure to HFD. Among the top induced genes, Kif20b, which encodes a kinesin protein, is overexpressed in HCC and essential for tumor cell proliferation.54,55 Dlgap5, a cell cycle regulated gene, has been identified as a prognostic indicator and potential therapeutic target for HCC.56,57
Our study is limited regarding the identification of cellular and molecular mechanisms by which endothelial c-Myc deficiency contributes to liver inflammation, fibrosis, or the risk of liver cancer. qPCR analysis of isolated liver endothelial cells showed a strong trend of increased Cdkn1a expression in EC-Myc KO group (Figure S2D), indicating a possible role of cellular senescence. There is a strong link between senescence, inflammation and cancer development.58 We have previously shown that c-Myc knockodown in HUVECs was specifically related to a senescence-associated pro-inflammatory phenotype.14 Single-cell analysis is an attractive approach that can be used in future studies to understand how loss of endothelial c-Myc triggers liver inflammation and fibrosis.
In conclusion, our findings provide evidence that endothelial c-Myc deficiency triggers systemic inflammation and suggest a novel protective role of c-Myc in diet-induced chronic liver disease. Targeting c-Myc downstream pathways in the endothelium may be a promising strategy for the treatment of NASH and other conditions associated with inflammation and fibrosis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Heart, Lung, and Blood Institute (NHLBI) at the National Institutes of Health (grant number 1R01HL128536-01A1) and the Florida Heart Research Institute (FHRI) (to C.O.R.). A. A. H. and R.A.F. were supported by the University of Miami and São Paulo Researchers in International Collaboration (SPRINT) (FAPESP grant number 2017/50022-1).
We would like to thank Dr. Ralf Adams for kindly providing us with the Cdh5(PAC)-CreERT2 mice through Ximbio; Dr. Prakash Ramachandran and Dr. Neil Henderson for sharing the Seurat object and tSNE embeddings for single-cell RNA-Seq analysis; Dr. Keith Webster for critical reading of the manuscript; the University of Miami Division of Veterinary Resources for assistance with animal care; the University of Miami Sylvester Comprehensive Cancer Center Flow Cytometry and Analytical Imaging Cores for assistance with FACS data collection and slide scanning; the University of Miami Dermatopathology Histology Laboratory for pathology services and the Mouse Metabolic Phenotyping Center (MMPC)—UC Davis (supported by NIH grant DK092993).
Funding information
HHS ∣ NIH ∣ National Heart, Lung, and Blood Institute (NHLBI), Grant/Award Number: 1R01HL128536-01A1; Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Grant/Award Number: 2017/50022-1; Florida Heart Research Institute
Abbreviations:
- CBC
complete blood count
- CMP
common myeloid progenitors
- CT
control
- CTD
low-fat control diet
- EC-Myc KO
endothelial c-Myc knockout
- GMP
granulocyte-monocyte progenitors
- H&E
hematoxylin and eosin
- HCC
hepatocellular carcinoma
- HFD
high-fat diet
- HUVECs
human umbilical vein endothelial cells
- LP
lymphoid progenitors
- LSECs
liver sinusoidal endothelial cells
- LT-HSC
long-term hematopoietic stem cells
- MEP
megakaryocyte-erythroid progenitors
- MPO
myeloperoxidase
- MPP
multipotent progenitors
- NASH
nonalcoholic steatohepatitis
- ST-HSC
short-term hematopoietic stem cells
Footnotes
DISCLOSURES
The authors have no conflicts to declare.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
REFERENCES
- 1.Rodrigues SF, Granger DN. Blood cells and endothelial barrier function. Tissue Barriers. 2015;3(1-2):e978720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Hinsbergh VW. Endothelium–role in regulation of coagulation and inflammation. Semin Immunopathol. 2012;34(1):93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pate M, Damarla V, Chi DS, Negi S, Krishnaswamy G. Endothelial cell biology: role in the inflammatory response. Adv Clin Chem. 2010;52:109–130. [PubMed] [Google Scholar]
- 4.Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. 2007;7(10):803–815. [DOI] [PubMed] [Google Scholar]
- 5.Jambusaria A, Hong Z, Zhang L, et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. eLife. 2020;9:e51413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao L, Liu Y, Wang N. New paradigms in inflammatory signaling in vascular endothelial cells. Am J Physiol Heart Circ Physiol. 2014;306(3):H317–H325. [DOI] [PubMed] [Google Scholar]
- 7.Dang CV, Resar LMS, Emison E, et al. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253(1):63–77. [DOI] [PubMed] [Google Scholar]
- 8.Pelengaris S, Khan M. The many faces of c-MYC. Arch Biochem Biophys. 2003;416(2):129–136. [DOI] [PubMed] [Google Scholar]
- 9.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18(20):5546–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5(10):1024–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudino TA, McKay C, Pendeville-Samain H, et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16(19):2530–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kokai E, Voss F, Fleischer F, et al. Myc regulates embryonic vascular permeability and remodeling. Circ Res. 2009;104(10):1151–1159. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigues CO, Nerlick ST, White EL, Cleveland JL, King ML. A Myc-Slug (Snail2)/Twist regulatory circuit directs vascular development. Development. 2008;135(11):1903–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florea V, Bhagavatula N, Simovic G, Macedo FY, Fock RA, Rodrigues CO. c-Myc is essential to prevent endothelial proinflammatory senescent phenotype. PLoS One. 2013;8(9):e73146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kortlever RM, Sodir NM, Wilson CH, et al. Myc cooperates with Ras by programming inflammation and immune suppression. Cell. 2017;171(6):1301–1315.e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Zhou Y, Ko KS, Yang H. Interactions between Myc and mediators of inflammation in chronic liver diseases. Mediators Inflamm. 2015;2015:276850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiner MK, Syrkina OL, Kolliputi N, Mark EJ, Hales CA, Waxman AB. Interleukin-6 overexpression induces pulmonary hypertension. Circ Res. 2009;104(2):236–244, 228 p following 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sipos F, Firneisz G, Muzes G. Therapeutic aspects of c-MYC signaling in inflammatory and cancerous colonic diseases. World J Gastroenterol. 2016;22(35):7938–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borrello MG, Degl'Innocenti D, Pierotti MA. Inflammation and cancer: the oncogene-driven connection. Cancer Lett. 2008;267(2):262–270. [DOI] [PubMed] [Google Scholar]
- 20.Sorensen I, Adams RH, Gossler A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 2009;113(22):5680–5688. [DOI] [PubMed] [Google Scholar]
- 21.de Alboran IM, O'Hagan RC, Gärtner F, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14(1):45–55. [DOI] [PubMed] [Google Scholar]
- 22.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. [DOI] [PubMed] [Google Scholar]
- 23.Torre D, Lachmann A, BioJupies MA. Automated generation of interactive notebooks for RNA-Seq data analysis in the cloud. Cell Syst. 2018;7(5):556–561.e553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qadir MMF, Álvarez-Cubela S, Klein D, et al. Single-cell resolution analysis of the human pancreatic ductal progenitor cell niche. Proc Natl Acad Sci USA. 2020;117(20):10876–10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575(7783):512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuzawa N, Takamura T, Kurita S, et al. Lipid-induced oxidative stress causes steatohepatitis in mice fed an atherogenic diet. Hepatology. 2007;46(5):1392–1403. [DOI] [PubMed] [Google Scholar]
- 31.Yu LX, Ling Y, Wang HY. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis Oncol. 2018;2(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Chen C, Wu L, et al. C-myc contributes to the release of Muller cells-derived proinflammatory cytokines by regulating lncRNA MIAT/XNIP pathway. Int J Biochem Cell Biol. 2019;114:105574. [DOI] [PubMed] [Google Scholar]
- 33.Jaruga B, Hong F, Sun R, Radaeva S, Gao B. Crucial role of IL-4/STAT6 in T cell-mediated hepatitis: up-regulating eotaxins and IL-5 and recruiting leukocytes. J Immunol. 2003;171(6):3233–3244. [DOI] [PubMed] [Google Scholar]
- 34.Pham B-N, Bernuau J, Durand F, et al. Eotaxin expression and eosinophil infiltrate in the liver of patients with drug-induced liver disease. J Hepatol. 2001;34(4):537–547. [DOI] [PubMed] [Google Scholar]
- 35.Marra F, Tacke F. Roles for chemokines in liver disease. Gastroenterology. 2014;147(3):577–594.e571. [DOI] [PubMed] [Google Scholar]
- 36.Lafdil F, Miller AM, Ki SH, Gao B. Th17 cells and their associated cytokines in liver diseases. Cell Mol Immunol. 2010;7(4):250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemmers A, Moreno C, Gustot T, et al. The interleukin-17 pathway is involved in human alcoholic liver disease. Hepatology. 2009;49(2):646–657. [DOI] [PubMed] [Google Scholar]
- 38.Paquissi FC. Immunity and fibrogenesis: the role of Th17/IL-17 axis in HBV and HCV-induced chronic hepatitis and progression to cirrhosis. Front Immunol. 2017;8:1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rau M, Schilling A-K, Meertens J, et al. Progression from non-alcoholic fatty liver to nonalcoholic steatohepatitis is marked by a higher frequency of Th17 cells in the liver and an increased Th17/resting regulatory T cell ratio in peripheral blood and in the liver. J Immunol. 2016;196(1):97–105. [DOI] [PubMed] [Google Scholar]
- 40.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18(7):1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li AY, Wang JJ, Yang SC, et al. Protective role of Gentianella acuta on isoprenaline induced myocardial fibrosis in rats via inhibition of NF-kappaB pathway. Biomed Pharmacother. 2019;110:733–741. [DOI] [PubMed] [Google Scholar]
- 42.Lemos DR, McMurdo M, Karaca G, et al. Interleukin-1beta activates a MYC-dependent metabolic switch in kidney stromal cells necessary for progressive tubulointerstitial fibrosis. J Am Soc Nephrol. 2018;29(6):1690–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai X, Li Z, Zhang Q, et al. CXCL6-EGFR-induced Kupffer cells secrete TGF-beta1 promoting hepatic stellate cell activation via the SMAD2/BRD4/C-MYC/EZH2 pathway in liver fibrosis. J Cell Mol Med. 2018;22(10):5050–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nevzorova YA, Hu W, Cubero FJ, et al. Overexpression of c-myc in hepatocytes promotes activation of hepatic stellate cells and facilitates the onset of liver fibrosis. Biochim Biophys Acta. 2013;1832(10):1765–1775. [DOI] [PubMed] [Google Scholar]
- 45.Matsumura I, Tanaka H, Kanakura Y. E2F1 and c-Myc in cell growth and death. Cell Cycle. 2003;2(4):333–338. [PubMed] [Google Scholar]
- 46.Zhang H, Xu A, Sun X, et al. Self-maintenance of cardiac resident reparative macrophages attenuates doxorubicin-induced cardiomyopathy through the SR-A1-c-Myc axis. Circ Res. 2020;127(5):610–627. [DOI] [PubMed] [Google Scholar]
- 47.Winkler M, Staniczek T, Kürschner SW, et al. Endothelial GATA4 controls liver fibrosis and regeneration by preventing a pathogenic switch in angiocrine signaling. J Hepatol. 2021;74(2):380–393. [DOI] [PubMed] [Google Scholar]
- 48.Wohlfeil SA, Häfele V, Dietsch B, et al. Hepatic endothelial notch activation protects against liver metastasis by regulating endothelial-tumor cell adhesion independent of angiocrine signaling. Cancer Res. 2019;79(3):598–610. [DOI] [PubMed] [Google Scholar]
- 49.Géraud C, Koch P-S, Zierow J, et al. GATA4-dependent organ-specific endothelial differentiation controls liver development and embryonic hematopoiesis. J Clin Invest. 2017;127(3):1099–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammoutene A, Rautou PE. Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. J Hepatol. 2019;70(6):1278–1291. [DOI] [PubMed] [Google Scholar]
- 51.Chen L, Li J, Zhang J, et al. S100A4 promotes liver fibrosis via activation of hepatic stellate cells. J Hepatol. 2015;62(1):156–164. [DOI] [PubMed] [Google Scholar]
- 52.Österreicher CH, Penz-Österreicher M, Grivennikov SI, et al. Fibroblast-specific protein 1 identifies an inflammatory subpopulation of macrophages in the liver. Proc Natl Acad Sci USA. 2011;108(1):308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balakrishnan M, Patel P, Dunn-Valadez S, et al. Women have a lower risk of nonalcoholic fatty liver disease but a higher risk of progression vs men: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2021;19(1):61–71.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Li Y, Zhang X, et al. Inhibition of kinesin family member 20B sensitizes hepatocellular carcinoma cell to microtubule-targeting agents by blocking cytokinesis. Cancer Sci. 2018;109(11):3450–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Zhou Y, Liu X, et al. MPHOSPH1: a potential therapeutic target for hepatocellular carcinoma. Cancer Res. 2014;74(22):6623–6634. [DOI] [PubMed] [Google Scholar]
- 56.Liao W, Liu W, Yuan Q, et al. Silencing of DLGAP5 by siRNA significantly inhibits the proliferation and invasion of hepatocellular carcinoma cells. PLoS One. 2013;8(12):e80789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsou A-P, Yang C-W, Huang C-Y, et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22(2):298–307. [DOI] [PubMed] [Google Scholar]
- 58.Lasry A, Ben-Neriah Y. Senescence-associated inflammatory responses: aging and cancer perspectives. Trends Immunol. 2015;36(4):217–228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.